Abstract
Studies attempting to identify and understand the function of mutated genes and deregulated molecular pathways in cancer have been ongoing for many years. The PI3K-PTEN-mTOR signaling pathway is one of the most frequently deregulated pathways in cancer. PIK3CA mutations are found 11%–33% of head and neck cancer (HNC). The hotspot mutation sites for PIK3CA are E542K, E545K and H1047R/L. The PTEN somatic mutations are in 9–23% of HNC, and they frequently cluster in the phosphatase domain of PTEN protein. PTEN loss of heterozygosity (LOH) ranges from 41%–71% and loss of PTEN protein expression occurs in 31.2% of the HNC samples. PIK3CA and PTEN are key molecules in the PI3K-PTEN-mTOR signaling pathway. In this review, we provided a comprehensive overview of mutations in the PI3K-PTEN-mTOR molecular circuitry in HNC, including PI3K family members, TSC1/TSC2, PTEN, AKT, and mTORC1 and mTORC2 complexes. We discussed how these genetic alterations may affect protein structure and function. We also highlight the latest discoveries in protein kinase and tumor suppressor families, emphasizing how mutations in these families interfere with PI3K signaling. A better understanding of the mechanisms underlying cancer formation, progression and resistance to therapy will inform selection of novel genomic-based personalized therapies for head and neck cancer patients.
Keywords: review, tumor sequencing, HNSCC, oncogenomics, mutations
Introduction
Head and neck squamous cell carcinoma (HNSCC) represents 90% of all malignant tumors in the head and neck region. It is formed by a solid mass of epithelial cells in various anatomical sites, such as oral mucosa, tongue, oropharynx, hypopharynx, and larynx. Although HNSCC has very standard histological features, the anatomical sites differ, and various risk factors (e.g., tobacco and HPV infection) contribute to its clinical behavior and response to therapy. This tumor is remarkably heterogeneous and success of future treatments relies on a better understanding of its diversity; this will allow us to identify groups of patients with similar HNSCC biological behavior and response to therapy. A major challenge in the scientific community is assessing the heterogeneity of HNSCC.
Large screening projects, such as the Head and Neck Genome Anatomy Project (HN-CGAP), started more than 10 years ago (http://cgap.nci.nih.gov) and were followed by the International Cancer Genome Consortium (ICGC) [1]. These joint efforts led to whole-exome sequencing analysis of HNSCC, resulting in identification of the most common genomic alterations [2, 3]. These results confirmed that HNSCC is extremely heterogeneous and there is no single genetic alteration or dysregulated molecular pathway responsible for its development and progression. Rather, it is evident that distinct genetic tumor signatures and dysregulated signaling pathways are shared within HNSCC subtypes, which may result in different clinical courses of the disease. In order to further advance therapy and improve survival of HNSCC patients, it is likely that patients with varying HNSCC should not be treated similarly.
Based on dysregulated pathways and genetic signatures in HNSCC, new therapies need to be developed that target specific subgroups of patients. For example, HPV-associated HNSCC became a subgroup of HNSCC with a distinct prognosis and response to therapy [4]. HPV-positive HNSCC displays several mutations but less than HPV-negative HNSCC. Indeed, the mutation rate in HPV-positive tumors is approximately half the mutation rate in HPV-negative HNSCC, suggesting that HPV oncogenes may play a role with acquired mutations to promote HPV-associated HNSCC [2]. Furthermore, additional studies revealed that PIK3CA or PIK3R1 were the only mutated genes in a subset of HPV-positive HNSCC [5].
The overall mutation rate of HNSCC is similar to other smoking-related tumors, such as small cell lung cancer and lung adenocarcinoma [2, 6]. HPV-negative HNSCC has a great diversity of genetic alterations in a variety of critical genes, which may underlie HNSCC formation, progression, and response to therapy. Mutations in the TP53 gene are common and occur in little over 50% of HNSCC, and mutations in Notch genes (NOTCH1, NOTCH2, and NOTCH3) are present in ~22% of HNSCC. Remarkably, PI3K/mTOR pathway genes, such as PTEN, TSC1 and PIK3CA, comprise 30.5% of the mutations found in HNSCC [3, 6]. Regardless of genomic heterogeneity, the phosphoinositide 3-kinase (PI3K) pathway is the most frequently somatically mutated pathway in HNSCC [5].
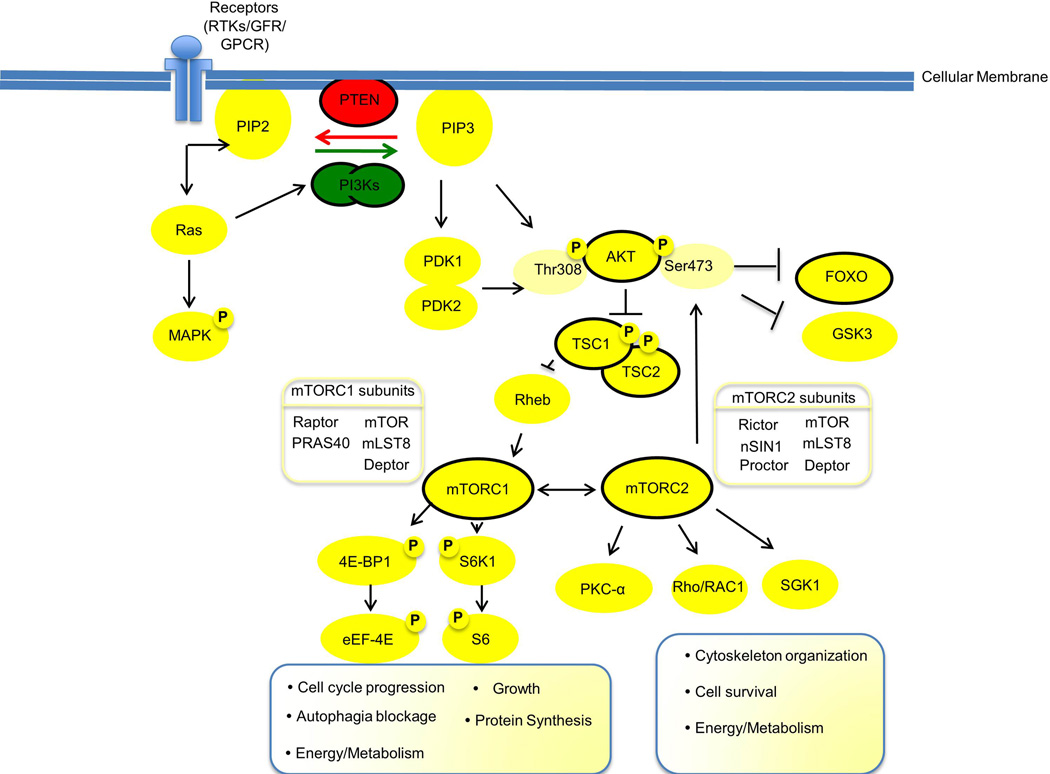
As seen in Figure 1, the PI3K pathway impacts most cellular functions involved in tumor behavior, including cell growth, local invasion, metastasis, survival, and resistance to therapy [7–9]. Genomic aberrations in HNSCC include posttranslational alterations and mutations in proteins downstream of PI3K and PTEN, including AKT, TSC1/TSC2, mTOR, RICTOR, and RAPTOR [2, 3, 5, 10]. Additional gene mutations and amplifications contribute to aberrant activation of the PI3K pathway. For example, increased Receptor Tyrosine Kinase (RTK) (e.g., EGFR, ErbBs, and MET) signaling induces PI3K/PTEN pathway activation [11]. Mutations in Ras genes (i.e., HRAS and KRAS) activate the PI3K pathway via p110α [12–15] and loss of p53 function promotes mTORC1 activation and regulation of PTEN transcription [16–18].
Figure 1.
The PI3K-PTEN-mTOR pathway. Activation of PI3K pathway via exogenous stimuli, endogenous signals and/or mutations promotes cancer formation, progression and resistance to therapy. The major signal modulators are PI3K in green and PTEN in red. The PI3Ks are shown as a simplified heterodimeric complex that combines the PI3K regulatory and catalytic subunits discussed in this review. As seen in the schematic representation, activated upstream cellular receptors (i.e. RTKs, GFRs, GPCRs) lead to activation of PI3Ks, which phosphorylate the membrane phospholipid substrate (i.e PIP2 into PIP3). PTEN is a phosphatase that opposes PI3K action; therefore, mutation or ablation of PTEN leads to PI3K/mTOR pathway over activation. Next, AKT is translocated to the cell membrane and is phosphorylated by PDK1 and PDK2. Once AKT is activated, it inhibits TSC1/TSC2 complex, which leads to RHEB inhibition. mTOR complexes (mTORC1 and mTORC2) collect the signals. mTORC1 activates downstream molecules. As a result, mTORC1 controls major processes, including cell cycle progression, growth, protein synthesis, lipid synthesis, and autophagy. mTORC2 functions are less understood, yet it is known to regulated, cell shape, cytoskeleton organization, cell survival, and cell metabolism. Therefore, PI3K/PTEN/mTOR pathway can control essential cancer cell processes, such as growth, invasion, metastases, and response to therapy. Black circles identify proteins that were found mutated in HNSCC. RTK: receptors of tyrosine kinase; GFR: growth factors receptors; PI3K: phosphatidylinositol 3-kinase; PIP2: Phosphatidylinositol 4,5-bisphosphateor PtdIns(4,5)P2; PIP3: Phosphatidylinositol (3,4,5)-trisphosphate or PtdIns(3,4,5)P3; PTEN: Phosphatase and tensin homolog; MAPK: Mitogen-activated protein kinases; FOXO: forkhead box O; GSK3: glycogen-synthase-kinase-3; PKC: protein kinase C; Rac1: ras-related C3 botulinum toxin substrate 1; SGK1: serum/glucocorticoid regulated kinase 1; S6K: ribosomal protein S6 kinase; TSC1/2: tuberous sclerosis 1/2; 4E-BP1: eukaryotic initiation factor 4E-binding protein 1; eEF-4E: Eukaryotic translation initiation factor 4E; Rheb: Ras homolog enriched in brain; mTORC1/2: rapamycin-sensitive mTOR complex 1 and 2.
This article reviews the profiling of genetic alterations in the PI3K/PTEN/mTOR pathway in HNSCC to understand how mutations alter HNSCC behavior and phenotype. This is an essential step towards personalized treatments of HNSCC using genomic-based therapy selection.
PTEN mutations
Phosphatase and Tensin homologue (PTEN; also known as MMAC1 and TEP1) is frequently mutated in multiple cancers [19, 20]. PTEN is well-known for its tumor suppressor role as a dual protein phosphatase. PTEN dephosphorylates phosphatidylinositol 3,4,5-trisphosphate (referred to as PIP3, PtdIns(3,4,5)P, or PI(3,4,5)P3) into phosphatidylinositol 4,5-bisphosphate (referred to as PIP2, PtdIns(4,5)P, or PI(4,5)P2), thereby broadly inhibiting oncogenic PI3K signaling. AKT relies on PIP3 for activation, an effect that is opposed by PTEN. Consequently, loss of PTEN function leads to increased oncogenic activity and enhanced PIP3 levels and PI3K/mTOR signaling. The phosphatase domain is the functional area of the PTEN protein responsible for modulating the levels of PIP3 (Figure 1A)[9].
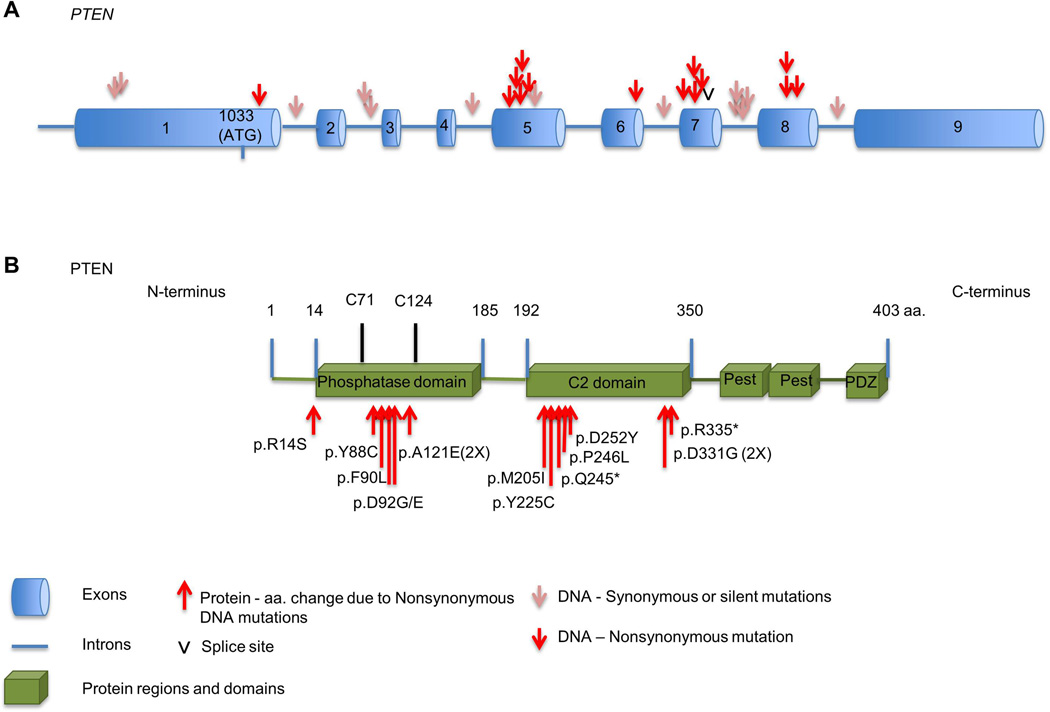
PTEN encodes for a protein with 403 amino acids and 2 major functional domains, the N-terminus and the C-terminus domains. Exons 1–5 encode the N-terminus, and exons 6–9 encode the C-terminus. Particularly, exon 5 encodes for the phosphatase domain [21] (Figure 2A).
Figure 2.
PTEN mutations in HNSCC. A. Analyses of the mutations sites are shown in the exons and introns of the PTEN gene. B. Nonsynonymous mutations affecting the PTEN protein are clustered mostly within functional area of the phosphatase domain, between the active site cysteine residues 71 (C71) and 124 (C124). Additional mutations were in the C2-domain between the amino acids 192 and 350.
Point mutations in the PTEN gene on 10q23.3 locus have been found in 9–23% of HNSCC (Table 1) [2, 3, 22–24]. Although not well understood, PTEN synonymous or silent mutations in HNSCC were identified in introns (i.e.,1, 2, 4, 6 and 7) and in exon 1 prior to the PTEN start codon (Figure 2A). To date, nonsynonymous mutations in PTEN have been found in exons 5–8. Recurrent mutations occur primarily in exon 5 codons, which results in p. D92E, p.D92G, and p. A121E at the phosphatase domain. In addition, recurrent mutations were also found in exon 8, resulting in p.D331G at the C2 domain (Figure 2A). Although the mutations appear dispersed mostly on exons 5–8 of the PTEN gene, the resulting amino acid changes are non-randomly distributed. Notably, the amino acids changes are concentrated in the functional areas of the PTEN protein. As seen in Figure 2B, nonsynonymous mutations in HNSCC cluster in the phosphatase domain, disrupting the functional area between active site cysteine residues 71 (C71) and 124 (C124). PTEN mutation in residue R14 has been described in HNSCC (Table 1). R14 mutation participates in PTEN decrease nuclear localization and it is important for PTEN membrane biding and catalytic activity [25, 26].
Table 1.
Mutations of PI3K pathway components in HNSCC: PTEN, TSC, AKT, and mTOR complexes.
| Gene Name |
Gene alias | UniProt | Protein Name |
Protein alias |
Mutation type and UTR/Intron change |
Amino Acid Change |
Ref. |
|---|---|---|---|---|---|---|---|
| PTEN | MMAC1, TEP1 | P60484 | PTEN | MMAC1 | Missense | p.A121E | 23 |
| Missense | p.D252Y | 2, 5 | |||||
| Missense | p.D331G | 23 | |||||
| Missense | p.D92E | 2; 5 | |||||
| Missense | p.D92G | 23 | |||||
| Missense | p.F90L | 23 | |||||
| Missense | p.M205I | 23 | |||||
| Missense | p.P246L | 2; 5 | |||||
| Nonsense | p.R335* | 2, 5 | |||||
| Missense | p.Y225C | 23 | |||||
| Missense | p.Y88C | 23 | |||||
| Nonsense | p.Q245* | 22 | |||||
| Missense | p.R14S | 5 | |||||
| intron7(del. TT) | 24 | ||||||
| c.e7_splice_site | 2; 5 | ||||||
| intron 1 | 23 | ||||||
| intron 2 | 23 | ||||||
| intron 2 | 23 | ||||||
| intron 4 | 22 | ||||||
| intron 6 | 23 | ||||||
| intron 7 | 23 | ||||||
| intron 7 (39 base insertion) | 22 | ||||||
| intron 7 (germiline) | 24 | ||||||
| UTR | 23 | ||||||
| UTR | 23 | ||||||
| TSC1 | TSC, KIAA0243 | Q92574 | Hamartin | TSC1 | Synonymous | p.P506P | 2 |
| Nonsense | p.R245* | 2; 5 | |||||
| TSC2 | TSC4 | P49815 | Tuberin | TSC2 | Nonsense | p.1415* | 5 |
| AKT2 | P31751 | AKT2 | PKB-β, PKA-2, RAC-PK-β | Missense | p.Y351C | 5 | |
| mTOR | FRAP, FRAP1, FRAP2, RAFT1, RAPT1 | P42345 | mTOR | Missense Missense |
p.L2260H p.R1161Q |
2; 5 2; 5 |
|
| RICTOR | KIAA1999 | Q6R327 | RICTOR | hAVO3 | Nonsense Missense |
p.E1633* p.D175H |
5 5 |
| RPTOR | KIAA1303, RAPTOR | Q8N122 | RAPTOR | Missense | p.P407L | 5 | |
Sporadic somatic mutations in PTEN are not an isolated event in HNSCC. Rather, they are one of the most common genetic alterations in human malignancies, particularly in solid tumors [19, 20]. However, the functional and biological roles of these mutations in HNSCC are largely unknown. Insights into PTEN structure and its role in various biological processes may help us understand how PTEN mutations and loss of expression contribute to HNSCC development, progression, and response to therapy.
The crystal structure of PTEN reveals important details about its three-dimensional folding that can add to our understanding of potential associations between distinct mutations and functional changes in HNSCC. The structure of the phosphatase domain displays an active pocket rich in basic residues that are essential for the ability of PTEN to dephosphorylate its substrates (i.e., acid phospholipids). The active pocket is also wide enough to fit the phosphorylated inositol moiety of PIP3. This active site has 3 catalytic loops: WPD-loop (residues 88–98), P-loop (residues 121–131) and TI loop (residues 160–171). Mutations in these phosphatase-binding loops are frequently found in human cancer, patients with hereditary PTEN related syndromes, and neurodevelopmental disorders, such as autism spectrum disorders (ASDs) [21, 27]. PTEN mutations in the phosphatase-binding loops in HNSCC are restricted to the WPD-loop (p.Y88C, p.F90L, p.D92G, p.D92E) and the P-loop (p.A121E) (Figure 3A and 3B). The PTEN catalytic signature (HCKAGKGR) is comprised of residues 123 to 130 and forms the bottom of the active pocket. The wall of the pocket is partly formed by the WPD-loop and the TI-loop [21, 27].
Figure 3.
Mutations of tumor suppressor PTEN in HNSCC. Crystal structure of PTEN protein is represented as ribbon diagrams. A. The functional active pocket of the phosphatase domain is shown. The mutated residues in the WPD-loop are highlighted in green (Y88), blue (F90), and white (D92). The mutated residue A121 is in yellow. B. Sequence of the WPD-loop and P-loop depicting the mutated residues within the walls of the active pocket present in the PTEN phosphatase domain. Mutated amino acid residues are in red. C. Mutated residues Q245 (in blue) and R335 (in cyan) in the C2 domain are nonsense mutations. Y225, P246, and D252 are shown in red, gray, and black respectively. M205 (in green) is in the Cβ 1/2 loop; and D331G (in yellow) is in the C2α loop.
The amino acids within the phosphatase domain are a common area for cancer-associated point mutations to arise, and they can lead to a reduction or abrogation of PTEN phosphatase activity. In order to understand the effects of PTEN mutations, extensive site-direct mutagenesis using comprehensive Ala–scanning mutagenesis (Ala-substitutions) was performed in the phosphatase binding loops of PTEN [27]. A humanized-yeast assay revealed that mutations in ASD influence the catalytic properties of PTEN. The Ala-substitutions in the WPD-loop resulted in partial enzymatic activity of PTEN. Interestingly, the p.D92E, p.D92G, and p.A121E mutations resulted in the inability of PTEN to catalyze PIP3 and P110α (catalytic subunit of class I PI3K) mediated growth. Conversely, p.Y88C and p.F90L did not alter PTEN lipid phosphatase activity, although p.Y88C partially affected growth in yeast [27]. Certain mutations, including p.Y88C, p.92AG, p.Y225C and a silent mutation (281A>T), were also paired with loss of heterozygosity (LOH) of chromosome 10 that involves the PTEN locus [23].
Located at the carboxy-terminus (C-terminus) is the C2 domain, a region important in protein-protein interaction, lipid membrane binding, and PTEN protein stability. The C2 domain (residues 192–350) binds to phospholipids at the cell membrane and allows PTEN to modulate cell migration. In HNSCC, p.M205I, p.Y225C, p.Q245*, p.P246L, p. D252Y, p.D331G, and p. R335* mutations are found in the C2 domain (Figure 3C and Table 1). There is little information regarding how mutations in the C2 domain affect PTEN function. The p.P246L and p.R335L mutations have also been identified in PTEN hereditary syndromes. Although located in the C2 domain, these two mutations partially interfere with PTEN lipid phosphatase activity and cell growth [27]. This may be related to the ability of the C2 domain to position the PTEN phosphatase active site towards to the cell membrane. Expression of the p.D331G mutation, located in the C2α loop, significantly reduces PTEN expression in U-87 MG glioblastoma cell lines. These results are consistent with the role of the C2 domain in PTEN protein stability [28].
C2 domain mutations found in HNSCC are closely related to mutations in endometrial carcinoma (D331G and S227F), lung cancer (G251C), and glioblastoma (P204S) [29–32]. P204S (present in the Cβ1/2 loop) and G125C mutations are associated with reduced PTEN expression and the inability to suppress cell growth [28]. Mutations in the C2α loop (i.e. D331G) and the Cβ1/2 loop (i.e. pM205I) likely affect PTEN protein stability and catalytic activity. Nonsense (i.e., p.Q245* and p.R335*) mutations, frameshift mutations, and mutations that produce splice variants also occur in the C2 domain and can prematurely terminate the open reading frame of PTEN to produce a truncated and nonfunctional protein. These newly described HNSCC PTEN mutations are located in important functional areas of PTEN and their effects on protein function and biological implications in HNSCC are unknown.
In addition to mutations, loss of heterozygosity (LOH) on chromosome 10 in the PTEN gene locus region is detected in HNSCC tissue samples at frequencies ranging from 41% to 71% [18, 22, 23, 33]. A second inactivation event (mutation or homozygous deletion) among the LOH of PTEN is only detected in up to 25% of HNSCC, suggesting that a single functional copy of PTEN in epithelial cells is insufficient for PTEN to function as a tumor suppressor (i.e.,PTEN haploinsufficiency). Additionally, LOH on chromosome 10 in the PTEN locus and loss of PTEN expression correlate to invasive and poorly differentiated carcinomas, lymph node involvement/metastases, and poor prognosis of HNSCC. Furthermore, loss of PTEN protein expression occurs in 31.2% (221/709) of human HNSCC samples [34].
Evidence from animal models implicates PTEN in epithelial lesions and benign tumors [35, 36]. To understand the biological relevance of PTEN alterations (i.e., mutations, LOH, and complete loss of function) and PI3K/mTOR activation in HNSCC initiation and progression, a novel oral specific HNSCC animal model was developed [34]. This genetically and environmentally defined HNSCC animal model has Pten LOH and full deletion of PTEN in the intraoral epithelia. Conditional deletion of Pten was achieved using the keratin promoter 14 CRE-lox system. Because tobacco is the most common etiological factor in HNSCC, animals were exposed to a tobacco surrogate (4-nitroquinoline-1-oxide or 4NQO) to increase their susceptibility to oral cancer. Deactivation and/or downregulation of PTEN due to mutations, epigenetic changes, or posttranslational regulation were recapitulated by hemizygous (partial) or homozygous (complete) deletion of the Pten gene. Pten ablation also resulted in increased activation of PI3K and other pathways. These mice showed rapid development of oral-specific premalignant lesions and carcinomas [34]. Notably, HNSCC with deregulated PTEN lack p53 and show overexpression of pAKT and mTOR. In addition, animals lacking PTEN expression have increased COX2 expression and angiogenesis in the surrounding stroma [34].
PI3K family gene mutations
PI3Ks belong to a family of intracellular lipid kinases that phosphorylate the 3′-hydroxyl group of phosphatidylinositol (also known as PI or PtdIns) and phosphoinositides. The lipid products of PI3K act as second messengers by binding and activating several signaling pathway molecules.
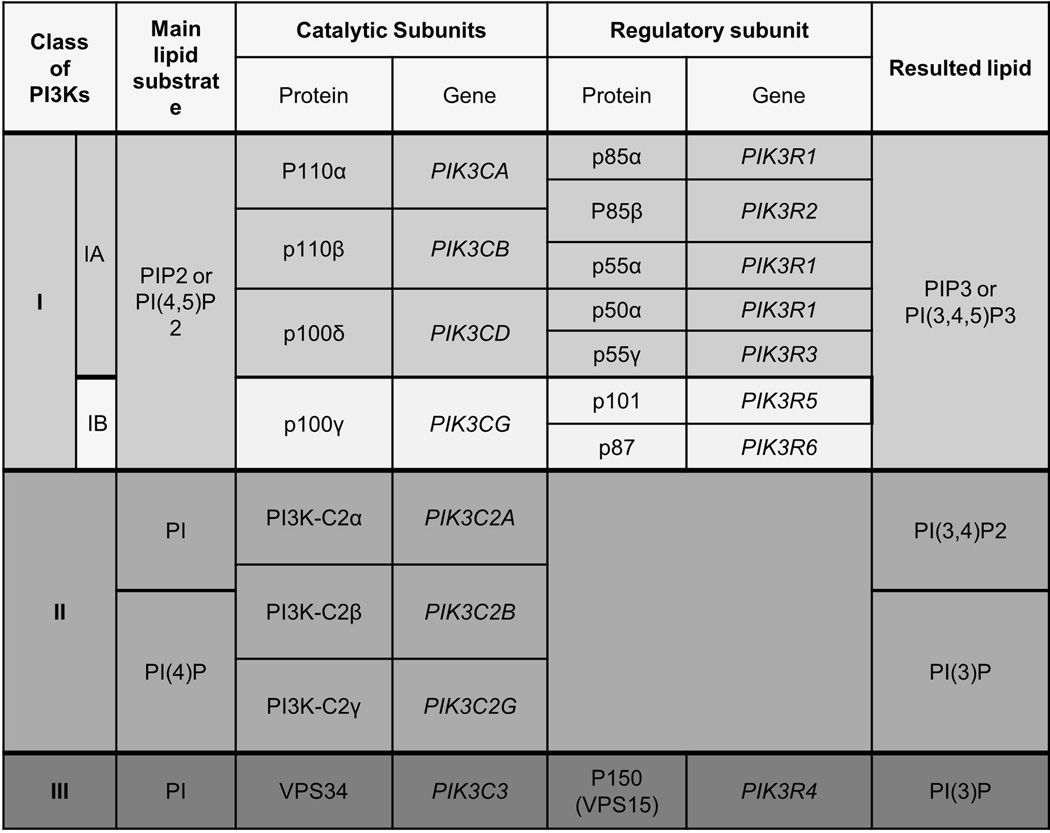
PI3K signaling is involved in the control of fundamental cellular functions and properties, including cell growth, proliferation, differentiation, survival, metabolism, vesicular trafficking, degranulation, cytoskeletal rearrangement, and motility (Figure 1) [8, 9, 37]. PI3Ks are divided into Class I, Class II, and Class III according to their structural features, in vitro lipid substrate specificity, tissue distribution, mechanism of activation, and function (Figure 6) [8, 12].
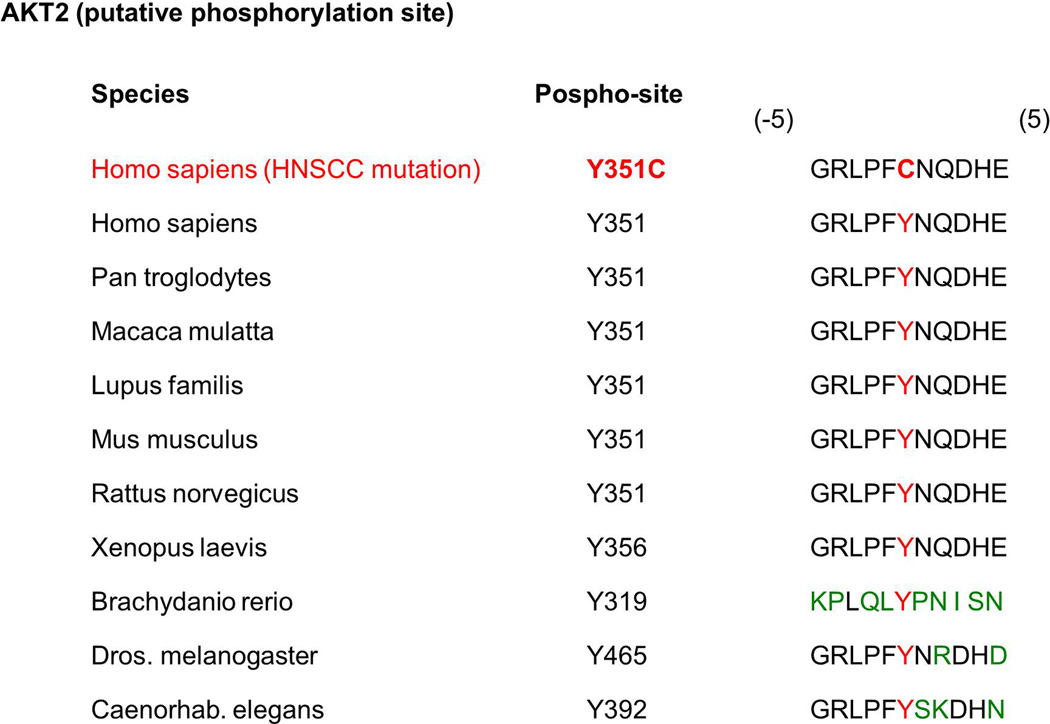
Figure 6.
AKT2 mutation in HNSCC. Protein sequences of the putative phosphorylation site (Y) for AKT2 are conserved across species. The amino acid residue change found in the HNSCC (Y351C) is shown in red.
The in vitro substrates for class I PI3Ks are PI, Phosphatidylinositol 4-phosphate (referred to as PI(4)P or PtdIns4P), and PIP2; and the primary in vivo substrate is PIP2. Therefore, class I PI3Ks primarily catalyzes the conversion of PIP2 to PIP3. PIP3 activates many downstream signaling proteins, including the protein serine-threonine kinase AKT. The lipid phosphatase PTEN dephosphorylates PIP3, thereby regulating PI3K signaling [12].
Class I PI3K isoforms are comprised of p110α (also called PIK3CA), p110β (also called PIK3CB), p110γ (also called PIK3CG), and p110δ (also called PIK3CD). In mammals, Class I PI3Ks is divided into two groups, Class IA and Class IB, based on structural and functional differences. Receptor tyrosine kinases (RTKs), such as epidermal growth factor receptor (EGFR), hepatocyte growth factor receptor (HGFR or MET), platelet-derived growth factor receptor (PDGFR), fibroblast growth factor receptor (FGFR), and insulin-like growth factor 1 receptor (IGF-1R), act as primary activators of Class IA PI3Ks. Hras also activates Class I PI3Ks isoforms [13, 14]. Hras is frequently mutated in HNSCC, specially in Asia [14], and its interaction with p110α is mediated by the RBD domain in p110α. The Class IA PI3Ks isoform, PIK3CA, is a critical effector for RAS-driven tumorigenesis [15]. Additionally, G-protein-coupled receptors (GPCRs) activate Class IB PI3Ks (GPCRs) [12, 37, 38].
Class IA is the most common PI3K class involved in human cancers [12]. Class IA PI3Ks are comprised of p110 catalytic subunits (i.e., p110α, p110β, and p110δ) that form heterodimeric protein complexes with p85 regulatory subunits. p110α, p110β, and p110δ are encoded by PIK3CA, PIK3CB, and PIK3CD genes, respectively. There are five isoforms of the p85 regulatory subunit, including p50α, p55α, p55γ, p85α, and p85β.p85α, and its splices variants p50α, p55α, are encoded by a single PIK3R1 gene, but p85β and p55γ isoforms are encoded by PIK3R2 and PIK3R3, respectively [12]. Binding of p85 subunits with p110 subunits promotes stabilization of p110 subunits. Once at the plasma membrane, p110 subunits phosphorylate its lipid substrates [12, 37, 39, 40].
Class IB PI3Ks are found exclusively in mammals. They are formed by dimerization between the p110δ and p101 (also called PIK3R5) or p87 (also called PIK3R6, p87PIKAP or p84). p101 is encoded by the PIK3R5 gene, p87 is encoded by the PIK3R6 gene, and the p110γ is encoded by the PIK3CG gene. GPCRs activate Class IB PI3Ks through a mechanism in which p101 recruits p110γ, permitting their activation and translocation to the plasma membrane by βγ subunits of trimeric G proteins [41–43]. The p110α and p110β are ubiquitously expressed, and p110δ and p110γ are primarily found in the hematopoietic system, especially in leukocytes [44]. This tissue-specific distribution suggests a more specific and essential function of PI3Kδ and PI3Kγ signaling in the microenvironment of tumors and in hematopoietic tumors and diseases.
Class II PI3Ks have three mammalian isoforms named PI3K-C2α (also called PIK3C2A), PI3K-C2β (also called PIK3C2B), and PI3K-C2γ (also called PIK3C2G), which are encoded by PIK3C2A, PIK3C2B and PIK3C2G genes, respectively [45–49]. PI3K-C2α and PI3K-C2β are ubiquitously expressed, and PI3K-C2γ is a liver-restricted protein [50]. Unlike class I PI3Ks, class II PI3Ks are monomers with only a catalytic domain [47]. Class II PI3Ks have well-recognized in vitro substrates, PI and PI(4)P, but the in vivo targets are still being identified. PI(4)P is used to generate PI(3,4)P2, and PI is used to generate PI3P [12].
Class III PI3K has a single member, the vacuolar protein-sorting 34 (VPS34, also called PIK3C3) that was first identified in mammals in 1995 [51]. It is highly conserved among yeast, plants, and mammals. The catalytic subunit VPS34 is encoded by the PIK3C3 gene. VPS34 forms a complex with serine threonine protein kinase p150 (also called VSP15 or PIK3R4), which is encoded by the PIK3R4 gene. The regulatory subunit is essential for activation and recruitment of VPS34 to intracellular membranes where it phosphorylates PI into PI3P, the only substrate of class III PI3Ks [12]. The most recognized function of class III PI3Ks is the regulation of vesicular trafficking in the endosomal/lysosomal system [52–56]. Class III PI3Ks also activate additional mechanisms in mammalian cells, such as endocytosis and phagocytosis. Additionally, VPS34 kinase may also play a role in autophagy and protein synthesis through an mTOR-dependent mechanism. PI3P, which is primarily localized in endosomes, phagosomes and intracellular organelles such as endoplasmic reticulum, can be converted to PI(3,5)P2 by the lipid FYVE finger-containing phosphoinositide kinase (PIKfyve, also called FAB1) [57–60].
Regardless of PI3K class distinctions, genetic alterations in PI3K genes and epigenetic aberrations in PI3K signaling are commonly found in cancer development and progression [61–64]. Multiple tumors, including colon, brain, gastric, breast and lung, were sequenced to determine which PI3Ks were mutated. PIK3CA was the only gene in the PI3K family with tumor-specific somatic mutations [64]. Several subsequent studies confirmed the presence of PIK3CA mutations in solid tumors [62, 64]. Sequencing of ~150 HNSCC tumor samples revealed mutations in the PI3K pathway in 30.5% of cases, making it the most frequently mutated oncogenic pathway in HNSCC [2, 3, 5]. HNSCC tumors harboring PI3K pathway mutations have greater genomic instability than tumors lacking mutations in PI3K genes. These data are supported by the finding that tumors with PI3K pathway mutations have a larger number of non-synonymous and cancer gene mutations than tumors without PI3K mutations. Additionally, tumors with PI3K mutations show increased frequency of mutations in DNA damage/repair genes [5].
Notably, advanced (i.e., Stage IV) ovarian, colorectal, and pancreatic tumors may exhibit concomitant mutational events in PI3K pathway genes and in other pathways, such as MAPK. These concomitant-activating mutations result in a poor response to chemotherapy [65, 66]. Simultaneous mutations in PI3K and MAPK pathways have not been reported in HNSCC [2, 3, 5, 65, 66], indicating that activation of concomitant signaling in these pathways is due to posttranslational activation and/or crosstalk between pathways in HNSCC. Furthermore, advanced stage HNSCC tumors have mutations in more than one PI3K pathway molecule, as evidenced by mutations in PI3C2G and PTEN; PIK3R1 and mTOR; PIK3CA and mTOR; PIK3CA and PTEN; PIK3C2G, PIK3R5 and PIK3CA [5] Therefore, combined aberrations in PI3K pathway molecules may be associated with progression of HNSCC, while mutually exclusive mutations and alterations within this pathway is related to HNSCC formation and initial progression. Indeed, amplification of PIK3CA is detected in oral dysplastic lesions, and amplification associated with mutations was associated to invasive HNSCC [67, 68]. Mutually exclusive mutations of PI3KCA and PTEN have also been observed in breast cancer and gastric carcinomas [69, 70].
More than 30% of human solid tumors, such as gastric, lung, breast, hepatocellular, colorectal, ovarian and glioblastoma carcinomas, exhibit mutations in PIK3CA [62, 64]. Moreover, PIK3CA is mutated in 11%–33% of HNSCC, making it one of the most commonly mutated genes in this disease [2, 3, 5, 71–73]. PIK3CA is located on chromosome 3q26.32 and encodes the p110α catalytic subunit of PI3K. In addition to the activating mutations, PI3KCA is also amplified in most human malignancies [62, 64], including HNSCCs [5, 67, 74, 75]. PIK3CA has been associated with advanced stage tumors, vascular invasion, and metastasis in HNSCC [67, 72, 74, 75]. Across cancer types, PIK3CA alterations impact cancer predisposition, progression, recurrence, metastasis, and prognosis [69, 76–81].
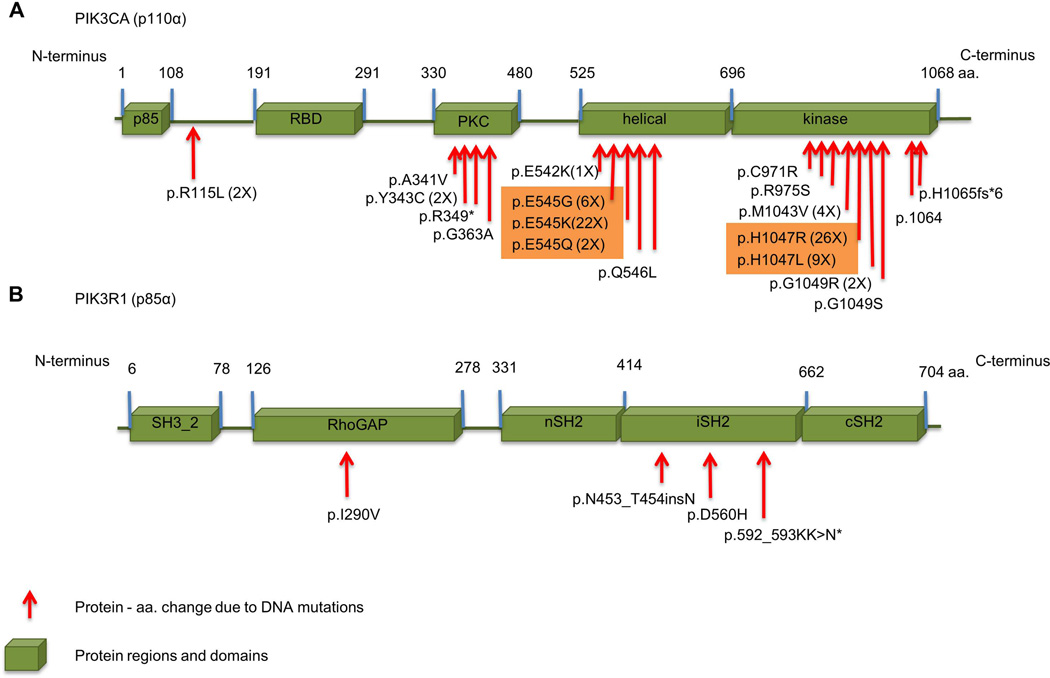
The PIK3CA mutations in human tumors are somatic, cancer-specific, heterozygous, and primarily missense [62, 64]. The pattern of PIK3CA mutation is normally characterized by more than 75% of mutations grouped in the helical (exon 9) and kinase domains (exon 20) of the gene. These domains are described as hot spots for PIK3CA mutations [64]. In HNSCC, hotspot mutation sites for PIK3CA mutations are E542K, E545K and H1047R/L [5, 64, 82–84], which comprise ~90% of PIK3CA mutations identified in HNSCC (Figure 5A and Table 2). Most commonly, the glutamic amino acid in positions 542 and 545 (i.e., E542 and E545) is substituted with a lysine (K), and histidine1047 (i.e., H1047) is frequently substituted with arginine (R).
Figure 5.
Mutations observed in the PI3KCA and p85α. The amino acids residues show mutations in the indicated protein domains. A. Hot spot mutations of PI3KCA in HNSCC are E545K/G/K and p.H1047R/L as indicted in orange boxes. B. The site of mutations of regulatory subunit PIK3R1 (also known as p85α) are found within the RhoGAP-binding domain and the iSH2 domain.
Table 2.
Mutations in the PI3K pathway components in HNSCC: PI3K family genes and proteins.
| Gene Name |
Gene alias |
UniProt | Protein Name |
Protein Alias |
Mutation Type | Amino Acid Change |
Ref. | Cell Line; Ref. |
|---|---|---|---|---|---|---|---|---|
| PIK3CA | P42336 | PIK3CA | p100α, PI3Kα | Frameshift-Ins | p.1064insA | 73 | ||
| Missense | p.A341V | 158 | ||||||
| Missense | p.C971R | 5 | ||||||
| Missense | p.E542K | 2, 5, 72, 158–160 | SCC-6; 167–168 | |||||
| Missense | p.E545G | 2 | HSC3; 73 | |||||
| Missense | p.E545K | 2, 5, 72,73, 159–165 | HSC4; 72–73 | |||||
| Missense | p.E545Q | 82 | SQ9G; 167 | |||||
| Missense | p.G1049R | 72, 163 | ||||||
| Missense | p.G1049S | 72 | ||||||
| Missense | p.G363A | 5 | ||||||
| Missense | p.H1047L | 2, 3 | ||||||
| Missense | p.H1047R | 2, 3, 5, 72, 160, 161,163 | Detroit562; SUNE1; HONE1, CNE2; CAL-33; HSC2; HOC92; 71–73, 159, 166, 169–171. | |||||
| Frameshift | p.H1065fs*6 | 73 | ||||||
| Missense | p.M1043V | 72, 160 | HOC719; 73 | |||||
| Synonymous | p.P381P | 159 | ||||||
| Missense | p.Q546L | 72 | ||||||
| Missense | p.R115L | 2 | ||||||
| Nonsense | p.R349* | 158 | ||||||
| Missense | p.R975S | 5 | ||||||
| Synonymous | pT1025T | 73 | ||||||
| Missense | p.Y343C | 159 | ||||||
| PIK3CB | PIK3C1 | P42338 | PIK3CB | p110β, PI3Kβ | Synonymous | p.P422P | 2 | |
| PIK3CD | O00329 | PIK3CD | p110δ, PI3Kδ | Missense | p.T423S | 5 | ||
| Missense | p.T458S | 2 | ||||||
| PIK3CG | P48736 | PIK3CG | p110γ, PI3Kγ | Missense | p.A156V | 2 | ||
| Missense | p.G491E | 2 | ||||||
| Missense | p.L843H | 2 | ||||||
| Missense | p.P526S | 2 | ||||||
| Missense | p.R1021S | 2 | ||||||
| Missense | p.S446F | 2 | ||||||
| Missense | p.R178C | 166 | CAL-27; 166 | |||||
| PIK3R1 | GRB1 | P27986 | PIK3R1 | p85α/p55α/p50α, PI3Kα | In frame_Ins | p.453_454insN | 2 | |
| Nonsense | p.592_593KK>N* | 5 | ||||||
| Missense | p.D560H | 2 | ||||||
| Missense | p.I290V | 2 | ||||||
| PIK3R5 | Q8WYR1 | PIK3R5 | p101, FOAP-2, p101-PIK3 | Missense | p.A341V | 5 | ||
| Missense | p.E125G | 5 | ||||||
| Missense | p.E322K | 5 | ||||||
| Nonsense | p.E60* | 5 | ||||||
| PIK3R6 | C17orf38 | Q5UE93 | PIK3R6 | p84, p87, p84 PIKAP, p87 PIKAP | Missense | p.R483H | 2 | |
| PIK3C2A | O00443 | PIK3C2A | PI3K-C2α | Splice Site SNP |
2 | |||
| Synonymous | p.L226L | 2 | ||||||
| PIK3C2B | O00750 | PIK3C2B | PI3K-C2β, C2-PI3K | Missense | p.R564C | 2 | ||
| PIK3C2G | O75747 | PIK3C2G | PI3K-C2γ | Deletion | p.P129del | 166 | SCC-15; Detroit562; SCC-25; 166 |
|
| Missense | p.S1272L | 5 | ||||||
| Frameshift_Ins | p.T62fs | 5 | ||||||
| Missense | p.V656L | 2 | ||||||
| PIKFYVE | PIP5K3, KIAA0981 | Q9Y2I7 | PIKfyve | PIP5K, PIP5K3, PIPkin-III | Missense Missense |
p.H1851Q p.R1766P |
2 2 |
|
PIK3CA mutations are activating mutations that often result in biological behavioral changes to the tumor. At all three hotspot mutation sites, aberrations lead to a gain of enzymatic function [64]. For example, in HNSCC, colorectal, breast and ovarian cancers, the primary functional consequence of PI3KCA hotspot mutations is elevated lipid kinase activity that results in constitutive AKT signaling, as revealed by increased phosphorylation of AKT and its downstream targets S6K and 4EBP [5, 73, 83, 85–88]. These hotspot mutations can also transform chicken-embryo fibroblasts, NIH 3T3 cells, and mammary epithelial cells [83, 85, 88]. These mutations cause increased proliferation rates and more severe metastatic phenotypes in HNSCC and breast and colorectal cancers, as evidenced by increased migration and invasion in vitro and in vivo [73, 86, 89].
Recently, four novels PIK3CA mutations, R115L, G363A, C971R, R975S, were reported in HNSCC tumors. Functional assays showed increased HNSCC cell growth when R975S, R115L and H1047R were overexpressed and AKT was phosphorylated at threonine 308 and serine 473 [5]. Xenografts of a HNSCC tumor sample containing the PIK3CA mutation (E542K) showed a better response to the BEZ235 dual inhibitor compared to HNSCC tumors expressing wildtype PIK3CA [5]. These findings support previous studies that showed tumors with altered and mutated PI3K family genes, including PI3KCA and PTEN, are exquisitely sensitive to PI3K/mTOR inhibitors [65, 90–95]. Therefore PI3K/mTOR inhibitors, used as a single or coadjuvant treatment, may be a novel targeted therapy for HNSCC treatment, especially HNSCC tumors with mutated and altered PI3K signaling molecules.
Janku et al., 2011 detected PIK3CA mutations in 11.5% of patients with different solid tumors, including endometrial, ovarian, colorectal, breast, cervical and HNSCC. Patients with PIK3CA mutations show a better response to PI3K/mTOR pathway inhibitors compared to patients without PIK3CA mutations [65]. However, as previously mentioned, malignant tumors expressing simultaneous mutations in MAPK and PI3K pathways have poor responses to PI3K/AKT/mTOR pathway inhibitors [65, 66], suggesting that inhibitors that target PI3K pathway effectors in combination with MEK inhibitors could be an effective molecular targeted therapy to treat HNSCC.
In addition to PIK3CA mutations, mutations in PIK3CB and PIK3CD, genes that encode the catalytic subunits p110β and p110δ, respectively, have been found in HNSCC. p110βharborsa synonymous mutation (p.P422P), and p110δ has missense mutations (p.T423S and p.T458S) (Table 2) [2, 5]. The significance of these mutations in PI3K signaling in HNSCC is unknown. Mutation in the PIK3CD gene was also described in neuroblastoma, and its biological function correlates to tumor development and progression [96].
Additional PI3K class I components are also mutated in HNSCC (Table 2). PIK3R1 has an in frame insertion (c.1914–1916 - TAT>TATAAT) and missense mutations (p.I290V and p.D560H). PIK3R1 encodes p85α, p55α and p50α [97, 98]. Mutations in PIK3R1 alone represent 7% of HNSCC mutations (3/41). Studies have reported p85α mutations in certain human tumors, including breast [99], colon [99, 100], ovarian [100], glioblastoma [101], and HNSCC [5], but their roles in tumor development and/or behavior are still unknown. Mice lacking PIK3R1 in the liver (L-Pik3r1KO mice) develop aggressive high-grade hepatocellular carcinomas and lung metastases [102]. p.D560Y induces anchorage-independent growth, increased proliferation and AKT activation in vitro. Mice implanted with BaF3 cells containing p85α D560Y develop metastases in the liver and spleen [97]. Potential effects of HNSCC mutations are suggested by where they arise in the p85α (Figure 5B). Mutation at D560 occurs in the p85α iSH2 domain, a protein domain that permits formation of a complex with p110α [103]. The p85 regulatory subunit is required to stabilize and inhibit the catalytic activity of p110 in basal conditions [40, 104]. The PIK3R1 mutation in colon carcinomas disrupts p85 inhibition of p110, resulting in constitutive activation of the PI3K pathway beyond p110α activation. Therefore, p85α mutants may promote AKT activation, cell survival, and oncogenesis in a p110-dependent manner [97]. Further PI3K activation via the p85αmutation may also regulate PTEN because reduced p85α expression is associated with decreased PTEN expression and activity [102, 105, 106].
Mutations in PIK3CG (p110γ), PIK3R5 (p101) and PIK3R6 (p84/p87PIKAP) were identified in HNSCCs (Table 2). [5]. These PI3K family members belong to Class IB PI3K, but the consequences of these mutations to HNSCC are still unknown. Notably, p110γ−/− mice develop tumors in the colon (colorectal adenocarcinomas) [107] and hematologic defects [108–110]. Although mutations in PIK3R5 and PIK3R6 have been identified in lung adenocarcinoma [111], their effects on tumorigenesis are unknown.
Mutations in PIK3C2A, PIK3C2B and PIK3C2G, members of the Class II PI3Ks, were also identified in HNSCC (Table 2). PIK3C2B is associated with prostate cancer risk [112], and surprisingly, PI3KC2B may promote resistance of leukemia cells to chemotherapeutics [113].
Lastly, the PIKfyve gene that encodes for PIKfyve protein is also mutated in HNSCC (Table 2). PIKFyve regulates key cellular processes, such as membrane trafficking, stress-induced signaling, cytoskeletal dynamics, nuclear transport, gene transcription and cell cycle progression [114]. PIKfyve regulation and functionality depends on stable complex formation between PIKFyve and the antagonistic PI(3,5)P2-specific phosphatase Sac3. Sac3 originated from the FIG 4 gene and orchestrates the coordinated regulation of PI(3,5)P2 synthesis and turnover [115]. In addition to HNSCC mutations, dysfunction and mutations in PIKfyve and Sac3 are also found cancers of the ovary, endometrium, and lung, among others (COSMIC database: http://www.sanger.ac.uk/cosmic) [71].
Figure 4.
Overview of the PI3K family members and their phospholipid substrates. PI3Ks constitute a large family of enzymes/oncogenes that is divided in the three classes as shown. Gene and protein names are paired by the catalytic and regulatory subunits that originate the PI3K heterodimers. Most common phospholipid substrate for each PI3K class is depicted.
Additional mutations in the PI3K/PTEN/mTOR pathway (AKT, mTOR, TSC1, TSC2, RICTOR, and RPTOR)
AKT2 mutations
AKT2 is mutated at p.Y351C in HNSCC (Table 1) [5]. Although Y351 is a putative phosphorylation site in AKT2 that is conserved among multiple species (Figure 6) (http://www.kinexus.ca), its contribution to AKT2 protein function and effect on cells is not understood. AKT2 overexpression also occurs in HNSCC [116].
AKT gene family members are AKT1, AKT2, and AKT3, which are ubiquitously expressed in most tissue types (www.genecards.org). These genes encode AKT1/PKB-α, AKT2/PKB-β, and AKT3/PKB-γ proteins isoforms, which share a high degree of amino acid similarity and are activated by similar pathways in a PI3K-dependent manner [117–119]. The well-known phosphorylation sites of AKT are threonine 308 (pAKTThr308) and serine 473 (pAKTSer473). These AKT phosphorylation sites, along with markers of mTOR activation (e.g. PRAS40), have prognostic value and have been implicated in predicting chemotherapeutic response in lung and breast cancer [94, 120, 121].
In HNSCC, AKT activation occurs in premalignant lesions and late stage carcinomas and is correlated with poor clinical outcome [10, 122–124]. Nonetheless, analyses of total AKT function is revealing specific roles for the isoforms [125]. In vivo studies using constitutive AKT1 (myrAKT) overexpression resulted in oral lesions that are blocked by senescence; subsequent ablation of Trp53 permits malignant progression [126]. In breast cancer mouse models, AKT1 participates in ErbB-2 induced tumorigenesis and but not in tumor invasion. Upregulation of AKT2 is involved in migration, invasion, metastases, and treatment resistance in neuroblastoma, breast, and colorectal cancers [125, 127–131]. Furthermore, in vivo studies showed that AKT2 directly affects glucose uptake [132]. Upregulation of AKT2 is induced during epithelial mesenchymal transition (EMT) by Twist [127]. Conversely, downregulation of AKT2 induces caspase-dependent apoptosis and suppresses EMT and migration [130, 133–135].
TSC1 and TSC2 mutations
The tumor suppressor genes TSC1 and TSC2 are on chromosome 9q34.13 and 16p13.3, respectively. TSC1 encodes Hamartin and TSC2 encodes Tuberin, which form heterodimers [136, 137]. The Hamartin/Tuberin formation is referred to as the TSC1/TSC2 complex, which is located downstream of AKT/PKB. AKT phosphorylates and inactivates TSC2, and inactivation of the TSC1/TSC2 complex leads to enhanced mTOR signaling through activation of mTORC1 [138, 139]. AKT can also signal to mTORC1 in a TSC1/TSC2-independent manner by phosphorylating the PRAS40, amTORC1 inhibitor [140, 141]. Pro-inflammatory cytokines important to HNSCC, such as TNFα, also activate mTOR via IKKβ phosphorylation of TSC1 [142]. In addition, Wnt signaling modulates mTOR via TSC1/TSC2 interaction and GSK3 [143].
Mutations in TSC1/TSC2 tumor suppressor genes that result in inactivation of the complex are commonly found in patients with tuberous sclerosis [144–146]. In HNSCC, unique mutations have been described in TSC1 (p.R245*) and TSC2 (p.S1514*) (Table 1) [2, 3] and DNA methylation and LOH in TSC1/TSC2 also occur [147]. How mutations in TSC1/TSC2 contribute to HNSCC may be revealed by the function of the TSC1/TSC2 complex. For example, the TSC1/TSC2 complex is the main regulator of mTORC1, and inactivation of TSC1/TSC2 promotes mTORC1 activation [148]. TSC2 overexpression inhibits growth of HNSCC cells [149].
Mutations in mTOR, Rictor, Raptor, and FOX gene families
The mTOR, RICTOR, and RPTOR genes are mutated in HNSCC (Table 1). Specifically, the mTOR mutations are p.L2260H and p.R1161Q, the RICTOR mutations are p.E1633*and p.D175H, and the RAPTOR mutation is p.P407L [2, 5]. RICTOR is also amplified in 38.7% of HNSCC, some of which also contain PI3KCA (45.2%) and EGFR (32.3%) amplification [82]. It is unclear how these newly described mutations and amplifications affect HNSCC. mTOR, RICTOR, and RPTOR genes encode for mTOR, RICTOR and RAPTOR proteins, respectively. The mTOR is the catalytic subunit of two distinctive molecular complexes known as mTOR complex 1 (mTORC1 and mTOR complex 2 (mTORC2) [150]. mTORC1 is comprised of RAPTOR, PRAS40, mTOR, mLST8, DEPTOR, and TTIL/TEL2 complex proteins. mTORC2 has seven known protein components, including RICTOR, mSIN, PROTOR1/2 mTOR, mLST8, DEPTOR, and TTIL/TEL2. mTORC1 and mTORC2 complexes can sense and respond to a variety of endogenous and exogenous signals, including growth factors, stress, oxygen, and amino acids, among others. mTORC1 is a well-known activator of translation regulators and initiation factors, such as p70/S6 and 4E binding protein 1 (4E-BP1) and Garb10 [151]. mTORC1 activation induces cell growth, cell cycle progression, protein and lipid syntheses, and autophagy [150]. In contrast to mTORC1, activation of mTORC2 seems to be independent of nutrients, but it does respond to growth factors stimuli such as insulin (citation). mTORC2 controls several molecules, including AKT, SGK1 (serum/glucocorticoid regulated kinase 1), and PKC-α (protein kinase C-α) [152–154]. mTORC2 phosphorylates AKT at Ser473 [155]. Notably, depletion of mTORC2 impairs AKT signaling, resulting in deficient activation of Forkhead Box proteins, such as FOXO1 and FOXO3A, but having no effect on TSC2 and GSK3 phosphorylation [152–154]. Finally, mTORC2 also controls paxillin and Rho GTPases via PKC-α [150]. Therefore, mTORC2 regulates cell shape, migration, cytoskeleton organization, survival, and metabolism [150]. FOX family genes, including FOXO1, FOXO3, FOXD1, FOXD3, FOXJ2, FOXK1, FOXI1, FOXN1, FOXN3, FOXP1, FOXP2, and FOXP4, are also mutated in HNSCC [2, 3]. FOX proteins belong to a large family of transcriptional regulators that affect the behavior of breast, prostate and colon tumors and may serve as a target for therapeutic intervention [61, 156, 157].
In Conclusion
There has been much progress in identifying mutated genes and proteins in HNSCC, but numerous questions remain regarding their physiological and pathological roles. In vitro and in vivo studies using genetic and pharmacological tools are necessary to better understand HNSCC tumor biology and to improve cancer therapy in order to increase patient survival, quality of life, and ultimately, disease resolution. Cancer, particularly of the head and neck, is a very complex genetic disease. The acquisition of oncogenic signals, such those driven by HPV infection and somatic mutations, affect cellular transformation, tumor development, progression, and metastases. Interestingly, distinct cell types, including cancer stem cell, may respond differently to mutations in p53, PI3KCA, PTEN and other proteins. Furthermore, not every group of head and neck cancer patients with distinctive group of mutations or deregulated signaling pathways will respond the same to therapy. Individualized therapies catering these different subgroups of HNSCC tumors may yield better outcomes. Much work is still needed to understand the contribution of these mutations and deregulated pathways to HNSCC and to determine the effects of inhibitors that target molecules in this pathway. This information will guide the discovery of novel drugs and treatment approaches for HNSCC.
Acknowledgments
This work was supported by the University of Michigan Comprehensive Cancer Center through grant P50-CA97248 (University of Michigan Head and Neck, Specialized Programs of Research Excellence) from the NIH/National Cancer Institute. F.S.G. was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Brasília, DF, Brazil).
References
- 1.International Cancer. Genome C, Hudson TJ, Anderson W, Artez A, Barker AD, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92:709–720. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 5.Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-13-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 7.Salmena L, Carracedo A, Pandolfi PP. Tenets of PTEN tumor suppression. Cell. 2008;133:403–414. doi: 10.1016/j.cell.2008.04.013. [DOI] [PubMed] [Google Scholar]
- 8.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 9.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 10.Molinolo AA, Amornphimoltham P, Squarize CH, Castilho RM, Patel V, et al. Dysregulated molecular networks in head and neck carcinogenesis. Oral Oncol. 2009;45:324–334. doi: 10.1016/j.oraloncology.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanhaesebroeck B, Stephens L, Hawkins P. PI3K signalling: the path to discovery and understanding. Nat Rev Mol Cell Biol. 2012;13:195–203. doi: 10.1038/nrm3290. [DOI] [PubMed] [Google Scholar]
- 12.Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol. 2010;11:329–341. doi: 10.1038/nrm2882. [DOI] [PubMed] [Google Scholar]
- 13.Chang F, Lee JT, Navolanic PM, Steelman LS, Shelton JG, et al. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia. 2003;17:590–603. doi: 10.1038/sj.leu.2402824. [DOI] [PubMed] [Google Scholar]
- 14.Murugan AK, Munirajan AK, Tsuchida N. Ras oncogenes in oral cancer: the past 20 years. Oral Oncol. 2012;48:383–392. doi: 10.1016/j.oraloncology.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129:957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 16.Pease EA, Tien M. Heterogeneity and regulation of manganese peroxidases from Phanerochaete chrysosporium. J Bacteriol. 1992;174:3532–3540. doi: 10.1128/jb.174.11.3532-3540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–460. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, et al. Regulation of PTEN transcription by p53. Mol Cell. 2001;8:317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Yen C, Liaw D, Podsypanina K, Bose S, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 20.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 21.Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 22.Okami K, Wu L, Riggins G, Cairns P, Goggins M, et al. Analysis of PTEN/MMAC1 alterations in aerodigestive tract tumors. Cancer Res. 1998;58:509–511. [PubMed] [Google Scholar]
- 23.Poetsch M, Lorenz G, Kleist B. Detection of new PTEN/MMAC1 mutations in head and neck squamous cell carcinomas with loss of chromosome 10. Cancer Genet Cytogenet. 2002;132:20–24. doi: 10.1016/s0165-4608(01)00509-x. [DOI] [PubMed] [Google Scholar]
- 24.Shao X, Tandon R, Samara G, Kanki H, Yano H, et al. Mutational analysis of the PTEN gene in head and neck squamous cell carcinoma. Int J Cancer. 1998;77:684–688. doi: 10.1002/(sici)1097-0215(19980831)77:5<684::aid-ijc4>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 25.Das S, Dixon JE, Cho W. Membrane-binding and activation mechanism of PTEN. Proc Natl Acad Sci U S A. 2003;100:7491–7496. doi: 10.1073/pnas.0932835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu F, Wagner S, Campbell RB, Nickerson JA, Schiffer CA, et al. PTEN enters the nucleus by diffusion. J Cell Biochem. 2005;96:221–234. doi: 10.1002/jcb.20525. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez-Escudero I, Oliver MD, Andres-Pons A, Molina M, Cid VJ, et al. A comprehensive functional analysis of PTEN mutations: implications in tumor- and autism-related syndromes. Hum Mol Genet. 2011;20:4132–4142. doi: 10.1093/hmg/ddr337. [DOI] [PubMed] [Google Scholar]
- 28.Georgescu MM, Kirsch KH, Kaloudis P, Yang H, Pavletich NP, et al. Stabilization and productive positioning roles of the C2 domain of PTEN tumor suppressor. Cancer Res. 2000;60:7033–7038. [PubMed] [Google Scholar]
- 29.Lin WM, Forgacs E, Warshal DP, Yeh IT, Martin JS, et al. Loss of heterozygosity and mutational analysis of the PTEN/MMAC1 gene in synchronous endometrial and ovarian carcinomas. Clin Cancer Res. 1998;4:2577–2583. [PubMed] [Google Scholar]
- 30.Forgacs E, Biesterveld EJ, Sekido Y, Fong K, Muneer S, et al. Mutation analysis of the PTEN/MMAC1 gene in lung cancer. Oncogene. 1998;17:1557–1565. doi: 10.1038/sj.onc.1202070. [DOI] [PubMed] [Google Scholar]
- 31.Kohno T, Takahashi M, Manda R, Yokota J. Inactivation of the PTEN/MMAC1/TEP1 gene in human lung cancers. Genes Chromosomes Cancer. 1998;22:152–156. doi: 10.1002/(sici)1098-2264(199806)22:2<152::aid-gcc10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 32.Zhou XP, Li YJ, Hoang-Xuan K, Laurent-Puig P, Mokhtari K, et al. Mutational analysis of the PTEN gene in gliomas: molecular and pathological correlations. Int J Cancer. 1999;84:150–154. doi: 10.1002/(sici)1097-0215(19990420)84:2<150::aid-ijc10>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 33.Gasparotto D, Vukosavljevic T, Piccinin S, Barzan L, Sulfaro S, et al. Loss of heterozygosity at 10q in tumors of the upper respiratory tract is associated with poor prognosis. Int J Cancer. 1999;84:432–436. doi: 10.1002/(sici)1097-0215(19990820)84:4<432::aid-ijc18>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 34.Squarize CH, Castilho RM, Abrahao AC, Molinolo A, Lingen MW, et al. PTEN Deficiency Contributes to the Development and Progression of Head and Neck Cancer. Neoplasia. 2013;15:461–471. doi: 10.1593/neo.121024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Squarize CH, Castilho RM, Gutkind JS. Chemoprevention and treatment of experimental Cowden's disease by mTOR inhibition with rapamycin. Cancer Res. 2008;68:7066–7072. doi: 10.1158/0008-5472.CAN-08-0922. [DOI] [PubMed] [Google Scholar]
- 36.Backman SA, Ghazarian D, So K, Sanchez O, Wagner KU, et al. Early onset of neoplasia in the prostate and skin of mice with tissue-specific deletion of Pten. Proc Natl Acad Sci U S A. 2004;101:1725–1730. doi: 10.1073/pnas.0308217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katso R, Okkenhaug K, Ahmadi K, White S, Timms J, et al. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2001;17:615–675. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- 38.Vanhaesebroeck B, Welham MJ, Kotani K, Stein R, Warne PH, et al. P110delta, a novel phosphoinositide 3-kinase in leukocytes. Proc Natl Acad Sci U S A. 1997;94:4330–4335. doi: 10.1073/pnas.94.9.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 40.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, et al. Regulation of the p85/p110 phosphatidylinositol 3'-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol. 1998;18:1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephens LR, Eguinoa A, Erdjument-Bromage H, Lui M, Cooke F, et al. The G beta gamma sensitivity of a PI3K is dependent upon a tightly associated adaptor, p101. Cell. 1997;89:105–114. doi: 10.1016/s0092-8674(00)80187-7. [DOI] [PubMed] [Google Scholar]
- 42.Wymann MP, Bjorklof K, Calvez R, Finan P, Thomast M, et al. Phosphoinositide 3-kinase gamma: a key modulator in inflammation and allergy. Biochem Soc Trans. 2003;31:275–280. doi: 10.1042/bst0310275. [DOI] [PubMed] [Google Scholar]
- 43.Krugmann S, Hawkins PT, Pryer N, Braselmann S. Characterizing the interactions between the two subunits of the p101/p110gamma phosphoinositide 3-kinase and their role in the activation of this enzyme by G beta gamma subunits. J Biol Chem. 1999;274:17152–17158. doi: 10.1074/jbc.274.24.17152. [DOI] [PubMed] [Google Scholar]
- 44.Fruman DA, Cantley LC. Phosphoinositide 3-kinase in immunological systems. Semin Immunol. 2002;14:7–18. doi: 10.1006/smim.2001.0337. [DOI] [PubMed] [Google Scholar]
- 45.Arcaro A, Volinia S, Zvelebil MJ, Stein R, Watton SJ, et al. Human phosphoinositide 3-kinase C2beta, the role of calcium and the C2 domain in enzyme activity. J Biol Chem. 1998;273:33082–33090. doi: 10.1074/jbc.273.49.33082. [DOI] [PubMed] [Google Scholar]
- 46.Domin J, Pages F, Volinia S, Rittenhouse SE, Zvelebil MJ, et al. Cloning of a human phosphoinositide 3-kinase with a C2 domain that displays reduced sensitivity to the inhibitor wortmannin. Biochem J. 1997;326(Pt 1):139–147. doi: 10.1042/bj3260139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacDougall LK, Domin J, Waterfield MD. A family of phosphoinositide 3-kinases in Drosophila identifies a new mediator of signal transduction. Curr Biol. 1995;5:1404–1415. doi: 10.1016/s0960-9822(95)00278-8. [DOI] [PubMed] [Google Scholar]
- 48.Ono F, Nakagawa T, Saito S, Owada Y, Sakagami H, et al. A novel class II phosphoinositide 3-kinase predominantly expressed in the liver and its enhanced expression during liver regeneration. J Biol Chem. 1998;273:7731–7736. doi: 10.1074/jbc.273.13.7731. [DOI] [PubMed] [Google Scholar]
- 49.Rozycka M, Lu YJ, Brown RA, Lau MR, Shipley JM, et al. cDNA cloning of a third human C2-domain-containing class II phosphoinositide 3-kinase, PI3K-C2gamma, and chromosomal assignment of this gene (PIK3C2G) to 12p12. Genomics. 1998;54:569–574. doi: 10.1006/geno.1998.5621. [DOI] [PubMed] [Google Scholar]
- 50.El Sheikh SS, Domin J, Tomtitchong P, Abel P, Stamp G, et al. Topographical expression of class IA and class II phosphoinositide 3-kinase enzymes in normal human tissues is consistent with a role in differentiation. BMC Clin Pathol. 2003;3:4. doi: 10.1186/1472-6890-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Volinia S, Dhand R, Vanhaesebroeck B, MacDougall LK, Stein R, et al. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 1995;14:3339–3348. doi: 10.1002/j.1460-2075.1995.tb07340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Odorizzi G, Babst M, Emr SD. Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem Sci. 2000;25:229–235. doi: 10.1016/s0968-0004(00)01543-7. [DOI] [PubMed] [Google Scholar]
- 53.Lindmo K, Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci. 2006;119:605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- 54.Wurmser AE, Gary JD, Emr SD. Phosphoinositide 3-kinases and their FYVE domain-containing effectors as regulators of vacuolar/lysosomal membrane trafficking pathways. J Biol Chem. 1999;274:9129–9132. doi: 10.1074/jbc.274.14.9129. [DOI] [PubMed] [Google Scholar]
- 55.Panaretou C, Domin J, Cockcroft S, Waterfield MD. Characterization of p150, an adaptor protein for the human phosphatidylinositol (PtdIns) 3-kinase. Substrate presentation by phosphatidylinositol transfer protein to the p150.Ptdins 3-kinase complex. J Biol Chem. 1997;272:2477–2485. doi: 10.1074/jbc.272.4.2477. [DOI] [PubMed] [Google Scholar]
- 56.Stack JH, Herman PK, Schu PV, Emr SD. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 1993;12:2195–2204. doi: 10.1002/j.1460-2075.1993.tb05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 58.Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 60.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marone R, Cmiljanovic V, Giese B, Wymann MP. Targeting phosphoinositide 3-kinase: moving towards therapy. Biochim Biophys Acta. 2008;1784:159–185. doi: 10.1016/j.bbapap.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 62.Bader AG, Kang S, Zhao L, Vogt PK. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer. 2005;5:921–929. doi: 10.1038/nrc1753. [DOI] [PubMed] [Google Scholar]
- 63.Samuels Y, Ericson K. Oncogenic PI3K and its role in cancer. Curr Opin Oncol. 2006;18:77–82. doi: 10.1097/01.cco.0000198021.99347.b9. [DOI] [PubMed] [Google Scholar]
- 64.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 65.Janku F, Tsimberidou AM, Garrido-Laguna I, Wang X, Luthra R, et al. PIK3CA mutations in patients with advanced cancers treated with PI3K/AKT/mTOR axis inhibitors. Mol Cancer Ther. 2011;10:558–565. doi: 10.1158/1535-7163.MCT-10-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Janku F, Lee JJ, Tsimberidou AM, Hong DS, Naing A, et al. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS One. 2011;6:e22769. doi: 10.1371/journal.pone.0022769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woenckhaus J, Steger K, Werner E, Fenic I, Gamerdinger U, et al. Genomic gain of PIK3CA and increased expression of p110alpha are associated with progression of dysplasia into invasive squamous cell carcinoma. J Pathol. 2002;198:335–342. doi: 10.1002/path.1207. [DOI] [PubMed] [Google Scholar]
- 68.Redon R, Muller D, Caulee K, Wanherdrick K, Abecassis J, et al. A simple specific pattern of chromosomal aberrations at early stages of head and neck squamous cell carcinomas: PIK3CA but not p63 gene as a likely target of 3q26-qter gains. Cancer Res. 2001;61:4122–4129. [PubMed] [Google Scholar]
- 69.Saal LH, Holm K, Maurer M, Memeo L, Su T, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 70.Byun DS, Cho K, Ryu BK, Lee MG, Park JI, et al. Frequent monoallelic deletion of PTEN and its reciprocal associatioin with PIK3CA amplification in gastric carcinoma. Int J Cancer. 2003;104:318–327. doi: 10.1002/ijc.10962. [DOI] [PubMed] [Google Scholar]
- 71.Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kozaki K, Imoto I, Pimkhaokham A, Hasegawa S, Tsuda H, et al. PIK3CA mutation is an oncogenic aberration at advanced stages of oral squamous cell carcinoma. Cancer Sci. 2006;97:1351–1358. doi: 10.1111/j.1349-7006.2006.00343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Murugan AK, Hong NT, Fukui Y, Munirajan AK, Tsuchida N. Oncogenic mutations of the PIK3CA gene in head and neck squamous cell carcinomas. Int J Oncol. 2008;32:101–111. [PubMed] [Google Scholar]
- 74.Fenic I, Steger K, Gruber C, Arens C, Woenckhaus J. Analysis of PIK3CA and Akt/protein kinase B in head and neck squamous cell carcinoma. Oncol Rep. 2007;18:253–259. [PubMed] [Google Scholar]
- 75.Estilo CL, P OC, Ngai I, Patel SG, Reddy PG, et al. The role of novel oncogenes squamous cell carcinoma-related oncogene and phosphatidylinositol 3-kinase p110alpha in squamous cell carcinoma of the oral tongue. Clin Cancer Res. 2003;9:2300–2306. [PubMed] [Google Scholar]
- 76.Renner O, Blanco-Aparicio C, Grassow M, Canamero M, Leal JF, et al. Activation of phosphatidylinositol 3-kinase by membrane localization of p110alpha predisposes mammary glands to neoplastic transformation. Cancer Res. 2008;68:9643–9653. doi: 10.1158/0008-5472.CAN-08-1539. [DOI] [PubMed] [Google Scholar]
- 77.Aleskandarany MA, Rakha EA, Ahmed MA, Powe DG, Paish EC, et al. PIK3CA expression in invasive breast cancer: a biomarker of poor prognosis. Breast Cancer Res Treat. 2010;122:45–53. doi: 10.1007/s10549-009-0508-9. [DOI] [PubMed] [Google Scholar]
- 78.Ogino S, Nosho K, Kirkner GJ, Shima K, Irahara N, et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol. 2009;27:1477–1484. doi: 10.1200/JCO.2008.18.6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Akagi I, Miyashita M, Makino H, Nomura T, Hagiwara N, et al. Overexpression of PIK3CA is associated with lymph node metastasis in esophageal squamous cell carcinoma. Int J Oncol. 2009;34:767–775. doi: 10.3892/ijo_00000202. [DOI] [PubMed] [Google Scholar]
- 80.Perez-Tenorio G, Alkhori L, Olsson B, Waltersson MA, Nordenskjold B, et al. PIK3CA mutations and PTEN loss correlate with similar prognostic factors and are not mutually exclusive in breast cancer. Clin Cancer Res. 2007;13:3577–3584. doi: 10.1158/1078-0432.CCR-06-1609. [DOI] [PubMed] [Google Scholar]
- 81.He Y, Van't Veer LJ, Mikolajewska-Hanclich I, van Velthuysen ML, Zeestraten EC, et al. PIK3CA mutations predict local recurrences in rectal cancer patients. Clin Cancer Res. 2009;15:6956–6962. doi: 10.1158/1078-0432.CCR-09-1165. [DOI] [PubMed] [Google Scholar]
- 82.Morris LG, Taylor BS, Bivona TG, Gong Y, Eng S, et al. Genomic dissection of the epidermal growth factor receptor (EGFR)/PI3K pathway reveals frequent deletion of the EGFR phosphatase PTPRS in head and neck cancers. Proc Natl Acad Sci U S A. 2011;108:19024–19029. doi: 10.1073/pnas.1111963108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A. 2005;102:802–807. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murugan AK, Munirajan AK, Tsuchida N. Genetic deregulation of the PIK3CA oncogene in oral cancer. Cancer Lett. 2013 doi: 10.1016/j.canlet.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 85.Ikenoue T, Kanai F, Hikiba Y, Obata T, Tanaka Y, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65:4562–4567. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- 86.Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, Cummins JM, Delong L, et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 87.Kuo KT, Mao TL, Jones S, Veras E, Ayhan A, et al. Frequent activating mutations of PIK3CA in ovarian clear cell carcinoma. Am J Pathol. 2009;174:1597–1601. doi: 10.2353/ajpath.2009.081000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 89.Pang H, Flinn R, Patsialou A, Wyckoff J, Roussos ET, et al. Differential enhancement of breast cancer cell motility and metastasis by helical and kinase domain mutations of class IA phosphoinositide 3-kinase. Cancer Res. 2009;69:8868–8876. doi: 10.1158/0008-5472.CAN-09-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Neshat MS, Mellinghoff IK, Tran C, Stiles B, Thomas G, et al. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc Natl Acad Sci U S A. 2001;98:10314–10319. doi: 10.1073/pnas.171076798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Podsypanina K, Lee RT, Politis C, Hennessy I, Crane A, et al. An inhibitor of mTOR reduces neoplasia and normalizes p70/S6 kinase activity in Pten+/− mice. Proc Natl Acad Sci U S A. 2001;98:10320–10325. doi: 10.1073/pnas.171060098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Janku F, Wheler JJ, Naing A, Falchook GS, Hong DS, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res. 2013;73:276–284. doi: 10.1158/0008-5472.CAN-12-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Di Nicolantonio F, Arena S, Tabernero J, Grosso S, Molinari F, et al. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. J Clin Invest. 2010;120:2858–2866. doi: 10.1172/JCI37539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wallin JJ, Guan J, Prior WW, Edgar KA, Kassees R, et al. Nuclear phospho-Akt increase predicts synergy of PI3K inhibition and doxorubicin in breast and ovarian cancer. Sci Transl Med. 2010;2:48ra66. doi: 10.1126/scitranslmed.3000630. [DOI] [PubMed] [Google Scholar]
- 95.Kendler KS. The genetic epidemiology of psychiatric disorders: a current perspective. Soc Psychiatry Psychiatr Epidemiol. 1997;32:5–11. doi: 10.1007/BF00800661. [DOI] [PubMed] [Google Scholar]
- 96.Caren H, Fransson S, Ejeskar K, Kogner P, Martinsson T. Genetic and epigenetic changes in the common 1p36 deletion in neuroblastoma tumours. Br J Cancer. 2007;97:1416–1424. doi: 10.1038/sj.bjc.6604032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jaiswal BS, Janakiraman V, Kljavin NM, Chaudhuri S, Stern HM, et al. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell. 2009;16:463–474. doi: 10.1016/j.ccr.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Berenjeno IM, Vanhaesebroeck B. PI3K regulatory subunits lose control in cancer. Cancer Cell. 2009;16:449–450. doi: 10.1016/j.ccr.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 99.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 100.Philp AJ, Campbell IG, Leet C, Vincan E, Rockman SP, et al. The phosphatidylinositol 3'-kinase p85alpha gene is an oncogene in human ovarian and colon tumors. Cancer Res. 2001;61:7426–7429. [PubMed] [Google Scholar]
- 101.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Taniguchi CM, Winnay J, Kondo T, Bronson RT, Guimaraes AR, et al. The phosphoinositide 3-kinase regulatory subunit p85alpha can exert tumor suppressor properties through negative regulation of growth factor signaling. Cancer Res. 2010;70:5305–5315. doi: 10.1158/0008-5472.CAN-09-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, et al. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 104.Kodaki T, Woscholski R, Hallberg B, Rodriguez-Viciana P, Downward J, et al. The activation of phosphatidylinositol 3-kinase by Ras. Curr Biol. 1994;4:798–806. doi: 10.1016/s0960-9822(00)00177-9. [DOI] [PubMed] [Google Scholar]
- 105.Chagpar RB, Links PH, Pastor MC, Furber LA, Hawrysh AD, et al. Direct positive regulation of PTEN by the p85 subunit of phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2010;107:5471–5476. doi: 10.1073/pnas.0908899107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cheung LW, Hennessy BT, Li J, Yu S, Myers AP, et al. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov. 2011;1:170–185. doi: 10.1158/2159-8290.CD-11-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sasaki T, Irie-Sasaki J, Horie Y, Bachmaier K, Fata JE, et al. Colorectal carcinomas in mice lacking the catalytic subunit of PI(3)Kgamma. Nature. 2000;406:897–902. doi: 10.1038/35022585. [DOI] [PubMed] [Google Scholar]
- 108.Hirsch E, Katanaev VL, Garlanda C, Azzolino O, Pirola L, et al. Central role for G protein-coupled phosphoinositide 3-kinase gamma in inflammation. Science. 2000;287:1049–1053. doi: 10.1126/science.287.5455.1049. [DOI] [PubMed] [Google Scholar]
- 109.Li Z, Jiang H, Xie W, Zhang Z, Smrcka AV, et al. Roles of PLC-beta2 and -beta3 and PI3Kgamma in chemoattractant-mediated signal transduction. Science. 2000;287:1046–1049. doi: 10.1126/science.287.5455.1046. [DOI] [PubMed] [Google Scholar]
- 110.Sasaki T, Irie-Sasaki J, Jones RG, Oliveira-dos-Santos AJ, Stanford WL, et al. Function of PI3Kgamma in thymocyte development, T cell activation, and neutrophil migration. Science. 2000;287:1040–1046. doi: 10.1126/science.287.5455.1040. [DOI] [PubMed] [Google Scholar]
- 111.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Koutros S, Schumacher FR, Hayes RB, Ma J, Huang WY, et al. Pooled analysis of phosphatidylinositol 3-kinase pathway variants and risk of prostate cancer. Cancer Res. 2010;70:2389–2396. doi: 10.1158/0008-5472.CAN-09-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Diouf B, Cheng Q, Krynetskaia NF, Yang W, Cheok M, et al. Somatic deletions of genes regulating MSH2 protein stability cause DNA mismatch repair deficiency and drug resistance in human leukemia cells. Nat Med. 2011;17:1298–1303. doi: 10.1038/nm.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shisheva A. PIKfyve: Partners, significance, debates and paradoxes. Cell Biol Int. 2008;32:591–604. doi: 10.1016/j.cellbi.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ikonomov OC, Sbrissa D, Fenner H, Shisheva A. PIKfyve-ArPIKfyve-Sac3 core complex: contact sites and their consequence for Sac3 phosphatase activity and endocytic membrane homeostasis. J Biol Chem. 2009;284:35794–35806. doi: 10.1074/jbc.M109.037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Iamaroon A, Krisanaprakornkit S. Overexpression and activation of Akt2 protein in oral squamous cell carcinoma. Oral Oncol. 2009;45:e175–e179. doi: 10.1016/j.oraloncology.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 117.Jones PF, Jakubowicz T, Hemmings BA. Molecular cloning of a second form of rac protein kinase. Cell Regul. 1991;2:1001–1009. doi: 10.1091/mbc.2.12.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brazil DP, Yang ZZ, Hemmings BA. Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem Sci. 2004;29:233–242. doi: 10.1016/j.tibs.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 119.Hers I, Vincent EE, Tavare JM. Akt signalling in health and disease. Cell Signal. 2011;23:1515–1527. doi: 10.1016/j.cellsig.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 120.Tsurutani J, Fukuoka J, Tsurutani H, Shih JH, Hewitt SM, et al. Evaluation of two phosphorylation sites improves the prognostic significance of Akt activation in non-small-cell lung cancer tumors. J Clin Oncol. 2006;24:306–314. doi: 10.1200/JCO.2005.02.4133. [DOI] [PubMed] [Google Scholar]
- 121.Yang SX, Costantino JP, Kim C, Mamounas EP, Nguyen D, et al. Akt phosphorylation at Ser473 predicts benefit of paclitaxel chemotherapy in node-positive breast cancer. J Clin Oncol. 2010;28:2974–2981. doi: 10.1200/JCO.2009.26.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Molinolo AA, Hewitt SM, Amornphimoltham P, Keelawat S, Rangdaeng S, et al. Dissecting the Akt/mammalian target of rapamycin signaling network: emerging results from the head and neck cancer tissue array initiative. Clin Cancer Res. 2007;13:4964–4973. doi: 10.1158/1078-0432.CCR-07-1041. [DOI] [PubMed] [Google Scholar]
- 123.Massarelli E, Liu DD, Lee JJ, El-Naggar AK, Lo Muzio L, et al. Akt activation correlates with adverse outcome in tongue cancer. Cancer. 2005;104:2430–2436. doi: 10.1002/cncr.21476. [DOI] [PubMed] [Google Scholar]
- 124.Yu Z, Weinberger PM, Sasaki C, Egleston BL, Speier WFt. Phosphorylation of Akt (Ser473) predicts poor clinical outcome in oropharyngeal squamous cell cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:553–558. doi: 10.1158/1055-9965.EPI-06-0121. [DOI] [PubMed] [Google Scholar]
- 125.Dillon RL, Muller WJ. Distinct biological roles for the akt family in mammary tumor progression. Cancer Res. 2010;70:4260–4264. doi: 10.1158/0008-5472.CAN-10-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Moral M, Segrelles C, Lara MF, Martinez-Cruz AB, Lorz C, et al. Akt activation synergizes with Trp53 loss in oral epithelium to produce a novel mouse model for head and neck squamous cell carcinoma. Cancer Res. 2009;69:1099–1108. doi: 10.1158/0008-5472.CAN-08-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cheng GZ, Chan J, Wang Q, Zhang W, Sun CD, et al. Twist transcriptionally up-regulates AKT2 in breast cancer cells leading to increased migration, invasion, and resistance to paclitaxel. Cancer Res. 2007;67:1979–1987. doi: 10.1158/0008-5472.CAN-06-1479. [DOI] [PubMed] [Google Scholar]
- 128.Qiao J, Lee S, Paul P, Qiao L, Taylor CJ, et al. Akt2 regulates metastatic potential in neuroblastoma. PLoS One. 2013;8:e56382. doi: 10.1371/journal.pone.0056382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rychahou PG, Kang J, Gulhati P, Doan HQ, Chen LA, et al. Akt2 overexpression plays a critical role in the establishment of colorectal cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:20315–20320. doi: 10.1073/pnas.0810715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dillon RL, Marcotte R, Hennessy BT, Woodgett JR, Mills GB, et al. Akt1 and akt2 play distinct roles in the initiation and metastatic phases of mammary tumor progression. Cancer Res. 2009;69:5057–5064. doi: 10.1158/0008-5472.CAN-08-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hutchinson JN, Jin J, Cardiff RD, Woodgett JR, Muller WJ. Activation of Akt-1 (PKB-alpha) can accelerate ErbB-2-mediated mammary tumorigenesis but suppresses tumor invasion. Cancer Res. 2004;64:3171–3178. doi: 10.1158/0008-5472.can-03-3465. [DOI] [PubMed] [Google Scholar]
- 132.Cho H, Mu J, Kim JK, Thorvaldsen JL, Chu Q, et al. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta) Science. 2001;292:1728–1731. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- 133.DeFeo-Jones D, Barnett SF, Fu S, Hancock PJ, Haskell KM, et al. Tumor cell sensitization to apoptotic stimuli by selective inhibition of specific Akt/PKB family members. Mol Cancer Ther. 2005;4:271–279. [PubMed] [Google Scholar]
- 134.Fujio Y, Mitsuuchi Y, Testa JR, Walsh K. Activation of Akt2 Inhibits anoikis and apoptosis induced by myogenic differentiation. Cell Death Differ. 2001;8:1207–1212. doi: 10.1038/sj.cdd.4400919. [DOI] [PubMed] [Google Scholar]
- 135.Yuan ZQ, Sun M, Feldman RI, Wang G, Ma X, et al. Frequent activation of AKT2 and induction of apoptosis by inhibition of phosphoinositide-3-OH kinase/Akt pathway in human ovarian cancer. Oncogene. 2000;19:2324–2330. doi: 10.1038/sj.onc.1203598. [DOI] [PubMed] [Google Scholar]
- 136.van Slegtenhorst M, Nellist M, Nagelkerken B, Cheadle J, Snell R, et al. Interaction between hamartin and tuberin, the TSC1 and TSC2 gene products. Hum Mol Genet. 1998;7:1053–1057. doi: 10.1093/hmg/7.6.1053. [DOI] [PubMed] [Google Scholar]
- 137.Plank TL, Yeung RS, Henske EP. Hamartin, the product of the tuberous sclerosis 1 (TSC1) gene, interacts with tuberin and appears to be localized to cytoplasmic vesicles. Cancer Res. 1998;58:4766–4770. [PubMed] [Google Scholar]
- 138.Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, et al. Tuberous sclerosis complex-1 and-2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci U S A. 2002;99:13571–13576. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pelletier CL, Maggi LB, Jr, Brady SN, Scheidenhelm DK, Gutmann DH, et al. TSC1 sets the rate of ribosome export and protein synthesis through nucleophosmin translation. Cancer Res. 2007;67:1609–1617. doi: 10.1158/0008-5472.CAN-06-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, et al. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 141.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 142.Lee DF, Kuo HP, Chen CT, Hsu JM, Chou CK, et al. IKK beta suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell. 2007;130:440–455. doi: 10.1016/j.cell.2007.05.058. [DOI] [PubMed] [Google Scholar]
- 143.Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 144.Kandt RS, Haines JL, Smith M, Northrup H, Gardner RJ, et al. Linkage of an important gene locus for tuberous sclerosis to a chromosome 16 marker for polycystic kidney disease. Nat Genet. 1992;2:37–41. doi: 10.1038/ng0992-37. [DOI] [PubMed] [Google Scholar]
- 145.European Chromosome 16 Tuberous Sclerosis C. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell. 1993;75:1305–1315. doi: 10.1016/0092-8674(93)90618-z. [DOI] [PubMed] [Google Scholar]
- 146.van Slegtenhorst M, de Hoogt R, Hermans C, Nellist M, Janssen B, et al. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science. 1997;277:805–808. doi: 10.1126/science.277.5327.805. [DOI] [PubMed] [Google Scholar]
- 147.Chakraborty S, Mohiyuddin SM, Gopinath KS, Kumar A. Involvement of TSC genes and differential expression of other members of the mTOR signaling pathway in oral squamous cell carcinoma. BMC Cancer. 2008;8:163. doi: 10.1186/1471-2407-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355:1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- 149.Kawaguchi S, Harada K, Supriatno, Yoshida H, Sato M. Overexpression of tuberous sclerosis complex 2 exerts antitumor effect on oral cancer cell lines. Oral Oncol. 2003;39:836–841. doi: 10.1016/s1368-8375(03)00106-4. [DOI] [PubMed] [Google Scholar]
- 150.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 153.Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 154.Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem J. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- 155.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 156.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 157.Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci Signal. 2009;2:pe24. doi: 10.1126/scisignal.267pe24. [DOI] [PubMed] [Google Scholar]
- 158.Chou CC, Chou MJ, Tzen CY. PIK3CA mutation occurs in nasopharyngeal carcinoma but does not significantly influence the disease-specific survival. Med Oncol. 2009;26:322–326. doi: 10.1007/s12032-008-9124-5. [DOI] [PubMed] [Google Scholar]
- 159.Qiu W, Schonleben F, Li X, Ho DJ, Close LG, et al. PIK3CA mutations in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12:1441–1446. doi: 10.1158/1078-0432.CCR-05-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Qiu W, Tong GX, Manolidis S, Close LG, Assaad AM, et al. Novel mutant-enriched sequencing identified high frequency of PIK3CA mutations in pharyngeal cancer. Int J Cancer. 2008;122:1189–1194. doi: 10.1002/ijc.23217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Suda T, Hama T, Kondo S, Yuza Y, Yoshikawa M, et al. Copy number amplification of the PIK3CA gene is associated with poor prognosis in non-lymph node metastatic head and neck squamous cell carcinoma. BMC Cancer. 2012;12:416. doi: 10.1186/1471-2407-12-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Morris LG, Taylor BS, Bivona TG, Gong Y, Eng S, et al. Genomic dissection of the epidermal growth factor receptor (EGFR)/PI3K pathway reveals frequent deletion of the EGFR phosphatase PTPRS in head and neck cancers. Proc Natl Acad Sci U S A. 2011;108:19024–19029. doi: 10.1073/pnas.1111963108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cohen Y, Goldenberg-Cohen N, Shalmon B, Shani T, Oren S, et al. Mutational analysis of PTEN/PIK3CA/AKT pathway in oral squamous cell carcinoma. Oral Oncol. 2011;47:946–950. doi: 10.1016/j.oraloncology.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 164.Bruckman KC, Schonleben F, Qiu W, Woo VL, Su GH. Mutational analyses of the BRAF, KRAS, and PIK3CA genes in oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2010;110:632–637. doi: 10.1016/j.tripleo.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fendri A, Khabir A, Mnejja W, Sellami-Boudawara T, Daoud J, et al. PIK3CA amplification is predictive of poor prognosis in Tunisian patients with nasopharyngeal carcinoma. Cancer Sci. 2009;100:2034–2039. doi: 10.1111/j.1349-7006.2009.01292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nichols AC, Yoo J, Palma DA, Fung K, Franklin JH, et al. Frequent mutations in TP53 and CDKN2A found by next-generation sequencing of head and neck cancer cell lines. Arch Otolaryngol Head Neck Surg. 2012;138:732–739. doi: 10.1001/archoto.2012.1558. [DOI] [PubMed] [Google Scholar]
- 167.Su Z, Dias-Santagata D, Duke M, Hutchinson K, Lin YL, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13:74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yarbrough WG, Whigham A, Brown B, Roach M, Slebos R. Phosphoinositide kinase-3 status associated with presence or absence of human papillomavirus in head and neck squamous cell carcinomas. Int J Radiat Oncol Biol Phys. 2007;69:S98–S101. doi: 10.1016/j.ijrobp.2007.04.082. [DOI] [PubMed] [Google Scholar]
- 169.vanHaaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ching-Shian Leong V, Jabal MF, Leong PP, Abdullah MA, Gul YA, et al. PIK3CA gene mutations in breast carcinoma in Malaysian patients. Cancer Genet Cytogenet. 2008;187:74–79. doi: 10.1016/j.cancergencyto.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 171.Or YY, Hui AB, To KF, Lam CN, Lo KW. PIK3CA mutations in nasopharyngeal carcinoma. Int J Cancer. 2006;118:1065–1067. doi: 10.1002/ijc.21444. [DOI] [PubMed] [Google Scholar]