Abstract
Uganda's forests are globally important for their conservation values but are under pressure from increasing human population and consumption. In this study, we examine how conversion of natural forest affects soil bacterial and fungal communities. Comparisons in paired natural forest and human-converted sites among four locations indicated that natural forest soils consistently had higher pH, organic carbon, nitrogen, and calcium, although variation among sites was large. Despite these differences, no effect on the diversity of dominant taxa for either bacterial or fungal communities was detected, using polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE). Composition of fungal communities did generally appear different in converted sites, but surprisingly, we did not observe a consistent pattern among sites. The spatial distribution of some taxa and community composition was associated with soil pH, organic carbon, phosphorus and sodium, suggesting that changes in soil communities were nuanced and require more robust metagenomic methods to understand the various components of the community. Given the close geographic proximity of the paired sampling sites, the similarity between natural and converted sites might be due to continued dispersal between treatments. Fungal communities showed greater environmental differentiation than bacterial communities, particularly according to soil pH. We detected biotic homogenization in converted ecosystems and substantial contribution of β-diversity to total diversity, indicating considerable geographic structure in soil biota in these forest communities. Overall, our results suggest that soil microbial communities are relatively resilient to forest conversion and despite a substantial and consistent change in the soil environment, the effects of conversion differed widely among sites. The substantial difference in soil chemistry, with generally lower nutrient quantity in converted sites, does bring into question, how long this resilience will last.
Introduction
Tropical rainforests (TRF) possess most of the world's terrestrial biodiversity and deforestation is the leading cause of biodiversity loss [1], [2]. Due to their high biodiversity and endemism, the tropical rainforests in Uganda's Nile river watershed are among the world's most important for their conservation values. But these areas are under pressure. The United Nations Population Division [3] predicts that the population of the Nile Basin states will increase by 57% from 2010 to 2030, reaching 647 million people. This rapid population growth, high levels of poverty and prevalent civil insecurity continue to exert severe pressure on natural resources in the region. Uganda in particular has one of the world's highest population growth rates (3.2% per year) [4]. Most of this growing population (nearly 80%) is dependent on agriculture leading to large scale and continuing conversion of natural habitats [5].
Soil communities form the foundation of any ecosystem, in terms of nutrient cycling and availability, so understanding how land conversion affects these communities is an important first step. The effect of land use change on soil microbial communities has been studied in South American and Southeast Asian forests [6], [7], but not in the biodiversity hotspots of the Nile river watershed. There is considerable global concern about the loss of biodiversity and the consequences for human well-being [8]. Microorganisms in particular play a vital role in many ecological processes and environmental services [9]: these roles are not always apparent or well characterized but if all microbes died the world would rapidly become buried in undecomposed dead material. Due to their significance in maintaining ecosystem function and productivity [9], [10], our study offers a vital exploratory appraisal of microbial community dynamics in natural TRF and human-converted sites. We don't know if there are reasons to be concerned unless we look. Developing such knowledge is critical at this point, because populations in the Nile river watershed are highly dependent on forests for basic requirements such as food and fuel wood, with the environment contributing between 40–60% of the gross domestic product (GDP) of the Nile riparian states [11].
Because of widespread loss of biodiversity, focus from species conservation within particular habitats has been shifted to conservation of communities [12], [13]. It is therefore important to explore and understand how composition and diversity changes across spatial scales in a given context [14]–[16]. Changes in ecosystems caused by conversion to intensive management can lead to biotic homogenization, the increase in community similarity over time and/or space and an implied loss of rare and vulnerable taxa when examined at larger scales [17]–[19]. Because microorganisms are the most diverse organisms on earth with most taxa and respective functions and behaviors as yet unknown, determining their sensitivities and biogeography remains a major challenge. But in the longer term such knowledge will help us better understand the sustainability of land-use systems and associated environmental values.
This study was therefore necessary as a first step in exploring these relationships, and to enhance understanding so as to contribute to the informed and appropriate stewardship of the region's natural resources. Our objective was to establish how forest conversion and soil factors affect soil bacterial and fungal diversity and community composition in the tropical rain-forests in the Nile river watershed of Uganda. We chose four forest sites found within protected areas, with paired treatments within each forest; (1) natural and (2) converted ecosystem sites. The natural forest ecosystem at each site had suffered minimal human disturbance, while converted areas had been transformed to cropland. These matched sites found in different locations and environmental conditions each experienced different land use histories, conservation circumstances and individual challenges for management.
In each matched set of natural and converted sites, we compared soil physical and chemical properties and microbial community diversity and composition using standard PCR-based genotyping techniques. We then calculated community similarity indices between sites. This approach would allow us to examine both environmental and biotic changes in the soil community associated with conversion. Disturbances of sufficient magnitude or duration may alter an ecosystem and force a different regime of predominant processes and structures that favor some populations over others [20].
We tested the null hypotheses that there was no difference in soil properties, band-types, and diversity between treatments. The influence of soil properties on microbial community diversity was measured by discriminant analysis and canonical correspondence analysis (CCA), [21], [22]. Because additive partitioning of diversity provides a useful framework for quantifying the spatial patterns of diversity across hierarchical spatial scales [23], [24], we partitioned total diversity (γ) in each ecosystem type (natural and converted) into additive components representing within-community diversity (α) and between-community diversity (β). Our objective was to identify the most important sources of total diversity so as to propose conservation measures for microbial communities in the TRF ecosystems of the Nile river watershed of Uganda.
Methods
Site description
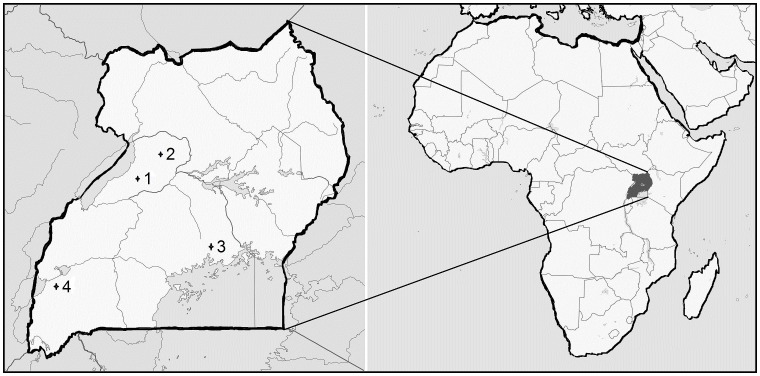
We selected four tropical rainforest (TRF) sites because of their relative size, biodiversity, socio-economic and scientific importance (Fig 1). Mabira forest is located between the highly populated and urbanized Kampala city on the western side; the extensive and mechanized Lugazi sugar and tea plantations on the Eastern; and Lake Victoria on the southern side. Budongo forest is located next to the extensive Kinyala sugar plantations on one side and a densely populated mainly subsistence population scattered around it. Maramagambo and Kaniyo Pabidi are located within Queen Elizabeth and Murchison Falls national parks (NP) respectively. These two NP forests had perhaps the best protection due to presence of Uganda Wildlife Authority (UWA) personnel. However, Maramagambo's location starting on the steep slopes of the rift valley subjected it to frequent storms with strong runoff flow that swept away most of its top soil (Table 1).
Figure 1. Map of Uganda showing the distribution of sampling sites; Budongo forest (1), Kaniyo Pabidi (2), Mabira forest (3), and Maramagambo forest (4).
Table 1. Summary of study site description.
| Forest Site | Location | Size (km2) | Altitude (masl) | Geology | Forest type | Habitat type | Ecosystem description |
| Budongo | 31°N 35° E 1°S 45° N | 793 | 700–1,270 | Weathered pre-cambium with ferrallitic sandy clay loams | Ironwood forest (Cynometra alexandri); Mixed forest (Maesopis), and colonizing forest (Entandrophragma) | Primary forest | Consists of a medium altitude moist semi-deciduous forest with areas of savanna and woodland. Converted areas consisted of deforested agricultural land being cultivated and planted with maize, beans, sweet potatoes and cassava. This land has existed as agricultural land for at least 15years. |
| Mabira | 33° 0.00' E 0° 30.00' N | 300 | 1,070–1,340 | Ferrallitic soils with mainly sandy clay loams | Mixed forest | Secondary forest, heavily influenced by humans | The forest is surrounded by a densely populated area. Converted areas were actively cultivated and used to grow maize, groundnuts, beans, yams, cassava, sweet potatoes and a few scattered plants of coffee and sugarcane. The converted land had existed as agricultural land for at least 10 years. |
| Maramagambo | 00° 33' 00" S and 29° 53' 00" E | 1,978 (QENP) | 910–1390 | Ferrallitic soils with undifferentiated dark horizons | Medium altitude, moist, semi-deciduous forest | Secondary forest influenced by wildlife and humans | Forms part of the Queen Elizabeth NP (QENP) which is 1,978 km2. Converted areas consisted of cultivated and grazed areas with gardens of sweet potatoes, beans, maize, and sorghum with areas commonly grazed by cattle and goats. |
| Kaniyo Pabidi | Lat 1.916667 Long 31.666667 | N/A | 700–1,270 | Freely drained ferruginous tropical soils | Mixed forest | Primary forest | Located north of Budongo forest and part of Murchison Falls N.P. Converted areas consisted of cultivated areas with crops like maize, beans, cassava and sweet potatoes. |
Soil sampling design
We collected 400 core soil samples within 40 plots (1000 m2 each) in four TRF sites (Fig 1). We sampled five plots from each site of the natural TRF and five plots from the converted TRF. We established the plots at least 100 m from the ecosystem edge and 500 m apart and collected 10 evenly placed core subsamples of top soil (0–15 cm) from each plot and homogenized them into one sample per plot. We then derived a 500 g composite sample from the mixture, sieved and packed it for physical and chemical analyses and DNA extraction.
Sample preparation
We sieved 100 g of the soil on-site through a 4 mm mesh, transported it to the laboratory on ice, and stored in a freezer at −40°C prior to nucleic acid extraction and analysis. We kept the rest of the soil for drying and physical and chemical analysis. We performed DNA extractions from 1 g of soil using the Ultra Clean soil DNA kit (Mo Bio Labs, Solana Beach, CA, USA) following the manufacturer's protocol. The purified DNA was detected by agarose gel electrophoresis, and the DNA was amplified by polymerase chain reaction (PCR).
Soil property analyses
We measured the soil pH in 2.5∶1 water to soil suspension using a pH meter (10 g soil+ 25 ml of distilled water, shaken for 30 min and read on a calibrated pH meter). We then used the Walkley and Black method [25] to analyze soil organic carbon (SOC) and the Kjeldahl method [26] to determine soil nitrogen. We measured the soil phosphorus by the Bray and Kurtz no. 1 method [27]. The photoelectric flame photometer was used to determine the soil potassium, sodium and calcium after extraction with neutral ammonium acetate. We used the atomic absorption spectrometer to measure the soil magnesium after extraction with neutral ammonium acetate. The Bouyoucos hydrometer method adopted from Gee and Bauder [28] was used to determine soil texture. The soil copper and iron were then determined using the atomic absorption spectrometer after extraction with EDTA.
PCR amplification and DGGE analysis
Polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) method has been used extensively in microbial ecology and is a robust and cost effective method for exploratory classification of microbial communities [29]. Following soil DNA extraction, we performed a PCR for each DNA extraction to amplify the 16S rRNA genes for bacteria and 18S rRNA genes for fungi using universal primers (Table 2).
Table 2. Sequences of primers used in study.
PCR reactions had a final volume of 25 µl containing a final concentration of 1× TaKaRa ExTaq PCR buffer with MgCl2, 300 pM of primers for bacteria. We then added 200 µM dNTPs, 2.5 U ExTaq DNA polymerase (TaKaRa Bio, Otsu, Japan) and milliQ H2O to complete the volume, BSA was also added for the fungal community analysis. We performed PCR cycles with an initial denaturing temperature of 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 sec, annealing temperature of 50 °C for 30 sec, extension of 72 °C for 1 min; and a final extension of 72 °C for 10 min. We checked the product of the PCR-rounds and quantified by agarose gel-electrophoresis.
We then performed 16S rRNA and 18S rRNA-DGGE analysis using a universal mutation detection system (Dcode Bio-Rad, Richmond, CA, USA) with a 6% and 8% acrylamide gel for bacteria and fungi respectively containing a gradient of 40–60% denaturant (100% denaturant contains 7 mol urea and 40% formamide). We applied 100 ng of PCR samples to the DGGE gel. DGGE was performed in 1 × TAE Buffer (40 mol Tris/acetate, pH 8; 1 mol ethylene diaminetetra acetic acid) at 60 °C and a constant voltage of 60 V for 16 hours. After staining with SYBR Green1, we recorded the DGGE gels as digital images and analyzed the DNA band numbers using image-processing software after subtracting background noise.
Data analysis
We used the Rolling disk method with Quantity One (Bio-Rad laboratories Inc.), which normalizes the band pattern from electrophoresis for identification of each band. We then converted the band patterns into binary data based on the presence or absence of each band for part of our analysis. The DGGE fingerprints were interpreted in terms of band richness (number of predominant DGGE bands/population). The pixel intensity of each band was detected by Quantity One software and is expressed as relative abundance (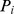 ) [30]. Shannon Index (H′) and Simpson index (D), the most widely used diversity indices were then calculated using the richness and relative abundance data following the equations:
) [30]. Shannon Index (H′) and Simpson index (D), the most widely used diversity indices were then calculated using the richness and relative abundance data following the equations:
| (1) |
| (2) |
Where R, the richness, is number of different bands each data set contains, 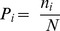 and
and 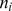 is the abundance of the ith band and N the total abundance of all bands in the sample.
is the abundance of the ith band and N the total abundance of all bands in the sample.
Band-type data of the DGGE fingerprints was then used to derive the alpha diversity (bands per sample and ecosystem type), beta diversity (total bands per site) [31]. Jaccard's similarity indices [32] between converted and natural TRF sites were determined using the equation:
Jaccard's Similarity Index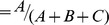
Where,
A = Total number of bands present in both converted (C) and natural (N) ecosystem samples (plots) (also β-diversity)
B = Number of bands present in C but not in N
C = Number of bands present in N but not in C
We determined the influence of site factors as revealed by soil physicochemical properties on the variation of soil microbial communities by applying discriminant analysis using Statistical Package for the Social Sciences (SPSS). This was done to assess the relative importance of each predictor variable (pH, SOC, N, P, K, Na, Ca, Mg, and soil texture). We also used the Mann-Whitney test to examine differences between soil properties in natural and converted ecosystems, and microbial communities in natural and converted ecosystems.
We tested the null hypothesis that diversity is uniform at all spatial scales by additive partitioning of total diversity (γ diversity). To determine contributions of α and β diversity to overall diversity across a range of spatial scales [14], [23], [33], an additive relationship between diversity components (i.e., β = γ - α) was derived (Fig 2.). The scale at which diversity is maximized was therefore identified [23], [34], to facilitate planning processes and management strategies to conserve natural levels of diversity accordingly [35]–[38].
Figure 2. Illustration of hierarchical spatial scales in our additive partitioning model.
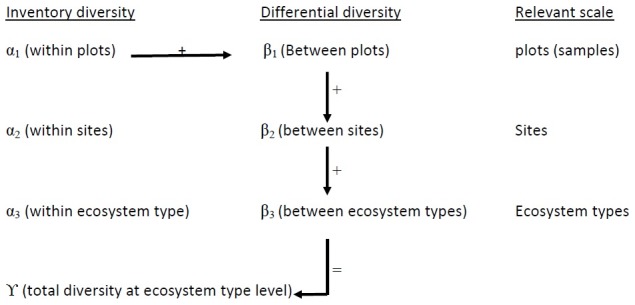
The α scale is the within-level, and β scale, the between-level components. Because a diversity at a given scale is the sum of the α and β diversity at the next lower scale, the total diversity (γ) can be described by the following formula: α1+β1+β2+β3 [14], [22].
We used PARTITION 3.0 software [39] to calculate average diversity at each scale and diversity was measured as band richness. Individual-based randomization procedure in the software was used to test whether the observed partitions of diversity within the ecosystem could have been obtained by a random allocation of lower-level samples nested among higher-level samples [34]. Null values of βi obtained from 1,000 randomizations were used to obtain a p-value for the observed βi at each hierarchical scale. Deviations of the observed diversity from the null expectation indicated a nonrandom spatial distribution of fungi or bacteria at a given scale.
Results
Soil property variations
Soil pH comparisons using a Mann-Whitney U test of significance, between five plots of natural and five plots of converted TRF ecosystems in each of the four forest sites, found significantly higher (less acidic) pH in three of the four sites at Budongo (p = 0.0107), Kaniyo Pabidi (p = 0.0112), and Mabira (p = 0.0269); and non-significant difference at Maramagambo (p = 0.1706). Percentage soil organic carbon (SOC) was significantly higher in natural than converted ecosystems in all four sites i.e. Budongo (p = 0.0119), Kaniyo Pabidi (p = 0.0212), Mabira (p = 0.0122) and Maramagambo (p = 0.0119) with combined %SOC in natural sites more than double of that in converted sites; whereas %soil nitrogen was only significantly higher in natural forests at Budongo (p = 0.0112) and Kaniyo Pabidi (p = 0.0119), and non-significant at Mabira (p = 0.6015) and Maramagambo (p = 0.0947) (Table 3).
Table 3. Mean (Standard deviation) for diversity indices of Bacterial (B) and Fungal (F) communities and soil properties in natural (N) and converted (C) ecosystems.
| Budongo | Kaniyo Pabidi | Mabira | Maramagambo | |||||
| Natural | Converted | Natural | Converted | Natural | Converted | Natural | Converted | |
| Shannon (B) | 2.49(0.68) | 2.73(0.10) | 3.25(0.21) | 3.12(0.25) | 3.06(0.18) | 3.19(0.18) | 3.04(0.16) | 2.93(0.07) |
| Simpson (B) | 0.89(0.10) | 0.93(0.01) | 0.95(0.13) | 0.95(0.02) | 0.95(0.01) | 0.95(0.01) | 0.95(0.01) | 0.94(0.01) |
| Shannon (F) | 2.44 (0.30) | 2.49 (0.17) | 2.83(0.19) | 2.77(0.20) | 2.57(0.62) | 2.64(0.30) | 2.10(0.50) | 1.93(0.67) |
| Simpson (F) | 0.90(0.29) | 0.90(0.03) | 0.93(0.01) | 0.93(0.02) | 0.90(0.06) | 0.92(0.03) | 0.84(0.08) | 0.82(0.12) |
| pH | 5.88(0.11)* | 5.08(0.20)* | 6.24(0.23)* | 5.38(0.27)* | 6.46(0.54)* | 6.18(0.42)* | 6.18(0.45) | 5.80(0.24) |
| OC (%) | 6.14(0.67)* | 1.59(0.17)* | 5.69(1.20)* | 3.65(0.54)* | 5.87(2.03)* | 3.53(1.19)* | 9.08(0.79)* | 3.98(1.09)* |
| N (%) | 0.43(0.05)* | 0.11(0.01)* | 0.43(0.08)* | 0.25(0.03)* | 0.21(0.10) | 0.22(0.06) | 0.28(0.04) | 0.19(0.08) |
| Ca (Cmoles/kg) | 8.00(2.56)* | 4.00(0.58)* | 13.54(3.36)* | 6.74(0.98)* | 8.14(1.19)* | 6.20(0.76)* | 12.12(2.13)* | 7.34(1.16)* |
*significant differences (p<0.05) between natural and converted ecosystems.
Ecosystem and site comparisons of microbial community diversity
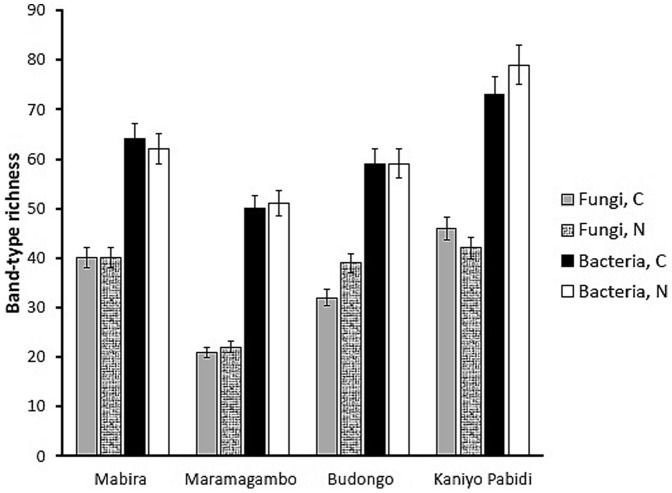
Bacterial (B) communities were significantly richer (p = 0.0304; Mann-Whitney U) in detectable bands than fungal (F) communities in both converted (C) and natural (N) ecosystems (converted: medians; F = 36, B = 61.5; natural: medians; F = 39.5, B = 60.5). While total band richness (B+F) did not differ between natural and converted forests we observed greater fungal richness in natural than in converted forests (medians: C = 36, N = 39.5; test stat = 18.5) and more bacterial bands in converted than in natural ecosystems (medians: C = 61.5, N = 60.5; test stat = 18.5). Kaniyo Pabidi was the most diverse site overall with the highest number of bacterial and fungal bands, while Maramagambo had the least band richness (Fig 3).
Figure 3. Band richness for fungal and bacterial communities in converted and natural ecosystems.
All richness values are total bands present in five samples of each ecosystem treatment (error bars are 5% confidence interval).
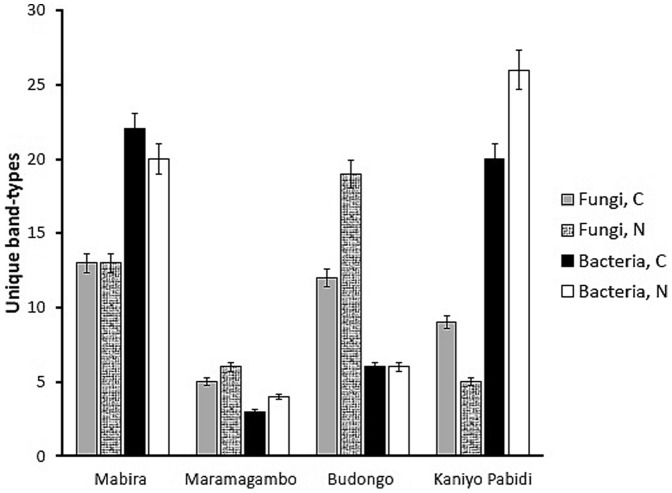
Natural sites harbored more bands unique to one site than converted sites for bacteria at Kaniyo Pabidi and Maramagambo and for fungi at Maramagambo and Budongo. Mabira and Kaniyo Pabidi had higher numbers of unique bacterial bands than at Maramagambo and Budongo. There were also more unique fungal bands at Mabira and Budongo than at Maramagambo and Kaniyo Pabidi (Fig 4).
Figure 4. Bacterial and fungal bands unique to converted (C) and natural (N) ecosystems at each site (error bars are 5% confidence interval).
We also found that Mabira and Maramagambo had the lowest bacterial Jaccard's community similarity indices [32] between natural and converted ecosystems, whereas Budongo and Mabira had the lowest fungal community similarity between natural and converted ecosystems (Table 4). Dissimilarity between natural and converted ecosystems was nonetheless non-significant in all sites for both fungal and bacterial communities. Also, there was generally greater dissimilarity between sites of fungal communities than in bacterial communities suggesting a higher susceptibility to habitat change among fungi than bacteria (Table 4).
Table 4. Jaccard's similarity indices between bacterial and fungal communities in natural (N) and converted (C) sites of Mabira (Mb), Maramagambo (Mg), Budongo (Bd) and Kaniyo Pabidi (Kp).
| F | U | N | G | I | |||||||
| Mb | Mg | Bd | Kp | Mb | Mg | Bd | Kp | ||||
| N | N | N | N | C | C | C | C | F | |||
| B | Mb | N | 1 | 0.713 | 0.769 | 0.732 | 0.671 | 0.685 | 0.696 | 0.768 | |
| A | Mg | N | 0.841 | 1 | 0.733 | 0.666 | 0.666 | 0.711 | 0.646 | 0.666 | U |
| C | Bd | N | 0.803 | 0.840 | 1 | 0.804 | 0.808 | 0.693 | 0.622 | 0.827 | |
| T | Kp | N | 0.731 | 0.765 | 0.698 | 1 | 0.774 | 0.670 | 0.794 | 0.785 | N |
| E | Mb | C | 0.667 | 0.788 | 0.739 | 0.741 | 1 | 0.663 | 0.710 | 0.761 | |
| R | Mg | C | 0.866 | 0.885 | 0.846 | 0.755 | 0.809 | 1 | 0.684 | 0.698 | G |
| I | Bd | C | 0.825 | 0.857 | 0.844 | 0.696 | 0.731 | 0.856 | 1 | 0.786 | |
| A | Kp | C | 0.805 | 0.869 | 0.716 | 0.683 | 0.759 | 0.776 | 0.733 | 1 | I |
| B | A | C | T | E | R | I | A |
Ecosystem classification and importance of predictor variables
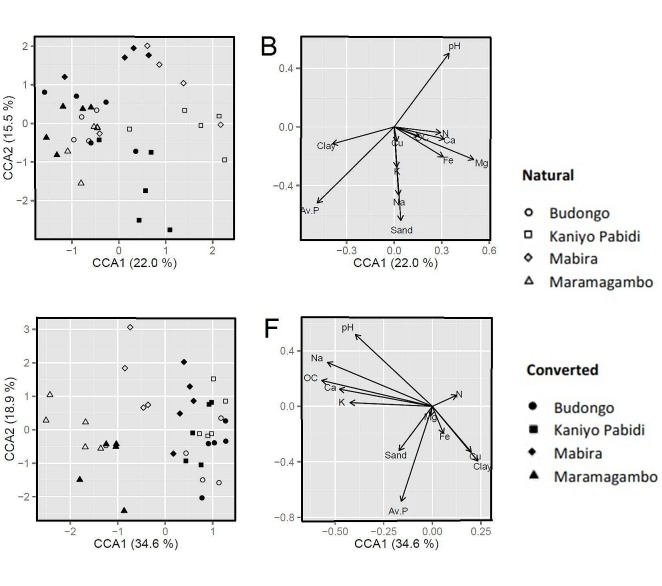
The CCA showed that, despite the relatively small amount of difference between sites, soil pH, average phosphorus, and texture (%sand) had strong influence on bacterial diversity in the TRF ecosystem (Fig 5); whereas organic carbon, sodium, pH and average phosphorus were strongly associated with fungal community variation in both natural and converted TRF ecosystems (Fig 5). The CCA also showed that bacterial communities in both Kaniyo Pabidi and Mabira were unique to bacterial communities in the other sites and there was high contrast between bacterial communities of converted and natural ecosystems at Kaniyo Pabidi. Fungal communities at Maramagambo and Mabira were also unique to those in other sites and there was high contrast between fungal communities at Mabira's natural and converted ecosystems. Furthermore, the CCA showed that fungal communities responded more to soil pH levels than bacterial communities (Fig 5), with site-specific patterns showing that bacteria and fungi were grouping according to sites.
Figure 5. CCA for bacterial (B) and fungal (F) relationships using relative abundance of bands and soil physicochemical properties in natural and converted ecosystems.
The symbols (left graphs) represent the similarity between each sample (plot) as defined by their diversity, and the vectors (right graphs) represent the structural matrix for soil properties and their influence on relative abundance of each band. The length of the vectors represents the relative strength of influence of the particular aspect of soil physicochemical property.
A discriminant analysis to predict whether bacterial or fungal communities were from natural or converted ecosystems found that only OC, Ca, N, and pH for bacterial communities; and OC, N, Ca, and pH for fungal communities (all ranked from most important to least important) were found to be significant predictors of soil physicochemical properties. All other variables were poor predictors in this context (Table 5).
Table 5. Structure matrix rank showing absolute size of correlation between discriminant analysis function from most important to least important predictor variable of site factors (soil physiochemical properties) and their influence on the variation of soil microbial communities.
| Bacteria | Fungi | ||
| Predictor Variables | Function 1 | Predictor variables | Function 1 |
| OC | 0.625* | OC | 0.625* |
| Ca | 0.471* | N | 0.493* |
| N | 0.421* | Ca | 0.473* |
| pH | 0.355* | pH | 0.340* |
| Mg | 0.298 | Mg | 0.281 |
| Cu | 0.197 | Cu | 0.221 |
| Na | 0.181 | Na | 0.183 |
| K | 0.169 | K | 0.160 |
| Av.P | −0.077 | Sand | 0.064 |
| Sand | 0.070 | Av.P | −0.060 |
| Simpson | −0.065 | Fe | 0.024 |
| Fe | 0.029 | Shannon | 0.012 |
| Shannon | −0.023 | Simpson | 0.011 |
(* = important predictor variable, with 0.30 used as the threshold).
Hierarchical scaling
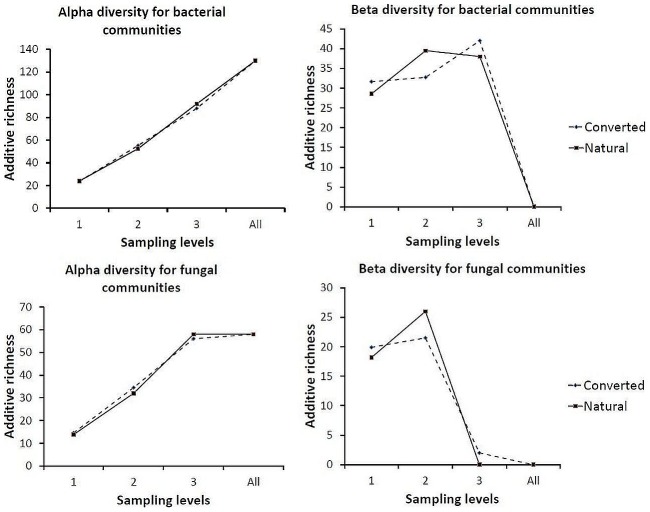
We found 58 and 56 fungal bands in natural and converted forests respectively, from 17 plots of natural ecosystems and 20 plots of converted forests. There were also 92 and 88 bacterial bands in natural and converted ecosystems respectively found in 20 plots of converted ecosystems and 17 plots of natural ecosystem. All these were within four sites. β-diversity varied more than α-diversity between natural and converted ecosystems for both bacteria and fungi. We found higher bacterial and fungal β-diversity in converted ecosystems than in natural ecosystems at lower hierarchical scales (β1); higher β-diversity in natural than converted at between-site scale (β2), and higher β-diversity in converted than in natural ecosystems at the between-ecosystem type scale (β3) (Fig 6).
Figure 6. Additive partitioning of bacterial and fungal diversity (expressed as additive richness) across alpha and beta hierarchical spatial scales at three sampling levels (plot, site and ecosystem type) in natural and converted TRF ecosystems.
We also found substantial contribution of observed β-diversity (β1, β2, and β3) to total band richness (γ-diversity), while α-diversity of both bacteria and fungi in converted and natural ecosystems were similar. Spatial partitioning of total diversity also consistently showed that the beta components (β1 and β2) were always greater than expected by chance, whereas the alpha component (α1) was always lower than expected. For both fungal and bacterial communities in natural and converted ecosystems, observed within plot diversity were substantially less than values expected from individual-based randomizations (Fig 7).
Figure 7. The additive partitioning of total bacterial and fungal community, γ -diversity into α and β components at three nested spatial scales, with each component expressing their relative contributions to γ –diversity; where γ –diversity is equal to α1+β1+β2+β3.
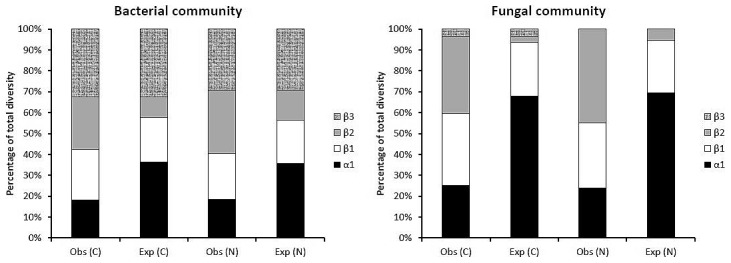
The observed (obs) partitions are compared with the expected (exp) values, as predicted by the null model based on 1000 iterations using individual-based randomization.
Discussion
Soil property variations and site differences
Studies in both tropical and temperate zones show that soils in converted or cropped areas normally have reduced soil aggregation, structural stability and organic matter, and an increase in bulk density when compared to forests [40], [41]. Habitat conversion may also alter soil properties such as nutrient levels, and abiotic conditions and may affect associations between organisms. In our study there are some local details that may influence our results.
Both Maramagambo and Kaniyo Pabidi are located within Queen Elizabeth NP and Murchison Falls NP respectively and are protected by Uganda Wildlife Authority (UWA) personnel. They are well protected and there is little evidence of recent encroachment. There is significant wildlife populations including elephants, buffaloes, zebras and the areas are frequented by tourists. Protection by UWA and presence of dangerous animals (such as buffalos and lions) reduce damaging human activity at Kaniyo Pabidi and Maramagambo which should enhance the difference between natural and converted ecosystems. Maramagambo's location, in contrast, means the forest is subjected to substantial natural disturbance from frequent storms and strong erosive runoff even within the natural forest, whereas tourist activity at Kaniyo Pabidi seemed to have little impact on soil properties. Converted areas at Kaniyo Pabidi were also sparsely populated with limited human impacts on the environment. Its sites were old and might have been cultivated for at least 20years.
In our study, Budongo is located next to a high, mainly subsistence population and resultant population activity. But even though encroachment, illegal hunting and logging in natural habitats in Budongo are not uncommon, there seems to be minimal impact of conversion on soil properties in our sample locations; whereas proximity of Mabira's natural forest to densely populated urban areas exposes it to increased human activities, likely reducing its difference with converted sites.
Microbial community variations
Soil properties determine many aspects of soil microbial community structure [42]–[44]. Carbon availability [45]–[47], nitrogen availability [45], [48], [49] and soil pH [44], [50], [51] can all influence microbial community composition and diversity. In addition, correlation studies have shown that plant species [52]–[54] and soil type [45], [54], [55] are associated with variation in microbial communities. It has also been shown that land use indirectly affects bacterial community structure by modification of soil properties [56] but similarity between converted and natural ecosystem bacterial communities may also suggest a high number of generalists.
Nacke et al., [56] found that bacterial community composition in forests and grassland was largely determined by tree species and soil pH. Jesus et al. [6] also showed that bacterial community structure is influenced by changes linked to soil acidity and nutrient concentration. Other studies also suggest that soil pH is a major factor influencing microbial community composition [50], [57]–[59]. This influence of soil pH has been recognized at different taxonomic levels [45], [60] with most microorganisms thriving within a limited pH range. This is because acids can denature proteins and large pH changes may inhibit growth in microorganisms. Fierer and Jackson, 2006 [44] found, in contrast, that net carbon mineralization rate (an index of C availability) was the best predictor of phylum-level abundances of dominant bacterial groups, and Bisset et al., [42] found that soil microbial communities were consistent with disturbance gradients within different agricultural treatments and relatively undisturbed non-agricultural sites.
Because of widespread forest conversion in Uganda as a result of increasing population pressure, estimated at between 1.1% and 3.15% per year [61]; natural ecosystems and their inhabitant biodiversity are at risk [62]. Loss of diversity increases the likelihood of losing important functional roles and associated ecosystem processes. At landscape scale, spatial and temporal variations of microbial communities in forest soils are influenced by numerous biotic and abiotic factors. These factors may include climate, soil types, and vegetation associations [50], [63], [64]. Owing to this study design, many of these factors were assumed to be similar between natural and converted ecosystems. For instance, the proximity of natural and converted ecosystem sites meant that climate and geology were, we assume, similar in the two treatments. Even though there could still be a number of underlying causes of community differences, two likely influences were assumed to be soil properties [45], [54], [55] and vegetation types [52]–[54]. Clearly both of these sets of factors change when forest is converted for agriculture or range lands.
Despite substantial reductions in SOC, N, Ca and pH in converted sites in this study, differences in microbial communities were small meaning that converted sites still had sufficient SOC, N and Ca to sustain the same microbial populations. The close proximity of the matched pairs could also lead to a source-sink relationship between the natural and converted forests, with the presence of unique taxonomic groups a likely indication of habitat preference (endemism) for some taxonomic groups. It may also be an indicator of relative habitat dissimilarity. The high numbers of unique bacterial bands at Mabira and Kaniyo Pabidi and unique fungal bands at Mabira and Budongo (Fig 4) thus suggests that ecosystem alteration at these sites was sufficient to force a different regime of processes and structures enabling a new set of taxonomic groups to predominate. Mabira had high numbers of both unique bacterial and unique fungal bands that can be attributed to the extent of disturbance at its sites (Mabira is the only peri-urban tropical rainforest site among the four selected sites).
The low numbers of unique bacterial and fungal bands at Maramagambo can be attributed to the high erosion at natural sites that reduced the contrast between the natural and converted sites. For the other sites, bacteria and fungi had different responses to ecosystem alterations. This could indicate separate influences on microbial distribution that exist when alteration is moderate. Similarity indices suggested that bacterial and fungal communities were determined by separate forces leading to distinct responses across the study locations.
Hierarchical scaling
Many studies have shown that specialist species are more negatively affected by current global changes than generalists [7], [65]. The process of biotic homogenization can involve the replacement of native biota with non-natives or the introduction of generalist species [66]. In this study, the net decrease in β-diversity from natural to converted TRF ecosystem at the between-site scale (β2) for both fungi and bacteria was an indication of biotic homogenization [18], [66]. This can result from ecosystem alterations which can in-turn alter ecosystem function and reduce ecosystem resilience to disturbance [65], [67].
We also showed that the β components of diversity (β1and β2; the average diversity between the plots and sites, respectively) were consistently higher than those expected by chance, whereas the local α1 diversity component (α1, the average diversity within the plots) was consistently lower than that expected (Fig 7). Such scale-dependent deviations of the observed diversity from the expected can be generally explained by aggregation at a relatively small “local” scale and, spatial differentiation of diversity at a larger “landscape” scale [33], [34], [68], [69].
Relatively lower diversity within converted ecosystems suggests that conversion of natural TRF ecosystems results in reduced diversity for both bacteria and fungi. This is compatible with recent studies that show that conversion of TRF ecosystems threatens microbial diversity [7] and because microorganisms, like all other organisms, have habitat preferences and may be affected by land-use changes [6], [64]. While we cannot be certain that such decline in diversity has led to a decline in any particular ecosystem functions or services, this is a possibility that deserves further evaluation, and we speculate that such loss of diversity will at the very least cause a reduction in functional redundancy and associated resilience.
Higher β-diversity of both bacterial and fungal communities at the between-plot scale (β1) in converted ecosystems indicates differentiation (reduced community similarity) in converted ecosystems at this hierarchical scale. Considering the multiple land-uses and cropping systems of converted areas, this was expected. There was also substantial contribution of β-diversity to total diversity (γ). This suggests the importance of nonrandom ecological processes at the between-plot and between-site scale in determining total richness and community composition [14], [34]. Differences between the observed and expected diversity components could be due to ecological processes that lead to a non-random dispersion of individuals. These processes could include intra-specific aggregation, habitat selection, and limited dispersal capacity [33].
Conclusion
There is international concern about the threat to natural habitats in the Nile river watershed and the consequential loss of important biodiversity. Whereas aspects of microbial biogeography and influence of forest conversion in Uganda's Nile river watershed is largely unknown, this study offers an important first glimpse into indicators of spatial diversity patterns of soil fungal and bacterial communities in the Uganda's Nile river watershed. Our observations of reduced soil microbial diversity, both bacterial and fungal, in converted ecosystems though unsurprising in itself causes us some concern and would justify further work to determine the significance of the diversity lost and the wider implications.
By focusing on diversity patterns across multiple hierarchical spatial scales, we were able to identify the scale at which regional microbial diversity is maximized. We showed that there was substantial contribution of β-diversity to total ecosystem diversity (γ) which includes taxa at the between-plot, site and ecosystem scales and unique taxa, highlighting the necessity to conserve marginal habitats and ecotones. Soil microbial communities in Uganda's Nile river watershed exhibit considerable resilience to forest conversion even though SOC, N, Ca and pH were all significantly altered. This result is surprising given that these physical and chemical properties typically strongly influence microbial diversity. Additionally, the variation among sites was quite large, indicating that soil communities in this region vary considerably on a regional spatial scale. Our results do not explain this variation. Most studies suggest that biogeographic barriers play little role in the geographic structure of soil communities. Rather than a consistent general pattern of microbial community change following forest conversion we find that responses are largely site-specific and widely variable.
Acknowledgments
We confirm that our field work did not involve any endangered or protected species and that we were granted access to protected areas by Uganda Wildlife Authority (UWA) and National Forestry Authority (NFA). Our sample site coordinates included: (1) Mabira; Lat, N00°24.495'; Long, E033°02.464' (Required permission obtained from NFA); (2) Maramagambo; Lat, S00°04.421'; Long, E030°02.305' (Required permission obtained from UWA); (3) Budongo; Lat, N01°41.852'; Long, E031°29.363' (Required permission obtained from UWA/NFA); (4) Kaniyo Pabidi; Lat, N01°55.128'; Long, E031°43.116' (Required permission obtained from UWA).
We would like to thank the Institute of Tropical Forest Conservation (ITFC) of Mbarara University of Science and Technology for supporting and co-advising this research. We also thank the Genetics Lab at Faculty of Science and the soil department lab at Faculty of Agriculture both at Makerere University, Kampala, for their lab facilitation during extractions and analyses. And finally we acknowledge the ideas and insights of Masatoshi Katabuchi, Dossa Gbadamassi, and our field and lab technicians and assistants Solomon Echel, Francis Alele Ozirit and Boniface Balikuddembe.
Supporting Information
Excel spreadsheet from Quantity One analysis of PCR-DGGE profiles of fungal and bacterial communities in natural and converted TRF ecosystems.
(XLSX)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information Files.
Funding Statement
This work was funded by Xishuangbanna Tropical Botanical Garden (XTBG) and the Chinese Academy of Sciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Turner IM (1996) Species loss in fragments of tropical rain forest: a review of the evidence. J Appl Ecol 33: 200–209. [Google Scholar]
- 2. Joseph Wright S, Muller-Landau HC (2006) The Future of Tropical Forest Species. Biotropica 38: 287–301. [Google Scholar]
- 3.United Nations (2013) World Population Prospects: The 2012 Revision. Highlights and Advance Tables. New York.
- 4.United Nations D of E and SAPD (2007) World Population Prospects: The 2006 Revision.
- 5. Kayanja FIB, Byarugaba D (2001) Disappearing forests of Uganda: The way forward. Curr Sci 81: 936–947. [Google Scholar]
- 6. Jesus da CE, Marsh TL, Tiedje, M J, de S Moreira FM (2009) Changes in land use alter the structure of bacterial communities in Western Amazon soils. ISME J 3: 1004–1011 Available: http://www.ncbi.nlm.nih.gov/pubmed/19440233 Accessed 15 November 2012. [DOI] [PubMed] [Google Scholar]
- 7.Rodrigues JLM, Pellizari VH, Mueller R, Baek K, Jesus EDC, et al. (2012) Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proc Natl Acad Sci U S A. Available: http://www.ncbi.nlm.nih.gov/pubmed/23271810. Accessed 29 December 2012. [DOI] [PMC free article] [PubMed]
- 8. Cardinale BJ, Duffy JE, Gonzalez A, Hooper DU, Perrings C, et al. (2012) Biodiversity loss and its impact on humanity. Nature 486: 59–67 Available: http://www.ncbi.nlm.nih.gov/pubmed/22678280 Accessed 25 May 2014. [DOI] [PubMed] [Google Scholar]
- 9. Venail PA, Vives MJ (2013) Positive effects of bacterial diversity on ecosystem functioning driven by complementarity effects in a bioremediation context. PLoS One 8: e72561 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3762786&tool=pmcentrez&rendertype=abstract Accessed 24 June 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Torsvik V, Øvreås L (2002) Microbial diversity and function in soil: from genes to ecosystems. Curr Opin Microbiol 5: 240–245 Available: http://www.ncbi.nlm.nih.gov/pubmed/12057676. [DOI] [PubMed] [Google Scholar]
- 11.Nile Basin Initiative (2012) State of the River Nile Basin. Entebbe (Uganda): Nile Basin Initiative Secretariat.
- 12. Olson DM, Dinerstein E, Powell GVN, Wikramanayake ED (2002) Conservation Biology for the Biodiversity Crisis. Conserv Biol 16: 1–3. [DOI] [PubMed] [Google Scholar]
- 13. Ricklefs RE (2004) A comprehensive framework for global patterns in biodiversity. Ecol Lett 7: 1–15 Available: http://doi.wiley.com/10.1046/j.1461-0248.2003.00554.x Accessed 21 January 2014. [Google Scholar]
- 14. Gering JC, Crist TO, Veech J a (2003) Additive Partitioning of Species Diversity across Multiple Spatial Scales: Implications for Regional Conservation of Biodiversity. Conserv Biol 17: 488–499 Available: http://doi.wiley.com/10.1046/j.1523-1739.2003.01465.x. [Google Scholar]
- 15. Whittaker RJ, Willis KJ, Field R (2001) Scale and species richness: towards a general, hierarchical theory of species diversity. J Biogeogr 28: 453–470 Available: http://doi.wiley.com/10.1046/j.1365-2699.2001.00563.x. [Google Scholar]
- 16.Demeny P (2003) Population Policy: A Concise Summary. Internatio. Demeny P, McNicoll G, editors New York: MacMillan Reference. doi:173.
- 17. Olden JD (2006) Biotic homogenization: a new research agenda for conservation biogeography. J Biogeogr 33: 2027–2039 Available: http://doi.wiley.com/10.1111/j.1365-2699.2006.01572.x Accessed 5 May 2014. [Google Scholar]
- 18. Olden JD, Poff NL (2004) Ecological processes driving biotic homogenization: testing a mechanistic model using fish faunas. Ecology 85: 1867–1875. [Google Scholar]
- 19. Olden JD, Poff NL (2003) Toward a mechanistic understanding and prediction of biotic homogenization. Am Nat 162: 442–460 Available: http://www.ncbi.nlm.nih.gov/pubmed/14582007. [DOI] [PubMed] [Google Scholar]
- 20. Folke C, Carpenter S, Walker B, Scheffer M, Elmqvist T, et al. (2004) Regime Shifts, Resilience, and Biodiversity in Ecosystem Management. Annu Rev Ecol Evol Syst 35: 557–581 Available: http://www.annualreviews.org/doi/abs/10.1146/annurev.ecolsys.35.021103.105711 Accessed 26 October 2012. [Google Scholar]
- 21. Ter Braak CJ F (1986) Canonical Correspondence Analysis: A New Eigenvector Technique for Multivariate Direct Gradient Analysis. Ecol Soc Am 67: 1167–1179. [Google Scholar]
- 22.Legendre P, Legendre L (1998) Numerical Ecology. 2nd ed. Amsterdam: Elsevier.
- 23. Lande R (1996) Statistics and partitioning of species diversity, and similarity among multiple communties. Oikos, Nord Soc 76: 5–13. [Google Scholar]
- 24. Godfray HCJ, Lawton JH (2001) Scale and species numbers. Trends Ecol Evol 16: 400–404 Available: http://www.ncbi.nlm.nih.gov/pubmed/11403873. [DOI] [PubMed] [Google Scholar]
- 25.Walkley A., Black I (1934) An examination of the Degtjareff method for determining organic carbon in soils: effect of variations in digestion conditions and of inorganic soil constituents. Soil Sci: 251–263.
- 26.Kjeldahl J (1883) A new method for the determination of nitrogen in organic matter. Zeitschreft fur Anal Chemie 22.
- 27. Bray RH, Kurtz LT (1945) Determination of total, organic, and available forms of phosphorus in soils. Soil Sci 59: 39–45. [Google Scholar]
- 28.Gee G., Bauder J. (1986) Particle-size analysis. p. 383–411. In A Klute (ed.) Methods of Soil Analysis, Part 1. Physical and Mineralogical Methods. Agronomy Monograph No. 9 (2ed). Am Soc Agron Sci Soc Am Madison, WI.
- 29. Cleary DFR, Smalla K, Mendonça-Hagler LCS, Gomes NCM (2012) Assessment of variation in bacterial composition among microhabitats in a mangrove environment using DGGE fingerprints and barcoded pyrosequencing. PLoS One 7: e29380 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3256149&tool=pmcentrez&rendertype=abstract Accessed 27 October 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reche I, Pulido-Villena E, Morales-Baquero R, Casamayor EO (2005) Does ecosystem size determine aquatic bacterial richness? Ecology 86: 1715–1722 Available: http://www.ncbi.nlm.nih.gov/pubmed/17489473. [Google Scholar]
- 31. Whittaker RH (1972) Evolution and Measurement of Species Diversity. Int Assoc Plant Taxon (IAPT) 21: 213–251. [Google Scholar]
- 32. Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat 44: 223–270. [Google Scholar]
- 33. Veech JA, Summerville KS, Crist TO, Gering JC (2002) The additive partitioning of species diversity: recent revival of an old idea. Nord Soc Oikos 99: 3–9. [Google Scholar]
- 34. Crist TO, Veech J a, Gering JC, Summerville KS (2003) Partitioning species diversity across landscapes and regions: a hierarchical analysis of alpha, beta, and gamma diversity. Am Nat 162: 734–743 Available: http://www.ncbi.nlm.nih.gov/pubmed/14737711. [DOI] [PubMed] [Google Scholar]
- 35. Chandy S, Gibson DJ, Robertson P a (2006) Additive partitioning of diversity across hierarchical spatial scales in a forested landscape. J Appl Ecol 43: 792–801 Available: http://doi.wiley.com/10.1111/j.1365-2664.2006.01178.x Accessed 22 January 2014. [Google Scholar]
- 36. Ribeiro DB, Prado PI, Brown Jr KS, Freitas AVL (2008) Additive partitioning of butterfly diversity in a fragmented landscape: importance of scale and implications for conservation. Divers Distrib 14: 961–968 Available: http://doi.wiley.com/10.1111/j.1472-4642.2008.00505.x Accessed 3 February 2014. [Google Scholar]
- 37. Wu F, Yang XJ, Yang JX (2010) Additive diversity partitioning as a guide to regional montane reserve design in Asia: an example from Yunnan Province, China. Divers Distrib 16: 1022–1033 Available: http://doi.wiley.com/10.1111/j.1472-4642.2010.00710.x Accessed 6 February 2014. [Google Scholar]
- 38.Sasaki T, Katabuchi M, Kamiyama C, Shimazaki M, Nakashizuka T, et al. (2012) Diversity partitioning of moorland plant communities across hierarchical spatial scales. Biodivers Conserv. doi:10.1007/s10531-012-0265-7.
- 39.Veech JA, Crist TO (2009) Partition: software for hierarchical partitioning of species diversity, version 3.0. Available: http://www.users.muohio.edu/cristto/partition.htm.
- 40. Monkiedje A, Spiteller M, Fotio D, Sukul P (2006) The effect of land use on soil health indicators in peri-urban agriculture in the humid forest zone of southern cameroon. J Environ Qual 35: 2402–2409 Available: http://www.ncbi.nlm.nih.gov/pubmed/17071911 Accessed 9 November 2012. [DOI] [PubMed] [Google Scholar]
- 41. Neris J, Jiménez C, Fuentes J, Morillas G, Tejedor M (2012) Vegetation and land-use effects on soil properties and water infiltration of Andisols in Tenerife (Canary Islands, Spain). Catena 98: 55–62 Available: http://linkinghub.elsevier.com/retrieve/pii/S0341816212001270 Accessed 28 December 2012. [Google Scholar]
- 42. Bissett A, Richardson AE, Baker G, Thrall PH (2011) Long-term land use effects on soil microbial community structure and function. Appl Soil Ecol 51: 66–78 Available: http://linkinghub.elsevier.com/retrieve/pii/S0929139311001922 Accessed 9 November 2012. [Google Scholar]
- 43. Garbisu C, Alkorta I, Epelde L (2011) Assessment of soil quality using microbial properties and attributes of ecological relevance. Appl Soil Ecol 49: 1–4 Available: http://linkinghub.elsevier.com/retrieve/pii/S0929139311001089 Accessed 9 November 2012. [Google Scholar]
- 44. Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A 103: 626–631 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1334650&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shange RS, Ankumah RO, Ibekwe AM, Zabawa R, Dowd SE (2012) Distinct soil bacterial communities revealed under a diversely managed agroecosystem. PLoS One 7: e40338 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3402512&tool=pmcentrez&rendertype=abstract Accessed 1 November 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang Y, Boyd E, Crane S, Lu-Irving P, Krabbenhoft D, et al. (2011) Environmental conditions constrain the distribution and diversity of archaeal merA in Yellowstone National Park, Wyoming, U.S.A. Microb Ecol 62: 739–752 Available: http://www.ncbi.nlm.nih.gov/pubmed/21713435 Accessed 9 November 2012. [DOI] [PubMed] [Google Scholar]
- 47. Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88: 1354–1364 Available: http://www.ncbi.nlm.nih.gov/pubmed/17601128. [DOI] [PubMed] [Google Scholar]
- 48. Saiya-Cork K, Sinsabaugh R, Zak D (2002) The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol Biochem 34: 1309–1315 Available: http://linkinghub.elsevier.com/retrieve/pii/S0038071702000743. [Google Scholar]
- 49. Steenwerth K, Jackson L, Calderon F, Scow K, Rolston D (2005) Response of microbial community composition and activity in agricultural and grassland soils after a simulated rainfall. Soil Biol Biochem 37: 2249–2262 Available: http://linkinghub.elsevier.com/retrieve/pii/S0038071705001549 Accessed 9 November 2012. [Google Scholar]
- 50. De Vries FT, Manning P, Tallowin JRB, Mortimer SR, Pilgrim ES, et al. (2012) Abiotic drivers and plant traits explain landscape-scale patterns in soil microbial communities. Ecol Lett 15: 1230–1239 Available: http://www.ncbi.nlm.nih.gov/pubmed/22882451 Accessed 2 November 2012. [DOI] [PubMed] [Google Scholar]
- 51. Hartman WH, Richardson CJ, Vilgalys R, Bruland GL (2008) Environmental and anthropogenic controls over bacterial communities in wetland soils. Proc Natl Acad Sci U S A 105: 17842–17847 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2584698&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cadotte MW, Cardinale BJ, Oakley TH (2008) Evolutionary history and the effect of biodiversity on plant productivity. Proc Natl Acad Sci U S A 105: 17012–17017 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2579369&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Garbeva P, Veen JA Van, Elsas JD Van (2004) Microbial Diversity in Soil: Selection of Microbial Populations by Plant and Soil Type and Implications for Disease Suppressiveness. Annu Rev Phytopathol 42: 243–270 doi:10.1146/annurev.phyto.42.012604.135455 [DOI] [PubMed] [Google Scholar]
- 54.Schulz S, Giebler J, Chatzinotas A, Wick LY, Fetzer I, et al. (2012) Plant litter and soil type drive abundance, activity and community structure of alkB harbouring microbes in different soil compartments. ISME J doi:10.103: 1763–1774. [DOI] [PMC free article] [PubMed]
- 55. Kuramae EE, Yergeau E, Wong LC, Pijl AS, van Veen J a, et al. (2012) Soil characteristics more strongly influence soil bacterial communities than land-use type. FEMS Microbiol Ecol 79: 12–24 Available: http://www.ncbi.nlm.nih.gov/pubmed/22066695 Accessed 7 November 2012. [DOI] [PubMed] [Google Scholar]
- 56. Nacke H, Thürmer A, Wollherr A, Will C, Hodac L, et al. (2011) Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS One 6: e17000 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3040199&tool=pmcentrez&rendertype=abstract Accessed 5 November 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Waldrop MP, Balser TC, Firestone MK (2001) Linking microbial community composition to function in a tropical soil. Soil Biol Biochem 32: 1837–1846. [Google Scholar]
- 58. Dinsdale E a, Edwards R a, Hall D, Angly F, Breitbart M, et al. (2008) Functional metagenomic profiling of nine biomes. Nature 452: 629–632 Available: http://www.ncbi.nlm.nih.gov/pubmed/18337718 Accessed 26 October 2012. [DOI] [PubMed] [Google Scholar]
- 59. Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75: 5111–5120 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2725504&tool=pmcentrez&rendertype=abstract Accessed 19 March 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Russo SE, Legge R, Weber K a, Brodie EL, Goldfarb KC, et al. (2012) Bacterial community structure of contrasting soils underlying Bornean rain forests: Inferences from microarray and next-generation sequencing methods. Soil Biol Biochem 55: 48–59 Available: http://linkinghub.elsevier.com/retrieve/pii/S0038071712002362 Accessed 8 November 2012. [Google Scholar]
- 61.Winterbottom B, Eilu G (2006) Uganda Biodiversity and Tropical Forest Assessment. Washington DC.
- 62. Laurance WF (1999) Reflections on the tropical deforestation crisis. Biol Conserv 91: 109–117. [Google Scholar]
- 63. Scheckenbach F, Hausmann K, Wylezich C, Weitere M, Arndt H (2010) Large-scale patterns in biodiversity of microbial eukaryotes from the abyssal sea floor. Proc Natl Acad Sci U S A 107: 115–120 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2806785&tool=pmcentrez&rendertype=abstract Accessed 1 November 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman J a, et al. (2006) Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4: 102–112 Available: http://www.ncbi.nlm.nih.gov/pubmed/16415926 Accessed 27 October 2012. [DOI] [PubMed] [Google Scholar]
- 65. McKinney M, Lockwood J (1999) Biotic homogenization: a few winners replacing many losers in the next mass extinction. Trends Ecol Evol 14: 450–453 Available: http://www.ncbi.nlm.nih.gov/pubmed/10511724. [DOI] [PubMed] [Google Scholar]
- 66. Olden JD, Rooney TP (2006) On defining and quantifying biotic homogenization. Glob Ecol Biogeogr 15: 113–120 doi:10.1111/j.1466-822x.2006.00214.x [Google Scholar]
- 67. Devictor V, Julliard R, Clavel J, Jiguet F, Lee A, et al. (2008) Functional biotic homogenization of bird communities in disturbed landscapes. Glob Ecol Biogeogr 17: 252–261 Available: http://doi.wiley.com/10.1111/j.1466-8238.2007.00364.x Accessed 21 January 2014. [Google Scholar]
- 68. Summerville KS, Boulware MJ, Veech J a, Crist TO (2003) Spatial Variation in Species Diversity and Composition of Forest Lepidoptera in Eastern Deciduous Forests of North America. Conserv Biol 17: 1045–1057 Available: http://doi.wiley.com/10.1046/j.1523-1739.2003.02059.x. [Google Scholar]
- 69. Weiher E, Howe A (2003) Scale-dependence of environmental effects on species richness in oak savannas. J Veg Sci 14: 917–920. [Google Scholar]
- 70. Obua J, Agea JG, Ogwal JJ (2010) Status of forests in Uganda. Afr J Ecol 48: 853–859. [Google Scholar]
- 71.National Environment Management Authority (NEMA) (2008) State of the Environment Report for Uganda.
- 72. Amann RI, Ludwig W, Schleifer K-H (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microb Rev 59: 143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Vainio EJ, Hantula J (2000) Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycol Res 104: 927–936. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Excel spreadsheet from Quantity One analysis of PCR-DGGE profiles of fungal and bacterial communities in natural and converted TRF ecosystems.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information Files.