Abstract
Histones play important roles in chromatin, due to various post-translational modifications and sequence variants, which are called histone proteoforms. Investigating modifications and variants is an on-going challenge. Previous methods are based on antibodies and because they usually detect only one modification at a time, they are not suitable to study the various combinations of modifications on histones. Fortunately, mass spectrometry has emerged as a high-throughput technology for histone analysis and does not require prior knowledge about any modifications. From the data generated by mass spectrometers, both identification and quantification of modifications and variants can be easily obtained. Based on this information, the functions of histones in various cellular contexts can be revealed. Therefore, mass spectrometry continues to play an important role in the study of histone proteoforms. In this review, we will discuss the analysis strategies of mass spectrometry, their applications on histones, and some key remaining challenges.
Keywords: histone proteoforms, post-translational modifications, variants, mass spectrometry, identification, quantification
1. Introduction
Histones play important roles in chromatin, due to the number of different histone proteoforms (e.g., various post-translational modifications (PTMs) and sequence variants) (1). First, there are numerous PTMs on histones, which include methylation, acetylation, phosphorylation, ubiquitination, and SUMOylation, etc. Each PTM is related to many distinct protein functions. Moreover, some PTMs have cross-talk with one another and function synergistically to regulate gene expression. Examples of histone PTMs can be found in the review (2).
Histones have five families, i.e. H1, H2A, H2B, H3, and H4. Each family has the canonical sequence and different sequence variants. The H1 variants include H1.0-H1.5, H1.t, H1.x, HILS1, and H1oo, etc. The H2A variants include H2A.J, H2A.V, H2A.X, H2A.Z, H2A.Bbd, and macroH2A, etc. The H2B variants include H2B1A, H2B1B, and H2B1C, etc. The H3 variants include H3.1-H3.3, H3.1t, and CENP-A, etc. Examples of histone variants can be found in the review (3). The diversity of histone proteoforms makes them a challenge to identify and characterize.
Traditionally, antibody-based methods (e.g., western blot) are used to analyze histone modifications (4). These methods have multiple disadvantages. First, antibodies are not available for every new PTM discovered. Second, PTMs on neighboring amino acids may prevent antibody binding, a phenomenon called epitope occlusion. Third, the quantification of PTMs via antibody-based methods is inaccurate at best. Fortunately, all these disadvantages can be overcome using mass spectrometry (MS). MS is a sensitive and efficient way to detect both previously identified and novel PTMs. Moreover, there are various MS-based methods to accurately quantify PTMs. MS methods also allow for identification and quantification of histone variants, which may be too similar in sequence to study using antibodies. Thus, MS is the key technology to analyze histone proteoforms. MS applications on histone proteoforms can be found in the review (5).
Although MS is an important technology, it still faces some challenges. In this review, we will cover the fundamentals of mass spectrometers, three MS strategies (i.e. bottom-up, top-down, and middle-down) for studying histones, and discuss some remaining challenges of MS.
2. Mass spectrometry for histone analysis
Mass spectrometry emerged more than a century ago and its application to biology, especially proteins, started as far back as 1958 (6). Since then many techniques have been developed to analyze proteins, including improvements in sample preparation, ionization, fragmentation, and detection. In this section, the fundamental methods and three strategies (i.e. bottom-up, top-down, middle-down) of MS will be introduced.
2.1. Fundamentals of mass spectrometry
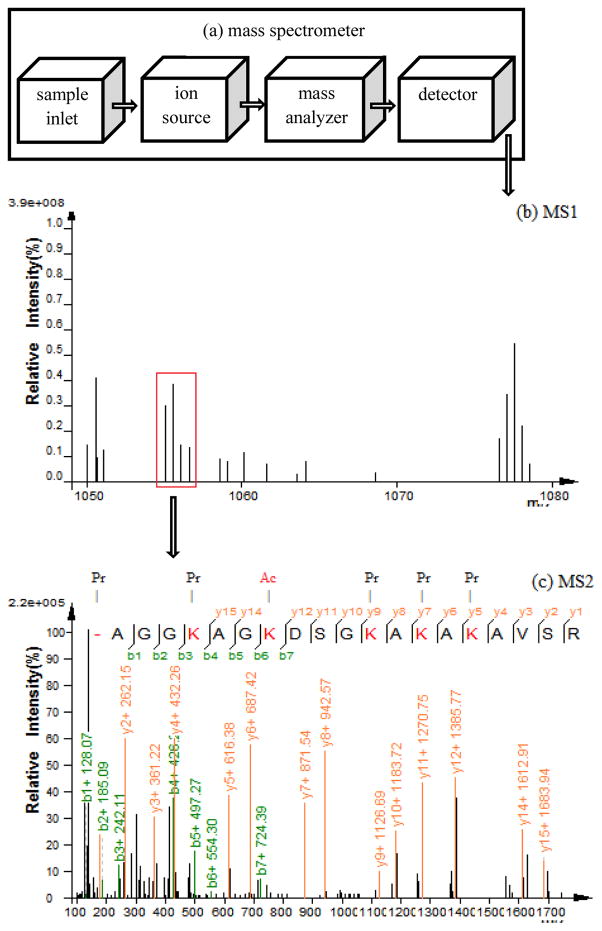
A typical mass spectrometer consists of four components: a sample inlet, an ion source, a mass analyzer, and a detector (7). Figure 1a shows a layout for these components. Samples undergoing mass spectrometric analysis go through a number of steps. First, they have to be introduced into the instrument. They can be eluted through liquid chromatography (LC) into the mass spectrometer or embedded in matrix on a target plate. Then the ion source converts sample molecules to ions, using electrospray ionization (ESI) or matrix-assisted laser desorption/ionization (MALDI) (8, 9). In the magnetic or electric field of the mass analyzer, ions can fly with different rates or rotate with different frequencies depending on their mass-to-charge ratio (m/z). When ions fly to a detector or rotate with stable frequency, the detector can obtain the ions or the analog signals, which can be converted to digital signals. The end result is a first level mass spectrum (MS1), which contains m/z values and intensities for sample ions (commonly called the precursor ions). Figure 1b shows an example MS1 spectrum.
Figure 1.
Fundamentals of mass spectrometry. (a) A mass spectrometer consists of four components: a sample inlet, an ion source, a mass analyzer, and a detector. (b) Precursor ions are scanned in MS1. (c) Some precursor ions are selected, fragmented, and scanned in MS2. Pr (Propionylation), Ac (Acetylation).
However, MS1 is not enough to distinguish some precursor ions. For example, PEPTIDE and PEPDITE have the same m/z values. To distinguish these precursor ions, we have to fragment them and obtain m/z values for their components. For example, PEPT and PEPD have different m/z values. Therefore, the second level of mass spectrum (MS2) is essential if other information is lacking (e.g., retention time in LC). To obtain an MS2 spectrum, a precursor ion is selected from the MS1, isolated, broken into fragment ions, analyzed and detected. Figure 1c shows an example MS2 spectrum.
To understand the complexity of the MS2 spectra, it is necessary to understand what those peaks represent. Although a single precursor mass is selected, there are many copies of the precursor ion available for fragmentation (shown in Figure 1b). Due to different fragmentation efficiencies, different copies of the precursor ion are fragmented at different sites along the amino acid sequence. Thus, the types and intensities of fragment ions are different (shown in Figure 1c). Additionally, one sequence can be fragmented into N-terminal and C-terminal parts. For example, collision induced dissociation (CID) produces b and y ions from the N-terminal and C-terminal parts, respectively, and electron transfer dissociation (ETD) and electron capture dissociation (ECD) produce c and z ions from the N-terminal and C-terminal parts, respectively (10, 11). The end result is an MS2 spectrum that contains the m/z values and intensities for different N- and C-terminal fragments of the precursor ion.
From the peaks in the MS2, the amino acid sequence can be obtained. The mass difference of two adjacent peaks is equal to the mass of one amino acid (e.g., y7 and y8 in Figure 1c). If all the fragment ions from one terminal are detected in the MS2, the sequence can be inferred by their mass differences. This is the basis for de novo sequencing, which infers the sequence from the MS2 directly (12–14). However, typically some fragment ions are missing or buried in the noise peaks due to the fragmentation efficiency. In this case, only parts of the sequence can be obtained. Moreover, if the N- and C-terminal fragment ions are missing (i.e. b1, y1, c1, and z1), the sequence terminal cannot be determined. To overcome these problems, database searching methods have been developed. A database search matches peaks from an MS2 with the theoretical fragment ions of sequences in a protein database to obtain the most likely sequence (15–19). If sequences in a protein database are correctly annotated, database searching is preferred over de novo sequencing.
The presence of PTMs further complicates MS2 analysis. All the fragment ions containing a PTM have a mass shift (equal to the mass of the PTM) compared to the unmodified fragment ions. If the PTM is not specified in the database search, both the modified fragment ions and the MS2 cannot be matched correctly. When the PTM is entered into the database, all potential sites are considered (i.e. acetylation can occur on any lysine in the sequence). The best matched result from such a search gives the peptide sequence and assigns the PTM to a single amino acid in that sequence. Figure 1c shows a spectrum for a histone peptide containing an acetylation site.
However, in complex analysis the types or sites of PTMs are usually unknown. Fortunately, there are two methods that can resolve this problem. The first method uses the mass difference of the modified and unmodified precursor ions (20). This method is fast because it only uses MS1and LC retention time information. This analysis requires that both unmodified and modified precursor ions are in the sample and that at least one of them is present in high abundance. Therefore, this method is not sensitive for low level PTMs. The second method is open searching, which opens the precursor mass tolerance to 300 Da and considers all amino acid sites within the sequence (21). This method is slow but sensitive. Therefore, once MS2 spectra are collected with modified peptides, both restricted (known) and unrestricted (unknown) PTMs can be identified and assigned to a specific site.
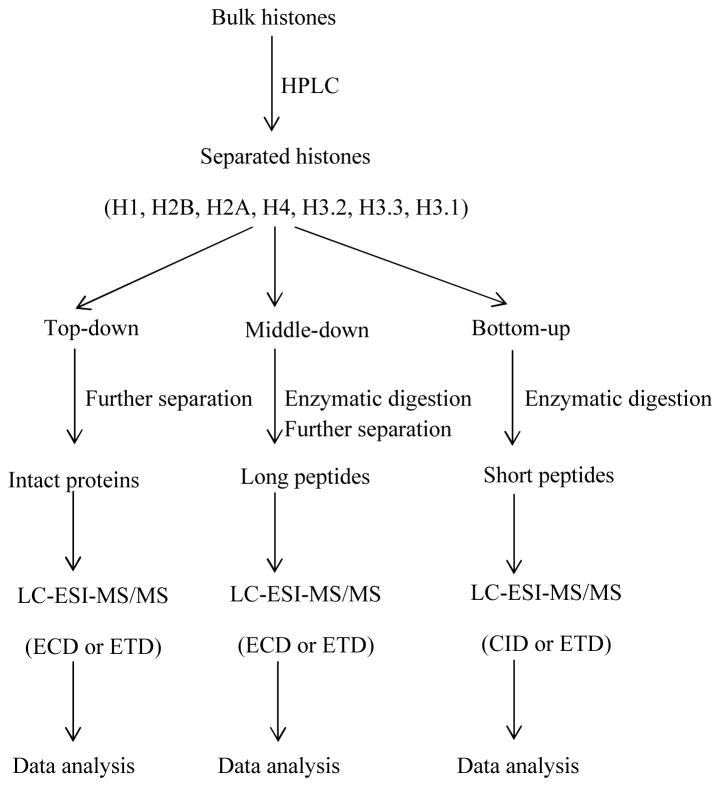
The above principles are about the collection and interpretation of mass spectra. There are three MS strategies for analyzing proteins: bottom-up, top-down and middle-down (shown in Figure 2).
Figure 2.
Mass spectrometry methods that can be used to analyze histone proteins. HPLC (high performance liquid chromatography), LC (liquid chromatography), ESI (electrospray ionization), MS/MS (tandem mass spectrometry), ECD (electron capture dissociation), ETD (electron transfer dissociation), CID (collision-induced dissociation).
2.2. Bottom-up mass spectrometry
In bottom-up MS, peptides are analyzed and used for protein identification. The procedure for bottom-up is as follows. First, proteins are extracted from cells. To reduce sample complexity, proteins can be separated by different techniques such as two-dimensional gel electrophoresis (2-DE) or high performance liquid chromatography (HPLC). Second, proteins are digested into peptides with proteases. Third, peptides are eluted by LC, ionized and scanned to generate MS1 spectra. Some peptides are selected and fragmented to generate MS2 spectra. After the mass spectra are generated, peptide sequences are assigned by database searching. The identified peptides can be assembled into proteins. Therefore, bottom-up is a peptide-centric MS technology.
To digest proteins into peptides, different enzymes can be used. The commonly used enzyme is trypsin, which cleaves at the C-terminal of lysine (K) and arginine (R) residues. However, the direct use of trypsin on histones is problematic. The N-terminal tails of histones are lysine and arginine rich so trypsin digestion results in small pieces that cannot be detected by MS. Though there are mis-cleavages, the number of mis-cleavages is small (e.g., one or two) and mis-cleavages are not reproducible. Other enzymes can be used but they are much less specific than trypsin and also result in non-reproducible digests. Therefore, histones should be derivatized before trypsin digestion.
There are two derivatization methods reported for histones. The first method uses acetic anhydride, which reacts with lysines and blocks trypsin digestion (22, 23). The second method uses propionic anhydride (24–27). Both chemical acetylation and propionylation occur on unmodified and mono-methylated lysines as well as the N-terminal amino acid. The propionylation not only blocks trypsin digestion at lysines, but also enhances hydrophobicity of histone peptides, thereby increasing the chromatographic resolution of different peptides. After the trypsin digestion, another propionyl group is added to the N-terminal of each peptide, further enhancing hydrophobicity. The increase in hydrophobicity and reproducible digestions resulting from propionylation make this the preferred derivatization method for histones.
2.3. Top-down mass spectrometry
Different from bottom-up, top-down analyzes whole proteins. The procedure of top-down is as follows. First, protein mixtures are separated. Second, they are eluted and introduced into the instrument. Third, proteins are scanned and MS1 spectra are generated. Some proteins are selected and fragmented to generate MS2 spectra. After the mass spectra are generated, they can be identified as proteins by database searching. Therefore, top-down is a protein-centric MS technology.
There are several differences between top-down and bottom-up methods. First, the molecular weights of precursor ions are significantly different. In bottom-up, the lengths of tryptic peptides are often between 6 and 20 amino acids. Thus, most peptides are around 2kDa. In top-down, proteins are often longer than 100 amino acids and larger than 10kDa. For example, the molecular weights of canonical histones are between 11kDa and 15kDa. The different molecular weights lead to different properties and challenges. Peptides are soluble while proteins are insoluble. Peptides and proteins require different separations. Peptides typically have low charge states (2+ or 3+ charge states) while proteins are highly charged (10+ to 100+ charge states).
Second, the fragmentation type is different. In CID, the fragmentation energy is high and labile modifications will be lost (e.g., phosphorylation). In ETD, the fragmentation method results in electron transfer and it is better to highly charged ions and labile modifications. In bottom-up, there are only a few PTMs on one peptide so both CID and ETD can be used. In top-down, histones are highly charged and there are multiple PTMs so ETD or related ECD is essential for studying PTMs on whole proteins.
Third, the data complexity is different. In bottom-up, it is easy to identify the monoisotopic peak and charge state for precursor ions in high resolution MS. Sometimes there are two or more peptides co-eluting, which can make analysis more complex. Fortunately, several methods have been developed to determine the monoisotopic peak and charge state for peptides (28–32). In top-down, it is difficult to detect the monoisotopic peak and charge state, even if there is only one precursor ion and the MS is high resolution. Usually, the monoisotopic peak is buried in the noise, and the resolution is not high enough to distinguish charge states. Manual interpretation of this data is difficult for these reasons. Though several computational methods have been developed to resolve these problems, their performance needs to be improved (33–35). Because the fragment ions in top-down are also highly charged, their monoisotopic peaks and charge states also need be determined. PTMs lead to similar and overlapping m/z values for precursor ions, which further complicates data analysis. The high number of PTMs on histones makes top-down analysis of histone proteoforms very complex.
For these reasons, top-down is more difficult than bottom-up. However, top-down has the advantage of viewing all the PTMs at the protein level. The whole protein can also be sequenced, making histone variants easy to study. To combine the advantages of top-down (global view of PTMs and variants) and bottom-up (easy to operate and sensitive), middle-down was developed.
2.4. Middle-down mass spectrometry
As implied from the name, middle-down is in between top-down and bottom-up. The procedure is similar to bottom-up. The difference is specific enzymes are used to obtain much longer peptides. For example, Glu-C is used to cleave histone H3 at the C-terminal of glutamic acid and obtain the 1–50 peptide. Similarly, Asp-N is used to cleave histone H4 at the N-terminal of aspartic acid and obtain the 1–23 peptide. Middle-down histone peptides have multiple PTMs and can be used for combinatorial PTM analysis. Since the peptides in middle-down are much longer than those in bottom-up, middle-down has similar problems as top-down and is less sensitive than bottom-up. Middle-down peptides have much wider charge state distributions than bottom-up peptides. Only one charge state is selected at a time for fragmentation. Thus, the signal in each charge state is lower in middle-down than in bottom-up.
The above three strategies all have their own advantages and disadvantages. Bottom-up is easy to operate and the most sensitive. However, it lacks the global view of PTMs and loses the relationship between peptides and proteins (i.e. it is difficult to distinguish proteins and their variants by peptides). Top-down is able to view all PTMs and protein variants but difficult to operate and less sensitive. Middle-down is in the middle. Nowadays, bottom-up is mature and has become the workhorse in mass spectrometric analysis, while top-down and middle-down are promising but require expertise.
3. Applications
Since histones are heavily modified and have large numbers of sequence variants, analysis of histone proteoforms is very difficult. Fortunately, MS can provide a vast amount of information in a high-throughput way and without prior knowledge. With different techniques, identification and quantification are the basic information obtained from MS. More information can be obtained, such as distinguishing histone variants, discovery of histone-binding proteins, and analysis of combinatorial histone PTMs. Thus, MS is a powerful tool for uncovering histone function. With the further development of MS, more functions and applications will be found in the future.
3.1. Identification of novel sites or types of histone PTMs
In contrast to antibody-based methods, MS does not need any prior knowledge of PTMs. One application of MS is to find more sites for the known types of PTMs (e.g., new acetylation sites within histones). In these cases, the PTMs can be identified by MS2 fragmentation and database searches. From the identification results, the spectra of the specific PTM can be separated. Generally, the identification results need to be manually checked because the site assignment of PTMs will lead to a combinatorial explosion of candidate sequences and a high false discovery rate. Thus, to confirm a PTM site several steps need be done. First, the match score should be high enough. Second, the sequence and the spectrum should match well and there should be fragment ions to support the PTM site assignment. Third, the chromatography of the precursor ion should be checked. Only after all of these properties are checked can the site be considered reliable. These tools are useful for a newly discovered PTMs as well, e.g., citrullination of arginine on histones (36). If the site appears unreliable, the sample preparation should be improved.
Another application is to find unexpected PTMs. As mentioned in section 2.1, open searching is usually used for novel PTM identification. Similar to restricted searching, the identification results from open searching also need be checked. At the beginning, only the PTM mass and potential site is known. The results can be compared with the PTM database Unimod (www.unimod.org). If the mass and site match well, this PTM is already known. Otherwise, this PTM may be novel. If the combinational mass of known PTMs is still not equal to this PTM, more techniques are needed to validate this PTM. Crotonylation of lysine on histones was found in this way (37).
If there is no enrichment, the concentration of novel PTM sites and types can be low. In these cases, the most sensitive bottom-up should be used to discover novel PTM sites or types.
3.2. Quantification of histone PTMs
Knowing the type and site of a PTM is important. More important is to know the level of PTMs, because the level can be related to function, e.g., gene regulation. There are two kinds of quantification methods: label-based methods and label-free methods. In label-free, no extra experiments are needed. However, the reproducibility is very important for reliable analysis. One label-free method is spectral counting, in which the number of identified spectra is used to quantify precursor ions (38–40). However, there are factors that affect the identification including ionization efficiency and precursor selection. In a word, spectral counting is simple but inaccurate.
Another label-free method relies on calculating the area under the precursor peak (41). For one sequence with different modifications, such as unmodified, mono-, di-, and tri-methylation, the area under each precursor peak is calculated. Then the proportion of each form is calculated by dividing by the total area. The reproducibility of this method is also important. In bottom-up, mis-cleavages will affect the calculation of the total area and lead to inaccurate proportions being assigned to each form.
There are two kinds of labeling methods: in vitro and in vivo. Chemical derivatization is an in vitro method, such as isobaric tags for relative and absolute quantitation (iTRAQ) (42–44). In iTRAQ, samples are treated separately. After the trypsin digestion, each sample interacts with different reagents. Each reagent contains the reporter group and the balance group. In the 4-plex reagents, the masses of reporter group are 114, 115, 116, and 117 daltons, and the corresponding masses of balance groups are 31, 30, 29, and 28 daltons. Then the four samples are equally mixed. In the MS1 spectra, the same peptides from different samples elute at the same time and are detected as the same m/z. All the peptides are selected and fragmented, during which the balance groups are lost and the reporter groups are detected. The relative ratios of the reporter groups in the MS2 spectra represent the relative quantification of the peptide from the four different samples.
Metabolic stable isotope labeling, such as stable isotope labeling by amino acids in cell culture (SILAC), is an in vivo labeling method (45). In SILAC, one sample is cultured in normal media (light), while the other is cultured in heavily labeled media (heavy), e.g., 13C615N2 on lysine, 13C615N4 on arginine. Since trypsin digests at the C-terminal of lysine and arginine, all tryptic peptides will be heavily labeled. After the cells are harvested, the light and the heavy samples are equally mixed. In MS1 spectra, the heavy peptide and the light peptide have a mass difference of 8 Da (one heavy lysine) or 10 Da (one heavy arginine) or other combinations. The heavy and the light pair are determined and the relative ratio can be calculated by the intensity of each peptide. After one of them is fragmented and an MS2 spectrum is generated, the peptide sequence will be identified.
In SILAC the heavy and light isotopes are mixed early during sample preparation, while in iTRAQ the heavy and light isotopes are mixed after the trypsin digestion. Therefore, it is easy to introduce biases with iTRAQ. Anyhow, both SILAC and iTRAQ need be calibrated to ensure proper relative quantification.
For bottom-up, the quantification of peptides can be accurate in label-based methods. However, it is difficult to obtain the accurate protein quantification because of shared peptides between protein variants and varying ionization efficiencies of different peptides. For top-down, there are no such problems.
3.3. Distinguishing histone variants
It has been found that each histone has its own family. In some families, the variants can have little difference, such as few amino acids. To distinguish them is a challenge that requires both separation and mass spectrometry. First, bulk histones can be separated into each family (as shown in Figure 2) by reverse phase high-performance liquid chromatography (RP-HPLC). Second, each family can be further separated by other methods, such as weak cation exchange hydrophilic interaction liquid chromatography (WCX-HILIC). Third, mass spectrometers are used to identify the family members. For bottom-up, it depends on the unique peptides to distinguish the variants. However, the unique peptides may be not identified for many reasons, such as ionization efficiency, precursor selection, fragmentation, and algorithms of identification. Thus, it is difficult to study most variants using bottom-up. Instead, top-down is usually used to investigate histone variants. Since top-down is not as sensitive as bottom-up, the sample volume should be higher for top-down than bottom-up. After the data are generated, some computational programs identify the variants and their PTMs. In summary, distinguishing histone variants is a big challenge for MS-based technologies and involves many steps including separation, MS, and data analysis. There are some papers delving into histone variants using top-down (46–48).
3.4. Discovery of histone-binding proteins
It has been found that histone PTMs can bind proteins to regulate genes (49–51). To discover histone-binding proteins, histone peptides are used as baits to essentially pull-down histone binders from cell lysates. The binders are both identified and quantified using mass spectrometric methods. In the forward experiment, the light lysate with the unmodified histone peptide serves as the control, while the heavy lysate with the modified histone peptide serves as the experimental. The control and the experimental are then equally mixed. If there is a protein binding to the modified histone peptide, the heavy-to-light ratio should be high. To further confirm, the backward experiment needs be done by switching the histone peptide baits so that the modified histone peptide is used with a light lysate. In the experiment, the heavy-to-light ratio should be low. The two samples require preparation for MS analysis. The proteins eluted from the histone peptides can be separated by gel electrophoresis. The gels are cut into bands. In-gel digestion is carried out and bottom-up data is obtained for each band. Computational methods are used to identify and quantify the histone-binding proteins. These experiments generate long lists of candidate proteins and therefore, candidate proteins need to be studied to assess biological function.
3.5. Analysis of combinatorial histone PTMs
In general, one single PTM can have functions, such as acetylation, methylation, and phosphorylation (52–54). Recently, it has been found that several histone PTMs function together, e.g., K27me1–3/S28ph on H3 (55). Since histones are heavily modified, there are several potential PTM combinations to be discovered, which brings challenges for current technologies. One potential technology is as follows. As mentioned above, the histone-binding proteins can be discovered by one PTM. Then other PTMs near the binding site can also be identified. When one of the latter PTMs is changed to the unmodified form, it can be checked whether the abundance of the binding protein has been changed. If so, the changed PTM is the combinatorial one with the PTM on the binding site. Therefore, this analysis is the most complex.
4. Challenges
Despite recent advances in MS techniques, there are still many challenges including sample preparation, mass spectrometry, and data analysis. When these challenges are overcome, MS will become more powerful for probing histone PTMs and their functions.
4.1. Sample preparation
As mentioned above, there are two significant differences between peptides and proteins: peptides are soluble while proteins can precipitate, and peptides are easily separated while proteins can be more difficult to separate. For these reasons, bottom-up has become a common approach and there are several protocols for sample preparation. Top-down needs to be improved before it can become a high-throughput and widely applied technology.
4.2. Mass spectrometry
The mass spectrometer is also not perfect. At least five aspects of MS can present problems: ionization efficiency, precursor ion selection, fragmentation, detection, and resolution. First, the ionization efficiency is different for all precursor ions. Some precursor ions ionize well and can be easily detected, while others ionize poorly. In one protein, the quantification of peptides is different due to different ionization efficiencies. Therefore, ionization efficiency can cause problems for identification and quantification.
Second, precursor ion selection is another problem. In data-dependent acquisition mode (DD), the top-n most intense peaks are selected for fragmentation. In DD mode, the isolation window of 2 Da is used and allows for one or more precursor ions to be selected for fragmentation. To prevent repeated selection of the same precursor ions, the dynamic exclusion windows can be set such that several seconds pass between selecting precursor ions with the same mass. However, this mode is not suitable for low-abundance precursor ions because they may be never selected for fragmentation. Some low-abundance precursor ions are important. For example, when some precursor ions contain PTMs, their abundance is often low if no enrichment or purification has been done. To overcome this problem, data-independent acquisition mode (DI) is used to fragment all precursor ions in a wider window (56–58). For example, the MS1 can be partitioned into windows of 25 m/z. With this approach, many precursor ions are fragmented at the same time, including the low-abundance precursor ions. The large number of precursor ions fragmented makes it difficult to identify the MS2 spectra. Thus, each DD and DI has its own pros and cons.
Third, fragmentation is not well understood and this has implications for identification and quantification. Some research has been started, such as charge-remote fragmentation (59), however, this is just the tip of the iceberg. Lacking of accurate predictions when generating theoretical spectra means that only the m/z values of theoretical ions are used in database searching. In contrast, the fragmentation pattern could be contained in a spectral library and have more information than m/z values. From this point of view, spectral library searching is more accurate than database searching (60).
Fourth, there are MS detection limitations. In sample mixtures some proteins and peptides are abundant, while others are low level. If the peaks are weak or buried in noise peaks, they will be difficult to detect. Therefore, it is important to separate the high- and low-abundance proteins or peptides. Another challenge can come from the isotope distribution, especially in top-down. Because the molecular weights of proteins are much larger than those of peptides, there are many more isotopic peaks, and the intensities are in normal distribution. Thus, the monoisotopic peak is much lower than the middle peaks. When the difference is as large as four orders of magnitude such as for 15kDa proteins, it is difficult to detect the monoisotopic peak. Therefore, detection is important for correctly assigning identity.
Lastly, MS resolution can be a limitation. Resolution is the ability to separate adjacent peaks. For bottom-up, the resolution is high enough to separate isotopes. The charge state is easily determined by the m/z intervals between isotopic peaks and is usually with 2+ or 3+. However, for top-down, the resolution is not high enough. For example, the m/z interval of 20+ is 0.05016, while the m/z interval of 21+ is 0.04777. The mass difference is 0.00239, which is 0.12 ppm at 20kDa. It is difficult to have such high resolution and mass accuracy. To obtain high resolution, scan speed is low and this decreases the speed of data acquisition. Therefore, there is a balance between resolution and speed.
4.3. Data analysis
Several computational problems exist that impact data analysis. First, the mass spectra need be preprocessed. Preprocessing includes noise deletion, monoisotopic peak and charge state detection, spectra filtration, and spectra clustering (61). The most important is monoisotopic peak and charge state detection. As mentioned above, it is easy to determine the monoisotopic peak and charge state for precursor ions in bottom-up. But in middle-down and top-down, the detection of the monoisotopic peak and charge state is much more difficult due to the limitations in MS detection and resolution. Moreover, the co-eluted precursor ions in DD and the co-fragmented precursor ions in DI make the MS2 spectra more complex. For the former, each precursor ion that is detected can be identified by the MS2. For the latter, the fragment ions can be correlated to the corresponding precursor ions by their similar chromatography.
Second, identification needs be improved. Though plenty of algorithms have been developed for bottom-up, the identification rate is still low - only 10–40% spectra can be identified. There are many reasons for the low identification rate, such as sample loss, MS loss, and imperfectness of algorithms (62). When the sample preparation and MS are done well, the identification rate can be as high as 80%. One example comes from the study of ABRF iPRG 2013 (63). The challenge is to increase the identification rate at a certain false discovery rate (FDR). Some exceptions should be considered in spectra identification, such as unexpected PTMs and semi- or non-specific digestion. These lead to combinatorial explosion of candidates and more false positives. How to fast and accurately process them is the key for accurate identification. Furthermore, there are methods to control the FDR. For each spectrum, the p-value can be calculated from the score distribution of candidates. For all spectra, FDR can be calculated by the mixture model of correct and incorrect matches or by target-decoy database searching (64, 65).
The site localization of PTMs is a special identification problem. If there is more than one potential PTM site on a peptide, this will cause problems for site localization (66). The fragment ions for the two sites should be checked. The probability for each potential site can be calculated from these distinguishable ions. If there is a significant difference in the probability of two sites, then the most probable site is the correct one. Otherwise, the site localization cannot be determined. This assumes that there is only one correct site. In more complex situations, several precursor ions with one sequence but different sites co-elute and are co-fragmented. In this case, all potential sites should be detected.
Third, quantification should be carefully checked because problems can arise from both experimental and computational work. When carrying out experiments, samples should be mixed equally. However, this is difficult to control so the distribution of quantification needs to be checked. The initial quantification can be normalized to the center of distribution. Moreover, interferences with quantification, such as noise peaks and co-eluted precursor ions can cause issues. Even in DD about 50% of MS2 are mixed spectra (67). When interference happens, the quantification may be inaccurate. Only isotopes that do not have interfering peaks can be used for accurate quantification. Therefore, to detect the non- or less-interfered isotopes is the key to improve the accuracy of quantification.
Lastly, there should be data analysis pipeline available. In one experiment, much raw data will be generated, including technical replicates. Identification and quantification are the basic analysis. However, there are several parameters to be set and steps to run the basic analysis, such as format conversion, database indexing, database searching, result filtering and quantification (61). It is easy to make mistakes when setting parameters and time-consuming to run each step manually. The ideal pipeline is to prepare the raw data, database file, and parameter file once and then run each step automatically to obtain the results. This will decrease labor associated with basic analysis allowing people to focus on deeper analysis, such as checking protein function.
5. Conclusions
A mass spectrometer detects the m/z values of ions. From the MS2 spectra, the sequences of peptides or proteins can be determined. Using label-free or label-based methods, quantification can be obtained – lending insight into protein function. The basic analysis of identification and quantification can be implemented with three strategies: bottom-up, top-down, and middle-down. Each strategy has its pros and cons. When applied to histones, these strategies provide a wealth of information. In contrast to antibody-based methods, no prior knowledge is needed for MS analysis. Therefore, MS is a powerful technology for studying histone proteoforms and their functions. MS can provide information about novel types or sites of PTMs including combinational PTMs, distinguish histone variants, and discover histone-binding proteins. To complete these tasks, techniques need be optimized including sample preparation, mass spectrometry methods, and data analysis platforms. Though these techniques are imperfect, they have helped to resolve many practical problems. In the future, these techniques will be improved and more findings will come.
Acknowledgments
BAG acknowledges funding from an NIH Innovator grant (DP2OD007447) from the Office of the Director, and the National Science Foundation (NSF) Early Faculty CAREER award.
References
- 1.Smith LM, Kelleher NL. Proteoform: a single term describing protein complexity. Nat Methods. 2013;10:186. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnaudo AM, Garcia BA. Proteomic characterization of novel histone post-translational modifications. Epigenetics Chromatin. 2013;6:24. doi: 10.1186/1756-8935-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnaudo AM, Molden RC, Garcia BA. Revealing histone variant induced changes via quantitative proteomics. Crit Rev Biochem Mol Biol. 2011;46:284. doi: 10.3109/10409238.2011.577052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung P. Generation and characterization of antibodies directed against di-modified histones, and comments on antibody and epitope recognition. Methods Enzymol. 2004;376:221. doi: 10.1016/S0076-6879(03)76015-7. [DOI] [PubMed] [Google Scholar]
- 5.Britton LM, Gonzales-Cope M, Zee BM, Garcia BA. Breaking the histone code with quantitative mass spectrometry. Expert Rev Proteomics. 2011;8:631. doi: 10.1586/epr.11.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersson C-O. Mass Spectrometric Studies on Amino Acid and Peptide Derivatives. Acta Chemica Scandinavica. 1958;12:1353. [Google Scholar]
- 7.Dass C. Fundamentals of Contemporary Mass Spectrometry. John Wiley & Sons; 2007. p. 5. [Google Scholar]
- 8.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, et al. Protein and Polymer Analyses up to m/z 100 000 by Laser Ionization Time-of flight Mass Spectrometry. Rapid Communications in Mass Spectrometry. 1988;2:151. [Google Scholar]
- 10.Wells JM, McLuckey SA. Collision-induced dissociation (CID) of peptides and proteins. Methods Enzymol. 2005;402:148. doi: 10.1016/S0076-6879(05)02005-7. [DOI] [PubMed] [Google Scholar]
- 11.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101:9528. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dancik V, Addona TA, Clauser KR, Vath JE, Pevzner PA. De novo peptide sequencing via tandem mass spectrometry. J Comput Biol. 1999;6:327. doi: 10.1089/106652799318300. [DOI] [PubMed] [Google Scholar]
- 13.Ma B, Zhang K, Hendrie C, Liang C, Li M, et al. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun Mass Spectrom. 2003;17:2337. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
- 14.Taylor JA, Johnson RS. Sequence database searches via de novo peptide sequencing by tandem mass spectrometry. Rapid Commun Mass Spectrom. 1997;11:1067. doi: 10.1002/(SICI)1097-0231(19970615)11:9<1067::AID-RCM953>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 15.Eng JK, McCormack AL, John R, Yates I. An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. J Am Soc Mass Spectrom. 1994;5:976. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 16.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 17.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 18.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, et al. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 19.Bern M, Cai Y, Goldberg D. Lookup peaks: a hybrid of de novo sequencing and database search for protein identification by tandem mass spectrometry. Anal Chem. 2007;79:1393. doi: 10.1021/ac0617013. [DOI] [PubMed] [Google Scholar]
- 20.Fu Y, Xiu LY, Jia W, Ye D, Sun RX, et al. DeltAMT: a statistical algorithm for fast detection of protein modifications from LC-MS/MS data. Mol Cell Proteomics. 2011;10:M110 000455. doi: 10.1074/mcp.M110.000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye D, Fu Y, Sun RX, Wang HP, Yuan ZF, et al. Open MS/MS spectral library search to identify unanticipated post-translational modifications and increase spectral identification rate. Bioinformatics. 2010;26:i399. doi: 10.1093/bioinformatics/btq185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith CM, Haimberger ZW, Johnson CO, Wolf AJ, Gafken PR, et al. Heritable chromatin structure: mapping “memory” in histones H3 and H4. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16454. doi: 10.1073/pnas.182424999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith CM, Gafken PR, Zhang Z, Gottschling DE, Smith JB, Smith DL. Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4. Anal Biochem. 2003;316:23. doi: 10.1016/s0003-2697(03)00032-0. [DOI] [PubMed] [Google Scholar]
- 24.Bonaldi T, Imhof A, Regula JT. A combination of different mass spectroscopic techniques for the analysis of dynamic changes of histone modifications. Proteomics. 2004;4:1382. doi: 10.1002/pmic.200300743. [DOI] [PubMed] [Google Scholar]
- 25.Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 26.Syka JE, Marto JA, Bai DL, Horning S, Senko MW, et al. Novel linear quadrupole ion trap/FT mass spectrometer: performance characterization and use in the comparative analysis of histone H3 post-translational modifications. J Proteome Res. 2004;3:621. doi: 10.1021/pr0499794. [DOI] [PubMed] [Google Scholar]
- 27.Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, et al. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc. 2007;2:933. doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoopmann MR, Finney GL, MacCoss MJ. High-speed data reduction, feature detection, and MS/MS spectrum quality assessment of shotgun proteomics data sets using high-resolution mass spectrometry. Anal Chem. 2007;79:5620. doi: 10.1021/ac0700833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park K, Yoon JY, Lee S, Paek E, Park H, et al. Isotopic peak intensity ratio based algorithm for determination of isotopic clusters and monoisotopic masses of polypeptides from high-resolution mass spectrometric data. Anal Chem. 2008;80:7294. doi: 10.1021/ac800913b. [DOI] [PubMed] [Google Scholar]
- 30.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 31.Yuan ZF, Liu C, Wang HP, Sun RX, Fu Y, et al. pParse: a method for accurate determination of monoisotopic peaks in high-resolution mass spectra. Proteomics. 2012;12:226. doi: 10.1002/pmic.201100081. [DOI] [PubMed] [Google Scholar]
- 32.Niu M, Mao X, Ying W, Qin W, Zhang Y, Qian X. Determination of monoisotopic masses of chimera spectra from high-resolution mass spectrometric data by use of isotopic peak intensity ratio modeling. Rapid Commun Mass Spectrom. 2012;26:1875. doi: 10.1002/rcm.6293. [DOI] [PubMed] [Google Scholar]
- 33.Senko MW, Beu SC, Mclafferty FW. Determination of monoisotopic masses and Ion populations for large biomolecules from resolved isotopic distributions. J Am Soc Mass Spectrom. 1995;6:229. doi: 10.1016/1044-0305(95)00017-8. [DOI] [PubMed] [Google Scholar]
- 34.Senko MW, Beu SC, Mclafferty FW. Automated assignment of charge states from resolved isotopic peaks for multiply charged ions. J Am Soc Mass Spectrom. 1995;6:52. doi: 10.1016/1044-0305(94)00091-D. [DOI] [PubMed] [Google Scholar]
- 35.Horn DM, Zubarev RA, McLafferty FW. Automated reduction and interpretation of high resolution electrospray mass spectra of large molecules. J Am Soc Mass Spectrom. 2000;11:320. doi: 10.1016/s1044-0305(99)00157-9. [DOI] [PubMed] [Google Scholar]
- 36.Anzilotti C, Pratesi F, Tommasi C, Migliorini P. Peptidylarginine deiminase 4 and citrullination in health and disease. Autoimmun Rev. 2010;9:158. doi: 10.1016/j.autrev.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Tan M, Luo H, Lee S, Jin F, Yang JS, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lundgren DH, Hwang SI, Wu L, Han DK. Role of spectral counting in quantitative proteomics. Expert Rev Proteomics. 2010;7:39. doi: 10.1586/epr.09.69. [DOI] [PubMed] [Google Scholar]
- 39.Heinecke NL, Pratt BS, Vaisar T, Becker L. PepC: proteomics software for identifying differentially expressed proteins based on spectral counting. Bioinformatics. 2010;26:1574. doi: 10.1093/bioinformatics/btq171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carvalho PC, Hewel J, Barbosa VC, Yates JR., 3rd Identifying differences in protein expression levels by spectral counting and feature selection. Genet Mol Res. 2008;7:342. doi: 10.4238/vol7-2gmr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DiMaggio PA, Jr, Young NL, Baliban RC, Garcia BA, Floudas CA. A mixed integer linear optimization framework for the identification and quantification of targeted post-translational modifications of highly modified proteins using multiplexed electron transfer dissociation tandem mass spectrometry. Mol Cell Proteomics. 2009;8:2527. doi: 10.1074/mcp.M900144-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gan CS, Chong PK, Pham TK, Wright PC. Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ) J Proteome Res. 2007;6:821. doi: 10.1021/pr060474i. [DOI] [PubMed] [Google Scholar]
- 43.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Aggarwal K, Choe LH, Lee KH. Shotgun proteomics using the iTRAQ isobaric tags. Brief Funct Genomic Proteomic. 2006;5:112. doi: 10.1093/bfgp/ell018. [DOI] [PubMed] [Google Scholar]
- 45.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 46.Boyne MT, 2nd, Pesavento JJ, Mizzen CA, Kelleher NL. Precise characterization of human histones in the H2A gene family by top down mass spectrometry. J Proteome Res. 2006;5:248. doi: 10.1021/pr050269n. [DOI] [PubMed] [Google Scholar]
- 47.Siuti N, Roth MJ, Mizzen CA, Kelleher NL, Pesavento JJ. Gene-specific characterization of human histone H2B by electron capture dissociation. J Proteome Res. 2006;5:233. doi: 10.1021/pr050268v. [DOI] [PubMed] [Google Scholar]
- 48.Thomas CE, Kelleher NL, Mizzen CA. Mass spectrometric characterization of human histone H3: a bird’s eye view. J Proteome Res. 2006;5:240. doi: 10.1021/pr050266a. [DOI] [PubMed] [Google Scholar]
- 49.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 50.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 51.LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol Cell. 2008;30:51. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 53.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 54.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 55.Garcia BA, Barber CM, Hake SB, Ptak C, Turner FB, et al. Modifications of human histone H3 variants during mitosis. Biochemistry. 2005;44:13202. doi: 10.1021/bi050906n. [DOI] [PubMed] [Google Scholar]
- 56.Venable JD, Dong MQ, Wohlschlegel J, Dillin A, Yates JR. Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat Methods. 2004;1:39. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 57.Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11:O111 016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geromanos SJ, Vissers JP, Silva JC, Dorschel CA, Li GZ, et al. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics. 2009;9:1683. doi: 10.1002/pmic.200800562. [DOI] [PubMed] [Google Scholar]
- 59.Cheng C, Gross ML. Applications and mechanisms of charge-remote fragmentation. Mass Spectrom Rev. 2000;19:398. doi: 10.1002/1098-2787(2000)19:6<398::AID-MAS3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 60.Lam H, Deutsch EW, Eddes JS, Eng JK, King N, et al. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics. 2007;7:655. doi: 10.1002/pmic.200600625. [DOI] [PubMed] [Google Scholar]
- 61.Domon B, Aebersold R. Challenges and opportunities in proteomics data analysis. Mol Cell Proteomics. 2006;5:1921. doi: 10.1074/mcp.R600012-MCP200. [DOI] [PubMed] [Google Scholar]
- 62.Nesvizhskii AI, Vitek O, Aebersold R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat Methods. 2007;4:787. doi: 10.1038/nmeth1088. [DOI] [PubMed] [Google Scholar]
- 63.Bandeira N, Chambers MC, Cottrell JS, Deutsch EW, Kapp EA, et al. iPRG-2013: Proteome Informatics Research Group Study: Using RNA-Seq Data to Refine Proteomic Data Analysis. J Biomol Tech. 2013;24:s23. [Google Scholar]
- 64.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 65.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 66.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 67.Houel S, Abernathy R, Renganathan K, Meyer-Arendt K, Ahn NG, Old WM. Quantifying the impact of chimera MS/MS spectra on peptide identification in large-scale proteomics studies. J Proteome Res. 2010;9:4152. doi: 10.1021/pr1003856. [DOI] [PMC free article] [PubMed] [Google Scholar]