Abstract
Many biological experiments are not compatible with the use of immunofluorescence or genetically-encoded fluorescent tags or FRET-based reporters. Conjugation of existing kinase inhibitors to cell-permeable fluorophores can provide a generalized approach to develop fluorescent probes of intracellular kinases. Here, we report the development of a small molecule probe of Src through conjugation of BODIPY to two well-established, dual Src-Abl kinase inhibitors, dasatinib and saracatinib. We show that this approach is not successful for saracatinib, but that largely dasatinib-BODIPY retains the biological activity of its parent compound and can be used to monitor the presence of Src kinase in individual cells by flow cytometry and to track the localization of Src by fixed and live-cell fluorescence microscopy. This strategy may enable generation of additional kinase-specific probes useful in systems not amenable to genetic manipulation or used together with fluorescent proteins to enable a multiplexed assay read-out.
Keywords: fluorescent probes, proteins, signal transduction, cell recognition, kinase inhibitor
Introduction
Many proteins are regulated by changes in abundance or subcellular localization, and the analysis of these changes has become a mainstay of modern cell biology. Antibody-based immunofluorescence and genetically encoded fluorescent reporters are currently the most widely utilized methods for monitoring a given protein of interest, but they are not entirely adequate for all applications. For example, immunofluorescence-based staining of intracellular proteins requires fixation and permeabilization of cells, which precludes use of this approach in fluorescence-activated cell sorting experiments in which it is desirable to capture subpopulations of live cells based on the abundance of a given intracellular protein marker. Likewise, the need for fixation prevents the use of immunofluorescence in live cell imaging experiments designed to monitor dynamic changes in protein localization. Although genetically-encoded fluorescent tags[1] and FRET-based reporters[2] can be used in live cell imaging, these approaches are only compatible with systems in which genetic manipulation is possible. Fluorescent, cell-permeable small molecules that are specific ligands of a protein of interest can provide a complementary tool for use in fluorescence microscopy.[3] High-throughput screening of combinatorially synthesized fluorophore libraries has successfully yielded specific probes of DNA, RNA, as well as specific proteins.[4] In addition, fluorescent probes of specific proteins have been developed by rational design efforts in which a known ligand of the protein of interest is conjugated to a cell-permeable fluorophore.[5] We previously demonstrated proof of concept of this approach with kinases[6] by conjugating BI2536, a selective inhibitor of polo-like kinases (PLKs), to BODIPY, a cell-permeable fluorophore. The resulting bi-valent ligand retained the biochemical and cellular activity of the parent compound in biochemical and cell-based assays; moreover, it co-localized with PLK1 during different stages of mitosis. Signal transduction studies that rely upon measurements of kinase activity and substrate phosphorylation made in cellular lysates do not permit detection of changes in intracellular kinase localization or analysis of the role of these changes in the regulation of kinase function. Probes like BI-BODIPY that report on kinase localization may complement this significant limitation and permit the study of dynamic changes in intracellular kinase localization without requiring genetic manipulation of the cells being studied.
To extend this approach, we here have focused on small molecule inhibitors of Src and Abl family kinases because these kinases have demonstrated biomedical significance; moreover small molecules that are specific ligands of these kinase families have been well-studied and validated in vivo. Dasatinib (Sprycel,BMS-354825)[7] is an FDA-approved inhibitor of the BCR-Abl kinase, a fusion protein resulting from the Philadelphia chromosomal translocation that is the cause of chronic myelogenous leukemia (CML) and acute lymphoblastic leukemia (ALL). An aminothiazole, dasatinib has potent activity against a number of additional kinases, with subnanomolar activity against members of the SRC-family (Src, Lck, Fyn, Yes, Fgr, Hck, Blk, Fgr, Frk) and double-digit nanomolar activity against c-Kit, PDGFR, and members of the Ephrin and Tec kinase families, amongst others.[7–8] Dasatinib’s high affinity for the kinase active site of its targets has facilitated its use as an affinity reagent[9] and prompted investigation of 18F-labeled derivatives as radioimaging probes.[10] Saracatinib is also a potent, dual Src-Abl kinase inhibitor with a pharmacophore structurally distinct from that of dasatinib. It has been tested in humans as a potential therapeutic against numerous tumor types[11] and is currently in trials as a treatment for Alzheimer’s disease as well as ovarian, pancreatic, and thymic cancers and osteosarcoma. Although it is less potent against Src and Abl kinases than dasatinib, it has a more narrowly focused kinase inhibitory profile.[12]
In order to develop these compounds as potential tools for monitoring the intracellular localization of their respective targets, we synthesized BODIPY-conjugated derivatives of both dasatinib and saracatinib. Although these conjugates might reasonably be expected to report on the localization of all of the targets of the parent inhibitors, we chose Src as the focus of our efforts to test these reagents in flow cytometry and fluorescence microscopy assays because it is known to undergo dramatic changes in subcellular localization during mitosis, cytoskeletal re-organization, and membrane trafficking.[13] Src localization with focal adhesions at peripheral membranes is correlated with kinase activity[14] whereas its colocalization with microtubule-organizing centers in the perinuclear region is inactive;[15] moreover, Src expression and/or activity is upregulated in multiple types of cancer. Using ectopic expression of low levels of a Src-mCherry fusion protein to independently monitor target abundance and localization, we here demonstrate that dasatinib-BODIPY but not saracatinib-BODIPY can be used as a probe to visualize Src.
Results and Discussion
Design and synthesis of dasatinib-BODIPY and saracatinib-BODIPY
Existing high resolution co-crystal structures of each compound complexed with relevant kinase domains were utilized in selecting appropriate sites for derivitization. In co-crystal structures of dasatinib with Abl[16] (PDB ID 2GQG) and Src[17] (PDB ID 3G5D), the aminothiazole motif binds to the kinase ‘hinge’ segment between the N- and C-terminal kinase lobes while the piperazine group is directed towards the solvent-exposed portion of the ATP-binding site. This suggested that a linker appended to the distal piperazine nitrogen might not interfere with recognition of the ATP-binding pocket. Similarly, the piperazine group of saracatinib is solvent-exposed in the co-crystal structure of it complexed with Src (PDB ID 2H8).[12] BODIPY (ex/em 503/512 nm)[18] was chosen as the fluorophore in these experiments due to its demonstrated cell-permeability, high extinction coefficient, and the compatibility of its excitation and emission spectra with commonly used fluorescence microscopes and flow cytometers. Syntheses of dasatinib and saracatinib conjugated to BODIPY (dasatinib-BODIPY and saracatinib-BODIPY, respectively; Figure 1) were modifications of published procedures.[8a, 10b] Details and compound characterization are provided as Supporting Information.
Figure 1. Synthesis schemes of dasatinib-BODIPY and saracatinib-BODIPY.
(A) Synthesis of dasatinib-BODIPY. Compound 1 was prepared following literature procedures[7] and was coupled with tert-butyl 4-(6-chloro-2-methylpyrimidin-4-yl) piperazine-1-carboxylate to yield 2. Deprotection of 2 followed by amide coupling of 3 with BODIPY acid gave the target, dasatinib-BODIPY. (B) Synthesis of saracatinib-BODIPY. Compound 4 was prepared following published procedures[12] and then was converted to compound 5 through treatment with ammonia in methanol solution. The Mitsunobu reaction of 5 with tert-butyl 4-(2-hydroxyethyl)-1-piperazinecarboxylate 6 afforded 7. Deprotection of compound 7 followed by the amide coupling with BODIPY acid gave the target, saracatinib-BODIPY.
Dasatinib-BODIPY and saracatinib-BODIPY are potent inhibitors of relevant kinase targets
To determine if conjugation to BODIPY perturbs interactions of the small molecule with its target kinases, we compared the kinase inhibitory activities of dasatinib-BODIPY and saracatinib-BODIPY with those of their respective parental compounds. Due to dasatinib’s well-documented multi-targeted inhibition of kinases, we first profiled dasatinib-BODIPY’s biochemical inhibition of ten known targets of dasatinib. Dasatinib-BODIPY appeared to have lost inhibitory activity against c-kit, PDGFR-alpha, and PDGFR-beta kinases (IC50 values greater than 100 μM) but exhibited potent inhibition of Src family kinase members, Abl, BTK, and ephrin B2 (EphB2) kinases (Table 1). Although decreases in inhibitory activity were observed (~2- to 10-fold for Src family members, 11- fold for BTK and EphB2, and 40-fold for Abl), the single-digit micromolar IC50 values observed against each of these kinases suggested that dasatinib-BODIPY still recognized and might be useful for intracellular detection of these kinases.
Table 1. Biochemical activity of dasatinib-BODIPY.
The kinase inhibitory activity of dasatinib-BODIPY was profiled in biochemical assays against ten known targets of dasatinib. Biochemical IC50 values were measured by Z′-Lyte kinase assay[20] a homogeneous, coupled-enzyme assay utilizing a FRET-peptide substrate such that kinase inhibition leads to proteolytic cleavage of the substrate and loss of the FRET signal.
| Kinase | Dasatinib (IC50 μM) | Dasatinib-BODIPY (IC50 μM) |
|---|---|---|
| ABL1 | 0.03 | 1.3 |
| BTK | 1.4 | 2.7 |
| EPHB2 | 0.4 | 4.4 |
| FYN | 0.8 | 7.0 |
| HCK | 0.3 | 1.5 |
| KIT | 0.8 | 138.0 |
| LYN | 0.6 | 2.1 |
| PDGFRA | 0.5 | 112.0 |
| PDGFRB | 0.6 | 100 |
| SRC | 0.2 | 2.1 |
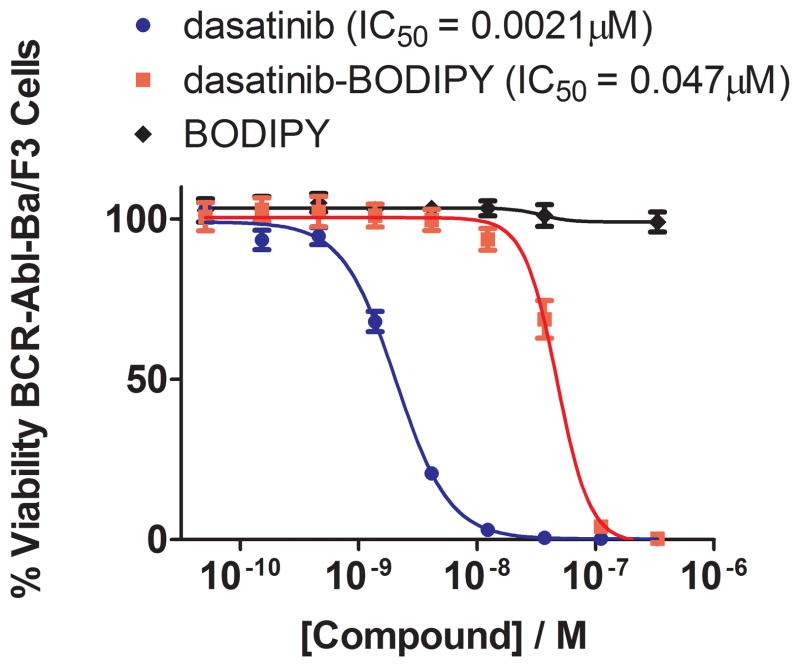
To test this possibility explicitly, we next performed experiments utilizing the Ba/F3 model to assess cell penetrance and intracellular kinase inhibition and hence intracellular binding of the kinase. Transformation of Ba/F3 cells by an oncogenic kinase relieves the dependence of this murine pro-B cell line on interleukin-3 (IL-3) for proliferation. Inhibition of the oncogenic kinase in turn blocks proliferation of the cells in the absence of IL-3 and provides a convenient readout for intracellular kinase inhibition.[19] We performed dose-response experiments to measure the anti-proliferative activity of dasatinib-BODIPY and saracatinib-BODIPY against BCR-Abl Ba/F3 cells. Consistent with the results of the biochemical kinase inhibition profiling (Table 1), dasatinib-BODIPY inhibits the proliferation of BCR-Abl Ba/F3 cells with activity comparable to that of dasatinib (EC50 values 47 nM and 2.1 nM, respectively) (Figure 2). This inhibition is not a consequence of non-specific cytotoxicity since dasatinib-BODIPY has no effect on the IL-3-dependent growth of parental Ba/F3 cells at concentrations below 5 μM (Supporting Figure 1). We likewise observed that saracatinib-BODIPY inhibits the proliferation of BCR-Abl transformed Ba/F3 cells in a dose-responsive manner, albeit with an approximately 10-fold decrease in potency relative to saracatinib itself (Supporting Figure 1) and without cytotoxic effects on parental Ba/F3 cells (Supporting Figure 1).
Figure 2. Cellular activity of dasatinib-BODIPY.
Inhibition of the proliferation of BCR-Abl Ba/F3 cells was used as a proxy to demonstrate cell penetrance as well as recognition and inhibition of BCR-Abl. Cell viability was measured in the presence of increasing concentrations of dasatinib, dasatinib-BODIPY, and free BODIPY to permit determination of IC50 values. Despite a 23-fold decrease in potency relative to dasatinib, dasatinib-BODIPY was a potent, sub-micromolar inhibitor of BCR-Abl Ba/F3 cell proliferation. This was due to the kinase-targeting moiety of dasatinib-BODIPY since free BOIDPY had no detectable effect on the viability of these cells.
Dasatinib-BODIPY labels Src in single cells as monitored by flow cytometry
The effects of dasatinib-BODIPY and saracatinib-BODIPY on the proliferation of BCR-Abl-dependent Ba/F3 cells demonstrate two characteristics essential for their use as probes of intracellular kinases: first, the compounds penetrate the plasma membrane; and second, the compounds engage a relevant kinase target. Despite their loss in potency relative to the parental compounds, we reasoned that this might not impede their use as probes because in many live cell applications the ideal probe would engage target(s) with affinity and specificity sufficient to permit their detection but without causing a significant biological perturbation. To evaluate dasatinib-BODIPY and saracatinib-BODIPY as probes of intracellular kinases explicitly, we engineered immortalized adherent hepatoma cell line Huh7 to express a Src-mCherry (ex/em 587/610 nm)[21] fusion protein, thereby enabling us to independently monitor the presence of Src via mCherry fluorescence. Huh7 cells transiently transfected with the Src-mCherry expression plasmid were incubated with 100 nM dasatinib-BODIPY or saracatinib-BODIPY. BI-BODIPY was used as a control since we have previously demonstrated that it can be used for intracellular staining of polo-like kinases,[6] and free BODIPY was employed as a negative control. Following washes with buffer, the cells were harvested as a single-cell suspension, fixed, and then analyzed by flow cytometry. Analysis of mCherry fluorescence versus forward scatter indicated variable expression of Src-mCherry in the transfected cell population (Figure 3A and Supporting Figure 2) with fluorescence intensities that were relatively modest to weak on a per cell basis, likely due to low expression of the Src-mCherry. We focused our analysis on the population of cells with the highest mCherry fluorescence (mCherryhi), reasoning that these cells would provide the best signal for correlating probe fluorescence with the presence of c-Src kinase.
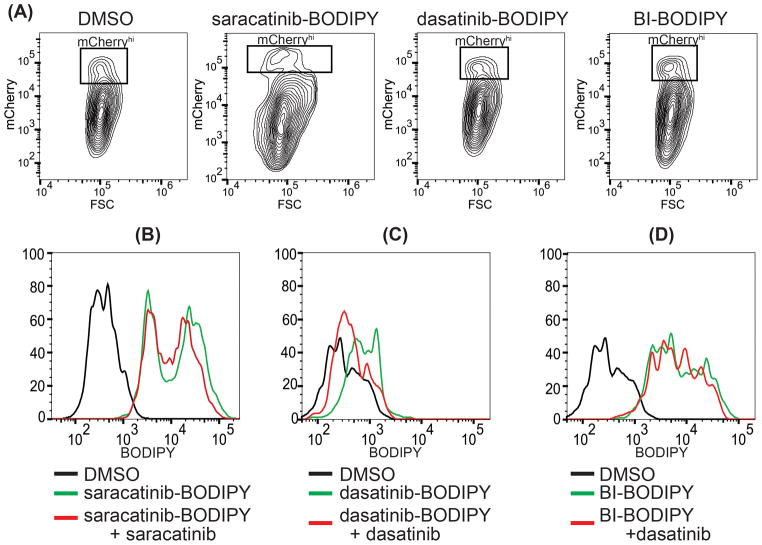
Figure 3. Evaluation of dasatinib-BODIPY and saracatinib-BODIPY as probes for flow cytometry.
Src-mCherry expressing Huh7 cells were treated with DMSO (negative control) or with 100 nM saracatinib-BODIPY, dasatinib-BODIPY, or BI-BODIPY. (A) mCherry fluorescence profile versus forward scatter showing the presence of a distinct high fluorescence population (mCherryhi) for each compound. This population was gated for further analysis. (B–D) For competition experiments, cells were labeled with 100 nM saracatinib-BODIPY, dasatinib-BODIPY, or BI-BODIPY in the presence or absence of 10 μM free saracatinib or dasatinib as competitor as indicated. Histogram plots for the BODIPY fluorescence intensity are shown. Unique peak profiles for saracatinib-BODIPY, dasatinib-BODIPY, and BI-BODIPY suggest that staining reflects binding of these probes to specific targets dictated by the kinase-targeting group and not by generic interactions of the fluorophore. (B) Competition with unlabeled saracatinib does not significantly reduce the fluorescence intensity of saracatinib-BODIPY. In contrast, unlabeled dasatinib significantly reduces the fluorescence intensity of (C) dasatinib-BODIPY but not (D) BI-BODIPY, suggesting that this competition is specific. Data are representative of four independent replicates.
Histogram plots of BODIPY fluorescence intensity versus cell number for the mCherryhi cells exhibited peak profiles for dasatinib-BODIPY-, saracatinib-BODIPY-, BI-BODIPY-labeled samples that were distinct from one another as well as distinct from the histogram plot for cells labeled with free BODIPY (Figure 3B–D and Supporting Figure 2B). This suggested that the staining is not due to generic interactions of the BODIPY moiety with cellular components but rather due to interactions of the kinase-targeting moieties with distinct kinases. While cells labeled with dasatinib-BODIPY exhibited only modest BODIPY fluorescence, this signal was significant over background and clearly higher than that of cells stained with free BODIPY (Figure 3C and Supporting Figure 2B–C).
To more directly test whether staining with saracatinib-BODIPY and dasatinib-BODIPY were due to their respective kinase-targeting moieties, we performed competition experiments using the unlabeled, parental compounds. Labeling of cells with saracatinib-BODIPY in the presence of excess saracatinib, we observed a slight shift in the histogram plot of BODIPY fluorescence intensity versus cell number (Figure 3B); however, through repeated experiments this very slight difference was found to not be statistically significant (Supporting Figure 2). This suggests that although saracatinib-BODIPY retains some kinase inhibitory activity, it does not selectively label Src-mCherry and likely gives signal in flow cytometry due to non-specific staining or binding to a target other than Src or the BCR-Abl kinase targeted in the cell-based kinase activity assay since Huh7 cells lack this kinase. Further experiments are needed to determine how coupling of saracatinib to BODIPY via the piperazine site affects the affinity, kinetics, and selectivity of the compound’s interactions with Src and other kinase targets. In contrast, incubation of cells with excess unlabeled dasatinib resulted in a reproducible and statistically significant loss in the BODIPY fluorescence of the dasatinib-BODIPY-labeled mCherryhi cells (Figure 3C and Supporting Figure 2D). The effect of the dasatinib competitor, moreover, appeared to be specific since excess dasatinib had no effect on BI-BODIPY-labeled cells (Figure 3D and Supporting Figure 2D). These observations demonstrate the specificity of dasatinib-BODIPY as a probe for Src-mCherry -- and presumably of Src -- in these experiments and illustrate the potential utility of dasatinib-BODIPY in flow cytometry applications to identify specific populations of cells based on the abundance of a kinase target of interest. We note that the relatively low signal- to-noise ratio for dasatinib-BODIPY-labeled cells may have been an artifact caused by low expression of the Src-mCherry reporter on which we gated cells for analysis. Given the many known kinase targets of dasatinib-BODIPY, we anticipate that conditions for staining and cytometric analysis can be optimized for the specific cell type, specific target, and target abundance in order to achieve more favorable signal to noise.
Dasatinib-BODIPY allows visualization of Src by fluorescence microscopy
To explore the potential use of dasatinib-BODIPY as an imaging probe in fluorescence microscopy experiments, we initially assessed its staining specificity in fixed cells. Src-mCherry-expressing Huh7 cells were first stained with dasatinib-BODIPY, BI-BODIPY, or free BODIPY then fixed and stained with DAPI to permit visualization of nuclei. Samples were then mounted and imaged by confocal fluorescence microscopy. Dasatinib-BODIPY-staining appeared as distinct puncta that appeared to colocalize with Src-mCherry whereas there was only incidental overlap of the BI-BODIPY stain with Src-mCherry and no signal observed for cells labeled with free BODIPY or for unlabelled cells incubated with DMSO (Figure 4A–C, Supporting Figure 3A and B). This suggested that the subcellular localization of dasatinib-BODIPY is mediated by its specific binding to Src-mCherry and is not driven by the BODIPY moiety. To examine dasatinib-BODIPY’s utility as a probe in live-cell imaging applications, we stained Huh7 cells expressing Src-mCherry with dasatinib-BODIPY or BI-BODIPY and then imaged these samples directly. Dasatinib-BODIPY and Src-mCherry appeared to colocalize significantly in live-cell images collected at thirty second intervals over a two-and-a-half minute time frame; moreover, dynamic changes in the localization of dasatinib-BODIPY appeared to coincide with those of Src-mCherry under basal unstimulated conditions during this experiment (Figure 4C and Supporting Movie 1). This colocalization of dasatinib-BODIPY with Src-mCherry appeared to be mediated by the kinase-targeting moiety since it was not observed with BI-BODIPY (data not shown).
Figure 4. Evaluation of dasatinib-BODIPY as a probe for Src in fluorescence microscopy.
Src-mCherry-expressing Huh7 cells were incubated with 100 nM free BODIPY, dasatinib-BODIPY, or BI-BODIPY for two hours and then processed for imaging (see Materials and Methods). A single plane from a representative Z-series is shown. Images are representative of three separate experiments. (A) Cells labeled with free BODIPY exhibit no signal above backround. Src-mCherry (red) in fixed cells appears to co-localize with (B) dasatinib-BODIPY (green) but not with (C) BI-BODIPY (green). Mander’s coefficients, a measurement of colocalization, averaged 0.858 and 0.567 for dasatinib-BODIPY and BI-BODIPY, respectively (see Supporting Figure 3C). (D) For live cell imaging, Src-mCherry-expressing Huh7 cells were incubated with 100 nM dasatinib-BODIPY for two hours and then washed twice with buffer and imaged. Images were captured every 30 seconds for 2.5 minutes. Images shown are a single confocal plane. Src-mCherry and dasatinib-BODIPY co-localized (mean Mander’s coefficient of 0.858; Supporting Figure 3C) and appeared to change localization together in the cytoplasm (see also Supporting Movie 1).
We confirmed these qualitative impressions of dasatinib-BODIPY’s colocalization with Src-mCherry by determining Mander’s coefficients for representative images as a metric of colocalization. With a coefficient of 1 representing complete colocalization and 0 representing the complete absence of colocalization of the BODIPY and mCherry signals, dasatinib-BODIPY-labeled cells had Mander’s coefficients ranging from 0.78 to 0.92 for both the fixed and live-cell experiments (mean 0.86), indicative of considerable colocalization of these two signals. In contrast, Mander’s coefficients for colocalization of BI-BODIPY with Src-mCherry ranged from 0.43 to 0.69 (mean 0.57), indicating that overlap of these two signals was considerably less and consistent with the idea that this control probe does not co-localize or interact with Src. Taken together, these experiments illustrate the use of dasatinib-BODIPY to selectively monitor the intracellular localization of a relevant kinase target in live cells and suggest that this compound may be useful in monitoring intracellular kinase dynamics. More broadly, these results suggest that cell-penetrant small molecule probes like dasatinib-BODIPY can fill an important unmet need in live-cell imaging experiments that are not amenable to immunofluorescence and/or the expression of fusion proteins.
Conclusion
Fluorescence detection provides one of the most powerful methods to study intracellular trafficking of proteins by fluorescence microscopy and to identify specific populations of cells expressing a protein of interest by flow cytometry. The majority of studies to date have utilized reporter proteins requiring genetic manipulation of the system being studied. Small molecule fluorophores with selectivity towards particular biomolecules of interest such as DNA or towards particular organelles (mitochondria, Golgi, etc.) have been used prior to the advent of molecular biology.[18] Many of these early probes were empirically discovered, and the molecular recognition and fluorescence properties are embedded in a single core structure.
A complementary approach is to create bi-valent ligands that contain separate molecular recognition and fluorescent elements. The primary advantage of the bivalent ligand approach is that it is modular and can exploit the tremendous number of selective small molecular binders that have been generated as a result of chemical biology and drug discovery research. We have focused our efforts on developing fluorescent ligands for protein kinases because of the large number of selective kinase ligands that have been developed and because there is a significant interest in being able to observe changes in intracellular kinase localization under physiological and pathophysiological conditions. We have demonstrated that it is possible to tether a cell permeable BODIPY fluorophore to the solvent exposed region of kinase inhibitors such as BI2356 and dasatinib in a manner that preserves biological activity and that enables fluorescent imaging using fluorescent microscopy or flow cytometry. We have also observed that with other kinase inhibitors, such as Mps1-IN-1[6] and saracatinib, that the approach is not successful either due to loss of binding potency and/or because the ligand binds too promiscuously to other intracellular targets resulting in a high background signal. As is well known in the field of making radioisotope imaging probes (such as 18F positron imaging probes), it must be empirically determined which ligands possess sufficient target to off-target binding to yield useful probes. We anticipate that a large number of useful kinase-specific fluorescent probes can be generated using the bi-valent ligand strategy by systematically mining the extensive known pharmacopeia of kinase inhibitors. These probes can then be used in systems that do not allow for genetic manipulation or used together with fluorescent proteins to enable a multiplexed assay read-out.
Experimental Section
Synthesis of dasatinib-BODIPY and saracatinib-BODIPY
Synthesis of dasatinib-BODIPY and saracatinib-BODIPY were performed based on published synthetic methods.[7, 12] Detailed synthetic methods and characterization are provided as Supporting Information.
Cell Culture
Human hepatoma cells (Huh7) were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) and were transfected with Lipofectamine 2000 (Life Technologies) according to manufacturer’s protocol. Parental Ba/F3 and BCR-Abl-transformed Ba/F3 cells were provided by J.D. Griffin (Dana Farber Cancer Institute, Boston). Both BCR-Abl Ba/F3 and parental Ba/F3 cell lines were cultured in RPMI 1640 (Life Science) with 10% FBS; parental Ba/F3 cell medium was supplemented with 10% WEHI-conditioned medium as a source of IL-3. All cells were grown in humidified incubators maintained at 5% CO2.
Ba/F3 Proliferation Assay
Parental and BCR-Abl BaF3 cells (0.3–0.6 × 106 per mL) were plated in duplicate in 96-well plates with increasing compound concentrations (0.005–10 μM). After incubation at 37 °C for 48 hours, cell viability was assessed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). Inhibition of cell proliferation was calculated as a percentage of growth based on DMSO-treated cells, and IC50 values were determined by using the four parameter dose response non-linear regression analysis included in the Graph Pad Prism 6 software.
Src-mCherry
c-Src-eGFP in the pEGFP-N1 vector was a kind gift from the lab of Marilyn Resh (Memorial Sloan-Kettering Cancer Center).[22] c-Src-eGFP was subcloned into plasmid p-mCherry-N1 (Promega) via EcoRI and BamHI restriction sites to produce the Src-mCherry expression plasmid
Microscopy
Fixed Cells
Huh7 cells on 12 mm coverslips were transfected with the Src-mCherry expression plasmid. Forty-eight hours post-transfection, cells were treated with 100 nM dasatinib-BODIPY or BI-BODIPY for two hours. Cells were then washed with PBS, fixed with 4% paraformaldehyde (PFA) for 15 minutes, washed again with PBS, and permeabilized with 0.1% (v/v) Triton X100 for 15 minutes. Cells were then stained with DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen) for 15 minutes and washed with PBS before mounting on glass slides with ProLong Gold (Invitrogen).
Live Cells
Huh7 cells were transfected with the Src-mCherry expression plasmid, and then 48 hours post-transfection incubated with 100 nM dasatinib-BODIPY or BI-BODIPY for two hours. Samples were then placed in an AttoFluor Cell Chamber (Invitrogen) and perfused with DMEM medium lacking phenol red. The cell chamber was then placed in a heated stage plate over the objectives and maintained at 37°C. The air above the cells was humidified and maintained at 37°C, 5% CO2 for the duration of the experiment. Samples were imaged every 30 seconds for 2.5 minutes. All images were acquired using a QuantEM (Photometrics) cooled charge-coupled device as part of a CSU-X1 spinning disk confocal system (Yokogawa Electric Corporation) using a 100X, 1.4NA objective (Zeiss). Samples were sequentially excited by 405nm (DAPI), 488 nm (BODIPY) and 561 nm (mCherry) lasers (Coherent), and signal was collected through emission filters 452/45 nm, 525/50 nm and 607/36 nm (Semrock), respectively. Initial data capture was performed using Slidebook software (Intelligent Imaging Innovations). Further processing was performed using Metamorph (Molecular Devices). Colocalization between BODIPY (green) compounds and Src-mCherry (red) was assessed by calculation of the Mander’s coefficient (ratio of intensity of green pixels coincident with red pixels to total green pixel intensity) using the ImageJ plug-in JACoP.[23] Each figure represents a single plane from the Z-stack.
Flow Cytometry
Forty-eight hours post-transfection with the Src-mCherry expression plasmid, Huh7 cells were incubated with DMSO or 10 μM dasatinib (competition samples) for one hour at 37 degree C, 5% CO2. Cells were subsequently incubated with 100 nM dasatinib-BODIPY or BI-BODIPY for two hours at 37 degrees C, 5% CO2. Cells were then washed with PBS, lifted from the culture dish with trypsin, washed with PBS, fixed in 2% PFA, and then analyzed with a LSR Fortessa (Becton Dickson). Samples were excited by 488 nM (BODIPY) and 561 nM (mCherry) lasers and signal was subsequently collected through 530/30 nM and 610/20 nM emission filters, respectively. Data were initially collected on FACSDiva software (Becton Dickson). Subsequent analyses were performed using FlowJo. Live cells were gated using forward scatter (FSC) and side scatter (SSC) parameters. Cells transfected with the Src-mCherry expression plasmid demonstrated a distinct population of mCherry bright cells. These cells were gated (mCherryhi) for the analysis of the BODIPY signal in samples labeled with dasatinib-BODIPY, saracatinib-BODIPY, BI-BODIPY, and negative controls. Individual histograms of the BODIPY fluorescence signal were produced using values from the BODIPY channel in the mCherryhi gate. Mean fluorescence intensity (MFI) was determined within FlowJo, and Graphpad Prism 6 was used to analysis significance of changes MFI.
Kinase profiling of dasatinib-BODIPY
The inhibitory activity of dasatinib-BODIPY against ten of its known substrates was performed by Z’lyte assay (Life Technologies), a coupled assay system in which the FRET signal from a substrate peptide reflects its phosphorylation by a recombinant kinase and kinase inhibition leads to decreased FRET signal.[20]
Supplementary Material
Acknowledgments
This work was supported by NIH/NCI grant U54 CA156732 (PIs: Emmons and Colon) via the University of Massachusetts Boston – Dana-Farber/Harvard Cancer Center (UMB-DFHCC) U54 Comprehensive Partnership to Reduce Cancer Health Disparities and by NIH/NIAID grant AI076442 (PI: P.L. Yang). Hong Zeng and Dianrong Zhu are gratefully acknowledged for their participation in synthetic work. We also acknowledge the Live Cell Imaging Core of the New England Regional Center of Excellence Biodefense and Emerging Infectious Diseases (U54 AI057159) and ICCB-Longwood Screening Facility for use of equipment and technical support. DNA sequencing was performed by the DNA Resource Core of the Dana-Farber/Harvard Cancer Center.
Footnotes
Supporting information for this article is available.
Contributor Information
Wei Zhang, Email: wei2.zhang@umb.edu.
Nathanael S. Gray, Email: nathanael_gray@dfci.harvard.edu.
Priscilla L. Yang, Email: priscilla_yang@hms.harvard.edu.
References
- 1.a) Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. J Am Chem Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]; b) Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 2.Zhou X, Herbst-Robinson KJ, Zhang J. Methods Enzymol. 2012;504:317–340. doi: 10.1016/B978-0-12-391857-4.00016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Bhardwaj A, Kaur J, Wuest F, Knaus EE. Chem Med Chem. 2014;9:109–116. doi: 10.1002/cmdc.201300355. [DOI] [PubMed] [Google Scholar]; b) Cao B, Hutt OE, Zhang Z, Li S, Heazlewood SY, Williams B, Smith JA, Haylock DN, Savage GP, Nilsson SK. Org Biomol Chem. 2014;12:965–978. doi: 10.1039/c3ob42332h. [DOI] [PubMed] [Google Scholar]
- 4.Vendrell M, Zhai D, Er JC, Chang YT. Chem Rev (Washington, DC, U S) 2012;112:4391–4420. doi: 10.1021/cr200355j. [DOI] [PubMed] [Google Scholar]
- 5.a) Jung D, Min K, Jung J, Jang W, Kwon Y. Mol BioSyst. 2013;9:862–872. doi: 10.1039/c2mb25422k. [DOI] [PubMed] [Google Scholar]; b) Liu CY, Zhang H, Christofi FL. Cell Tissue Res. 1998;293:57–73. doi: 10.1007/s004410051098. [DOI] [PubMed] [Google Scholar]; c) Tiyanont K, Doan T, Lazarus MB, Fang X, Rudner DZ, Walker S. Proc Natl Acad Sci U S A. 2006;103:11033–11038. doi: 10.1073/pnas.0600829103. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zambaldo C, Sadhu KK, Karthikeyan G, Barluenga S, Daguer JP, Winssinger N. Chemical Science. 2013;4:2088–2092. [Google Scholar]
- 6.Zhang Z, Kwiatkowski N, Zeng H, Lim SM, Gray NS, Zhang W, Yang PL. Mol BioSyst. 2012;8:2523–2526. doi: 10.1039/c2mb25099c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das J, Chen P, Norris D, Padmanabha R, Lin J, Moquin RV, Shen Z, Cook LS, Doweyko AM, Pitt S, Pang S, Shen DR, Fang Q, de Fex HF, McIntyre KW, Shuster DJ, Gillooly KM, Behnia K, Schieven GL, Wityak J, Barrish JC. J Med Chem. 2006;49:6819–6832. doi: 10.1021/jm060727j. [DOI] [PubMed] [Google Scholar]
- 8.a) Chen Z, Lee FY, Bhalla KN, Wu J. Mol Pharmacol. 2006;69:1527–1533. doi: 10.1124/mol.105.020172. [DOI] [PubMed] [Google Scholar]; b) Hantschel O, Rix U, Schmidt U, Burckstummer T, Kneidinger M, Schutze G, Colinge J, Bennett KL, Ellmeier W, Valent P, Superti-Furga G. Proc Natl Acad Sci U S A. 2007;104:13283–13288. doi: 10.1073/pnas.0702654104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, Sweetman G, Bauer A, Bouwmeester T, Hopf C, Kruse U, Neubauer G, Ramsden N, Rick J, Kuster B, Drewes G. Nat Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]; b) Rix U, Hantschel O, Durnberger G, Remsing Rix LL, Planyavsky M, Fernbach NV, Kaupe I, Bennett KL, Valent P, Colinge J, Kocher T, Superti-Furga G. Blood. 2007;110:4055–4063. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- 10.a) Benezra M, Hambardzumyan D, Penate-Medina O, Veach DR, Pillarsetty N, Smith-Jones P, Phillips E, Ozawa T, Zanzonico PB, Longo V, Holland EC, Larson SM, Bradbury MS. Neoplasia. 2012;14:1132–1143. doi: 10.1593/neo.121750. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Dunphy MP, Zanzonico P, Veach D, Somwar R, Pillarsetty N, Lewis J, Larson S. Molecular Imaging and Biology. 2012;14:25–31. doi: 10.1007/s11307-010-0462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Veach DR, Namavari M, Pillarsetty N, Santos EB, Beresten-Kochetkov T, Lambek C, Punzalan BJ, Antczak C, Smith-Jones PM, Djaballah H, Clarkson B, Larson SM. J Med Chem. 2007;50:5853–5857. doi: 10.1021/jm070342g. [DOI] [PubMed] [Google Scholar]
- 11.a) Gangadhar TC, Clark JI, Karrison T, Gajewski TF. Invest New Drugs. 2013;31:769–773. doi: 10.1007/s10637-012-9897-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Laurie SA, Goss GD, Shepherd FA, Reaume MN, Nicholas G, Philip L, Wang L, Schwock J, Hirsh V, Oza A, Tsao MS, Wright JJ, Leighl NB. Clinical Lung Cancer. 2014;15:52–57. doi: 10.1016/j.cllc.2013.08.001. [DOI] [PubMed] [Google Scholar]; c) Nam HJ, Im SA, Oh DY, Elvin P, Kim HP, Yoon YK, Min A, Song SH, Han SW, Kim TY, Bang YJ. Mol Cancer Ther. 2013;12:16–26. doi: 10.1158/1535-7163.MCT-12-0109. [DOI] [PubMed] [Google Scholar]
- 12.Hennequin LF, Allen J, Breed J, Curwen J, Fennell M, Green TP, Lambert-van der Brempt C, Morgentin R, Norman RA, Olivier A, Otterbein L, Ple PA, Warin N, Costello G. J Med Chem. 2006;49:6465–6488. doi: 10.1021/jm060434q. [DOI] [PubMed] [Google Scholar]
- 13.Bjorge JD, Jakymiw A, Fujita DJ. Oncogene. 2000;19:5620–5635. doi: 10.1038/sj.onc.1203923. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan KB, Swedlow JR, Varmus HE, Morgan DO. J Cell Biol. 1992;118:321–333. doi: 10.1083/jcb.118.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.a) Kaplan KB, Bibbins KB, Swedlow JR, Arnaud M, Morgan DO, Varmus HE. EMBO J. 1994;13:4745–4756. doi: 10.1002/j.1460-2075.1994.tb06800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sandilands E, Frame MC. Trends Cell Biol. 2008;18:322–329. doi: 10.1016/j.tcb.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Tokarski JS, Newitt JA, Chang CY, Cheng JD, Wittekind M, Kiefer SE, Kish K, Lee FY, Borzillerri R, Lombardo LJ, Xie D, Zhang Y, Klei HE. Cancer Res. 2006;66:5790–5797. doi: 10.1158/0008-5472.CAN-05-4187. [DOI] [PubMed] [Google Scholar]
- 17.Getlik M, Grutter C, Simard JR, Kluter S, Rabiller M, Rode HB, Robubi A, Rauh D. J Med Chem. 2009;52:3915–3926. doi: 10.1021/jm9002928. [DOI] [PubMed] [Google Scholar]
- 18.Haugland RP. Molecular Probes Handbook - a Guide to Fluorescent Probes and Labeling Technologies. 11. Life Technologies; 2010. [Google Scholar]
- 19.a) Melnick JS, Janes J, Kim S, Chang JY, Sipes DG, Gunderson D, Jarnes L, Matzen JT, Garcia ME, Hood TL, Beigi R, Xia G, Harig RA, Asatryan H, Yan SF, Zhou Y, Gu XJ, Saadat A, Zhou V, King FJ, Shaw CM, Su AI, Downs R, Gray NS, Schultz PG, Warmuth M, Caldwell JS. Proc Natl Acad Sci U S A. 2006;103:3153–3158. doi: 10.1073/pnas.0511292103. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Warmuth M, Kim S, Gu XJ, Xia G, Adrian F. Current Opinion in Oncology. 2007;19:55–60. doi: 10.1097/CCO.0b013e328011a25f. [DOI] [PubMed] [Google Scholar]
- 20.Rodems SM, Hamman BD, Lin C, Zhao J, Shah S, Heidary D, Makings L, Stack JH, Pollok BA. Assay Drug Dev Technol. 2002;1:9–19. doi: 10.1089/154065802761001266. [DOI] [PubMed] [Google Scholar]
- 21.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 22.Donepudi M, Resh MD. Cell Signal. 2008;20:1359–1367. doi: 10.1016/j.cellsig.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolte S, Cordelieres FP. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.