Abstract
Given the importance of deregulated phosphoinositide (PI) signaling in leukemic hematopoiesis, genes coding for proteins that regulate PI metabolism may have significant and as yet unappreciated roles in leukemia. We performed a targeted knockdown screen of PI modulator genes in human AML cells and identified candidates required to sustain proliferation or prevent apoptosis. One of these, the lipid kinase phosphatidylinositol-5-phosphate 4-kinase, type II, alpha (PIP4K2A) regulates cellular levels of phosphatidylinositol-5-phosphate (PtsIns5P) and phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2). We found PIP4K2A to be essential for the clonogenic and leukemia-initiating potential of human AML cells, and for the clonogenic potential of murine MLL-AF9 AML cells. Importantly, PIP4K2A is also required for the clonogenic potential of primary human AML cells. Its knockdown results in accumulation of the cyclin-dependent kinase inhibitors CDKN1A and CDKN1B, G1 cell cycle arrest and apoptosis. Both CDKN1A accumulation and apoptosis were partially dependent upon activation of the mTOR pathway. Critically, however, PIP4K2A knockdown in normal hematopoietic stem and progenitor cells, both murine and human, did not adversely impact either clonogenic or multilineage differentiation potential, indicating a selective dependency which we suggest may be the consequence of the regulation of different transcriptional programmes in normal versus malignant cells. Thus, PIP4K2A is a novel candidate therapeutic target in myeloid malignancy.
Keywords: leukemia, MLL, AML, phosphoinositide, PIP4K2A, knockdown screen
INTRODUCTION
Phosphatidylinositol (PtdIns) is a membrane phospholipid which may be phosphorylated at the 3, 4 and/or 5 positions of its inositol ring to generate seven phosphoinositide (PI) species with distinct and important cellular functions.1 PIs interact with specific protein modules such as PH, FYVE, PX, PHD or ENTH domains in both nuclear or cytoplasmic proteins leading to functional consequences such as protein relocalization, activation or conformational change.1-3 A key example is PtdIns(3,4,5)P3 which is generated through phosphorylation of PtdIns(4,5)P2 by phosphoinositide-3-kinase (PI3K). PtdIns(3,4,5)P3 activates signaling cascades through recruitment to the membrane of the serine threonine kinase AKT and other proteins, resulting in enhanced proliferation, transcription, translation and cell survival.1,4,5 Other examples include PtdIns4P which regulates the trans-Golgi network; PtdIns3P which regulates endosomal trafficking and autophagy; PtdIns(3,5)P2 which regulates endosome structure and PtdIns(4,5)P2 which regulates actin filament elongation, interaction of the plasma membrane with the cytoskeleton, phagocytosis, clathrin-mediated endocytosis and exocytosis.1 PtdIns(4,5)P2 is also important because it is hydrolysed by phospholipase C to generate diacylglycerol (DAG), which activates protein kinase C, and inositol 1,4,5-triphosphate (Ins(1,4,5)P3), which triggers calcium release from the endoplasmic reticulum.1,6
Enzymes which regulate the conversion of PIs from one species to another are important in both normal and leukemic haematopoiesis. For example, PI3K isoforms are required for normal B-cell development, erythropoiesis and platelet function, and the PtdIns(3,4,5)P3 phosphatases PTEN and INPP5D are required to maintain long-term hematopoietic stem cells (HSC).5,6 Constitutive activation of PI3K signaling is associated with hematologic malignancy. In addition to loss of long term HSCs, deletion of Pten or Inpp5d results in development of a myeloproliferative disorder (MPD) which, in the former case, evolves to acute myeloid leukemia (AML).7-9 Thus strict control of PtdIns(3,4,5)P3 levels in HSC is essential to prevent their conversion into leukemic stem cells (LSC). In human AML constitutive activation of PI3K signaling is observed, at least by comparison with normal human CD34+ hematopoietic stem and progenitor cells (HSPC), because AKT is typically phosphorylated in 80-90% of patient samples, versus none for CD34+ HSPC.10-13 PI3K is assumed to be activated downstream of signaling proteins such as FLT3, KIT and RAS, all of which are recurrently mutated in AML, or by activation of autocrine signaling pathways.4,5 Somewhat in contrast to these findings, FOXO transcription factors exhibit nuclear rather than cytoplasmic localization due to low level AKT signaling in certain murine models of AML, and in some primary AML samples, with nuclear FOXO contributing to the myeloid differentiation block.14
In view of the importance of PtdIns(3,4,5)P3 signaling in hematologic malignancies, genes coding for proteins that regulate the interconversion of PI species may have significant and as yet unappreciated roles in leukemia. Given the enzymatic activities of many of these, some may represent novel therapeutic targets. To identify PI modulators with candidate roles in leukemia we performed a lentivirally delivered short hairpin RNA (shRNA) knockdown (KD) screen in human AML cells. Genes targeted by the library included those coding for PI kinases, PI phosphatases, phospholipases, inositol kinases and phosphotransfer proteins. We focused our validation studies on one of the identified candidates, the gene coding for the lipid kinase PIP4K2A. PIP4K2A has functional roles in breast and colorectal cancer cells,15,16 and we found it to be selectively required for the proliferation and survival of AML cells, including primary human cells, but not normal HSPC.
RESULTS
Lentiviral KD screen
The 339 pLKO-puro shRNA lentiviral vectors in the library together targeted 103 genes with putative roles in PI metabolism for KD (Supplementary Table 1). The screening approach is outlined in Supplementary Figure 1a. The readout was fold change in cellular biomass over three days for cells infected with each of the KD vectors by comparison with cells infected with non-targeting control lentiviruses. For the first screen we used human THP1 AML cells which exhibit a t(9;11) translocation, the cytogenetic hallmark of MLL-AF9 (observed in 3-5% of patients with AML).17 Optimization experiments demonstrated typical lentiviral transduction efficiencies for THP1 cells of 90-100% (data not shown). The majority of shRNA constructs had little impact (Supplementary Figure 1b). However, 18.8% of constructs reduced cellular biomass to less than 50% of the control value and 5.9% of constructs increased it to more than 150% of the control value. This indicated that the approach was able to identify KD constructs with either an adverse or a positive impact on leukemia cell growth. To confirm that the screening method was robust and reproducible we performed a second screen of THP1 AML cells using a separately manufactured batch of lentiviral supernatant. The results from this screen were highly correlated with those from the first screen (Supplementary Figure 1c). Next, we performed two additional full screens using cell lines representative of other molecular subtypes of AML: Kasumi1 (carrying a t(8;21) translocation, the cytogenetic hallmark of a RUNX1-RUNX1T1 fusion) and U937 (carrying a t(10;11), the hallmark of a PICALM-MLLT10 fusion). The distribution of results from the Kasumi1 screen was similar to that observed in the THP1 cell screens (Supplementary Figure 2). By contrast, the distribution of results from the U937 screen was more broad, with 18.8% of KD constructs promoting growth relative to control cells (Supplementary Figure 2). This may indicate a differential dependency of this line on PI signaling or potentially a growth inhibitory effect of the control vector not seen in other lines.
To identify candidate PI modulator genes that promote AML cell proliferation or survival, we analyzed the identities of the 70 constructs from each screen which reduced cellular expansion relative to control cells by the greatest amount, looking for instances where two or more constructs targeted the same gene. The genes identified from each individual screen are shown in Table 1, as are the 10 genes scored as candidate regulators of AML proliferation or survival in multiple screens. These included genes coding for PI-3-kinase catalytic or regulatory subunits (PIK3C2A, PIK3R3 and PIK3R6), members of the inositol polyphosphate 5-phosphatase family (INPP5B, INPP5J and SYNJ2), phospholipase C β2, a gene coding for an inositol monophosphatase domain containing protein (IMPAD1), a gene coding for a PI transfer domain containing protein (PITPNM2) and the PI phosphate kinase PIP4K2A. Some of these have previously been reported to have functional roles in cancer, affirming the screening approach. For example, (i) PIK3R3 is amplified by copy number gain and its expression is up regulated in ovarian cancer, and its KD induces apoptosis of ovarian cancer cell lines;18 (ii) PLCB2 is highly expressed in breast cancer and promotes mitosis and migration of breast cancer cells;19,20 and (iii) expression of PIP4K2A has been linked to metastasis in breast cancer and is required for the proliferation or survival of KRAS mutated colorectal cancer cells, and tumor growth in TP53 null mice.15,16,21 Most of the identified genes, however, have not previously been demonstrated to have any role in either normal or leukemic hematopoiesis and, consistent with the rationale for the screening approach, some code for potential targets for small molecule inhibitors (e.g. PIK3C2A, INPP5B, PIP4K2A).
Table 1.
Candidate positive regulators of AML cell survival or proliferation identified by the screening strategy.
| Cell line | Candidate positive regulators of AML cell proliferation or survival |
|---|---|
|
| |
| THP1 a (mean of two screens) | AKT2, IMPAD1, INPP5B, INPP5J, PIK3R3 (4), PIK3R6 (3), PIP4K2A (4), PITPNM2, PLCG2, SYNJ2 |
|
| |
| Kasumi1 a | IMPAD1 (3), INPP5B, IP6K3, ITPKA, MTMR3 (3), PIK3C2A, PIK3R5, PIK3R6, PIP4K2A, PITPNM1, PITPNM2, PLCB2, PLCH1, SBF2, SYNJ2, TPTE |
|
| |
| U937 a | INPP5B (4), INPP5J, MTMR2 (3), MTMR14, PI4K2B (3), PIK3C2A, PIK3R3 (3), PIK3R6 (3), PIP4K2A (4), PIP5K1C, PLCB2, PRKAA1, PTPMT1, RPTOR |
|
| |
| Genes scoring as candidate regulators in more than one AML cell screen b | IMPAD1, INPP5B (3), INPP5J, PIK3C2A, PIK3R3, PIK3R6 (3), PIP4K2A (3), PITPNM2, PLCB2, SYNJ2 |
Numbers in brackets indicate the number of constructs, where greater than two, targeting the same gene among the 70 constructs from each screen which reduced cellular expansion relative to control cells by the greatest amount.
Numbers in brackets indicate the number of screens, where greater than two, where the gene was scored as a candidate positive regulator of AML cell proliferation.
To identify PI modulator genes that restrain AML cell growth, we analyzed the identities of the constructs from each screen which increased cellular expansion relative to control cells by equal to or greater than 30%, looking for instances where two or more constructs targeted the same gene. Six genes were identified by two or more screens (Table 2). These included the AMP-activated protein kinase catalytic subunit genes PRKAA1 and PRKAA2, CDP-diacylglycerol-inositol 3-phoshatidyltransferase (CDIPT), transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 (TPTE2), inositol-trisphosphate 3-kinase B (ITPKB) and a gene coding for an actin filament crosslinking protein (MARCKS). As for the genes identified in the screens as candidate positive regulators of AML cell proliferation (Table 1), these six genes have likewise not previously been demonstrated to have a role in normal or leukemic hematopoiesis. Consistent with potential roles as tumor suppressors, both PRKAA1 and PRKAA2 are recurrently mutated in cancer cell lines, including those derived from hematopoietic tissue.22 Likewise TPTE2, a close homolog of the tumor suppressor PTEN, and ITPKB are recurrently mutated in cancer (for example, in 5% and 2.5% respectively of lung cancers).23 Together the genes identified by our screening strategy (Tables 1 and 2) represent a significant resource for future more detailed investigations of positive and negative regulators of AML cell proliferation and/or survival.
Table 2.
Candidate negative regulators of AML cell survival or proliferation identified by the screening strategy.
| Candidate negative regulators of AML cell proliferation or survival | |
|---|---|
|
| |
| Genes scoring as candidate regulators in more than one AML cell screen a b | CDIPT, ITPKB, MARCKS, PRKAA1, PRKAA2, TPTE2 (3) |
Numbers in brackets indicate the number of screens, where greater than two, where the gene was scored as a candidate negative regulator of AML cell proliferation.
Data from THP1 cells is the mean of the two screens.
The clonogenic and leukemia-initiating potential of THP1 AML cells depends on PIP4K2A
Illustrative of this, we focused our further investigations on phosphatidylinositol-5-phosphate 4-kinase, type II, alpha (PIP4K2A) for four reasons. First, it is highly expressed in hematopoietic cell lines relative to cell lines representative of other malignancies (Supplementary Figure 3a).22 Second, it is expressed at significantly higher levels in AML with a poor cytogenetic risk profile by comparison with both intermediate risk AML and good risk AML (Supplementary Figure 3b).24,25 Third, its expression correlates with resistance to chemotherapy in pediatric acute lymphoblastic leukemia.26 Finally, as a lipid kinase, it is potentially a druggable target. PIP4K2A phosphorylates PtdIns5P on the fourth hydroxyl of the inositol ring to generate PtdIns(4,5)P2 which acts as a substrate for PI3K or phospholipase C to generate the essential second messengers PtdIns(3,4,5)P3 and Ins(1,4,5)P3 respectively.27 While PIP4K2A is known to be expressed abundantly in erythrocytes28 and megakaryocytes,29 and to regulate megakaryocyte size during terminal differentiation,29 to date there are no data as to any functional role for PIP4K2A in malignant hematopoiesis.
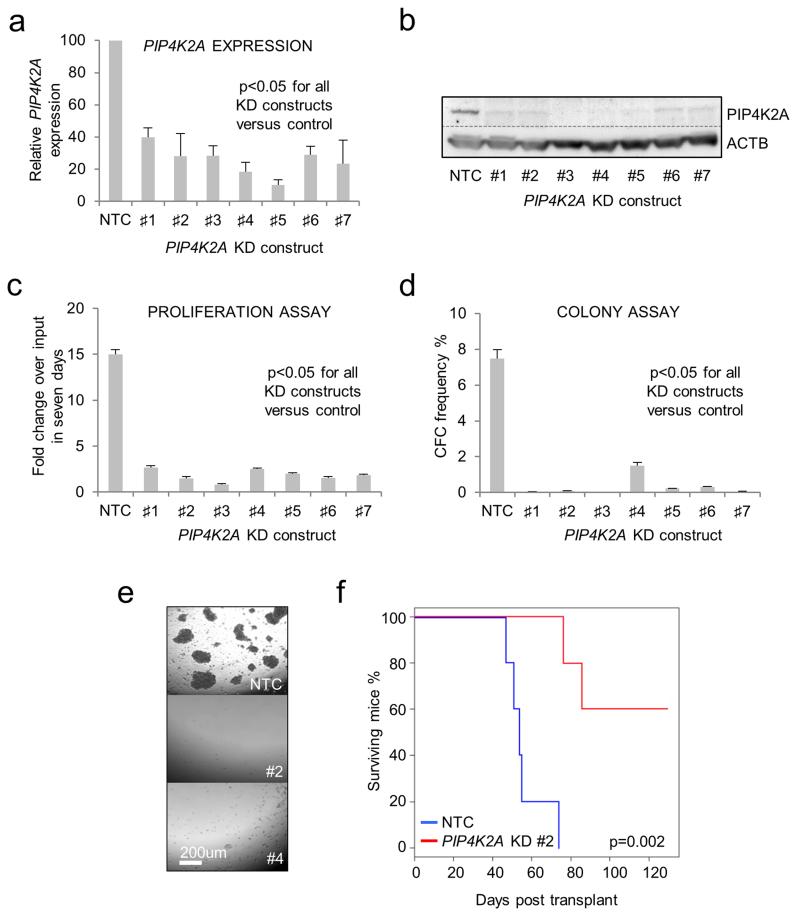
To confirm and extend our observations, we performed proliferation and colony forming assays with control or PIP4K2A KD human THP1 AML cells using four KD vectors from the original screen (KD#1-4) and three additional vectors (KD#5-7) (to provide further evidence that the observed phenotype was specific to PIP4K2A) (Figures 1a, b). In keeping with a role for PIP4K2A in proliferation or survival in multiple AML cell lines (Supplementary Figure 4 and Supplementary Table 1), all KD constructs significantly inhibited THP1 cell expansion in liquid culture (Figure 1c) and virtually abolished clonogenic potential in semisolid culture (Figures 1d, e). Likewise, while xenotransplantation of control THP1 AML cells induced short latency leukemia, with animals exhibiting a characteristic syndrome of pale bone marrow (BM), hind limb paralysis and, occasionally, chloromas of liver and kidney, animals transplanted with PIP4K2A KD THP1 cells did not develop disease (Figure 1f). While two mice in the KD cohort did die before the termination of the experiment, no evidence of leukemic engraftment was detected at autopsy and death was not due to AML.
Figure 1.
Loss of clonogenic and leukemia-initiating potential in PIP4K2A KD human THP1 AML cells. Human THP1 AML cells were infected with lentiviral vectors targeting PIP4K2A for KD, or a non-targeting control (NTC), with puromycin as the selectable marker. Cells were treated with puromycin for 48 hours to kill untransduced cells. Typical transduction efficiencies were 90-100%. (a) Bar chart shows mean±SEM expression of PIP4K2A relative to control cells, as determined by quantitative PCR, 72 hours following lentiviral infection and initiation of KD (n=4). (b) Representative western blot shows expression of PIP4K2A and ACTB 72 hours following initiation of KD. Bar charts show mean±SEM (c) fold expansion in cell number over input, as determined by hemocytometer counting, in a seven day liquid culture proliferation assay (n=3), and (d) colony forming cell (CFC) frequencies (n=3), of control and PIP4K2A KD cells. Assays were initiated 72 hours following lentiviral infection with cell viability confirmed by trypan blue dye exclusion. (e) Representative image shows THP1 AML cell colonies enumerated 10 days following assay initiation. (f) Survival curve of mice xenotransplanted with 104 control or PIP4K2A KD AML cells. Viability of transplanted cells was confirmed by Trypan Blue dye exclusion. For (a, c & d) significance was assessed by one way ANOVA followed by Fisher’s least significant difference post hoc test; for (f) by log rank test.
PIP4K2A KD induces increased levels of intracellular PtdIns5P
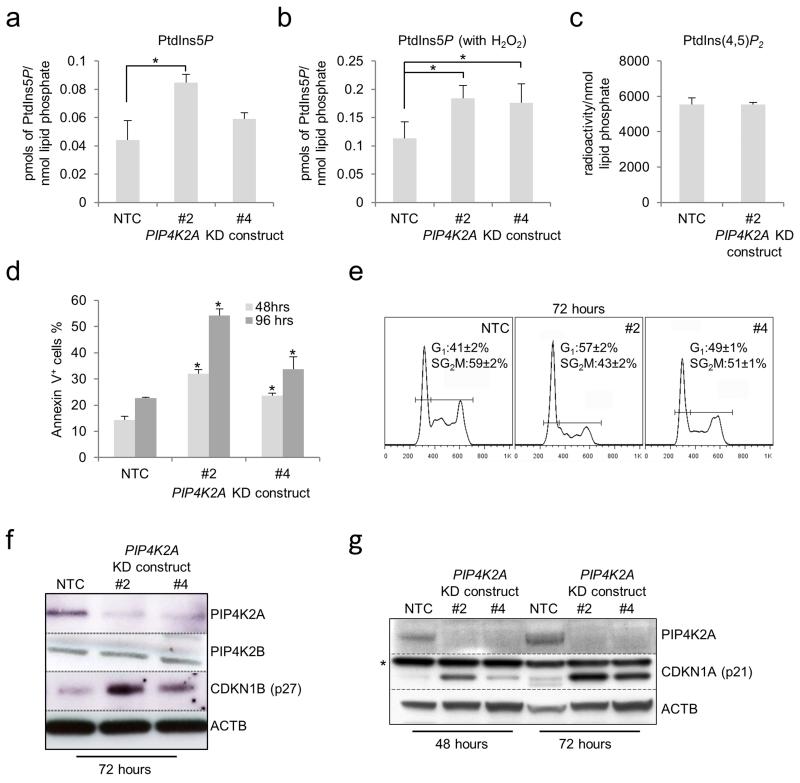
Following PIP4K2A KD in THP1 AML cells there was a significant increase in whole cell levels of PtdIns5P (Figure 2a, b), but not PtdIns(4,5)P2 (Figure 2c), with the increase in the former being accentuated by concomitant exposure of cells to oxidative stress (which promotes cellular production of PtdIns5P) through addition of H2O2 (Figure 2b).30 The changes in lipid levels were associated with induction of apoptosis and G1 cell cycle arrest (Figures 2d, e). In keeping with these phenotypic observations, KD (which did not affect PIP4K2B expression (Figure 2f)) was associated with induction of the cell cycle inhibitors CDKN1A and CDKN1B (Figure 2f, g). Therefore, PIP4K2A regulates levels of PtdIns(5)P in AML cells, as well as cell cycle progression and prevention of apoptosis.
Figure 2.
Consequences of PIP4K2A KD in human THP1 AML cells. Human THP1 AML cells were infected with lentiviral vectors targeting PIP4K2A for KD, or a non-targeting control (NTC), with eGFP as the selectable marker. Bar charts show mean±STDEV (a) whole cell PtdIns5P levels, (b) whole cell PtdIns5P levels following 40 minutes of treatment of cells with 1mM H2O2 and (c) whole cell PtdIns(4,5)P2 levels in THP1 cells 48 hours following lentiviral infection with the indicated control or KD constructs (n=3). * indicates p<0.05, as assessed by one way ANOVA followed by Fisher’s least significant difference post hoc test. (d) Bar chart shows mean±SEM percentage of control or PIP4K2A KD cells exhibiting annexin V binding at the indicated time points following lentiviral infection (n=3). In each case >95% of cells were eGFP positive. * indicates p<0.05 by comparison with control cells at the same time point, as determined by one way ANOVA followed by Fisher’s least significant difference post hoc test. (e) Representative cell cycle profiles (excluding sub-G1 apoptotic cells) from control and PIP4K2A KD cells 72 hours following initiation of KD. Numbers indicate mean±SEM percentage of cells in each phase of the cell cycle from three separate experiments (p<0.05 for NTC versus #2 or #4, as determined by one way ANOVA followed by Fisher’s least significant difference post hoc test). (f & g) Representative western blots show expression of the indicated proteins in control and PIP4K2A KD cells 48 or 72 hours following initiation of KD. * indicates a non-specific band.
Increased MTOR protein and activity upon PIP4K2A KD
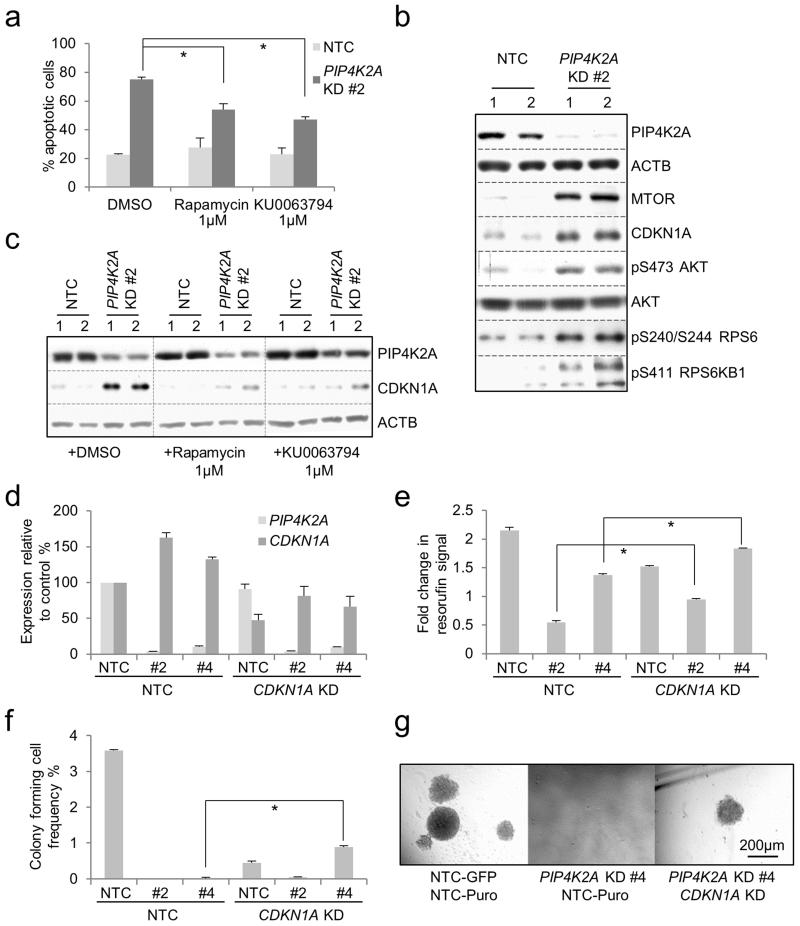
To identify cellular pathways mediating apoptosis, we treated PIP4K2A KD or control THP1 AML cells with pharmacological inhibitors. While MAP2K1/2 (Mek1/2) (U0126 10μM or PD184352 3μM), MAPK14/11/12/13 (p38 MAPK) (SB203580 10μM and BIRB0796 1μM) or AKT (MK2206 2μM) inhibition had no effect (data not shown), two structurally distinct inhibitors of MTOR (rapamycin 1μM and KU0063794 1μM) significantly retarded the onset of apoptosis (Figure 3a). Indeed, following PIP4K2A KD, in addition to up regulation of CDKN1A (Figures 2g, 3b), there was increased expression of MTOR protein, as well as increased phosphorylation of targets downstream of MTOR such as RPS6KB1 (p70S6 kinase) and RPS6 (ribosomal protein S6) (Figure 3b). This suggests that depletion of PIP4K2A increases both the amount and activity of MTOR, and that this contributes to apoptosis. There was also an increase in S473 phosphorylation of AKT1 (Figure 3b), but not in phospho-MAPK1/3 (p42/p44 MAPK) or phospho-MAPK14 (data not shown). The accumulation of CDKN1A was likewise dependent on MTOR, because it was largely ablated by both rapamycin and KU0063794 (Figure 3c). To determine whether CDKN1A accumulation contributed to the loss of proliferative potential of PIP4K2A KD AML cells, we performed double KD experiments. Following PIP4K2A KD alone there was an increase in CDKN1A transcripts (Figure 3d) suggesting that the increase in protein is in part driven by increased transcription. In keeping with a requirement for CDKN1A for the maintenance of leukemia-initiating cells,31 single CDKN1A KD to ~50% of control levels (Figure 3d) reduced proliferative and, more significantly, clonogenic potential (Figures 3e-g). Concomitant PIP4K2A and CDKN1A KD reduced the PIP4K2A dependent increase in CDKN1A transcripts without changing the efficiency of PIP4K2A KD, and led to a partial rescue of both proliferation and clongenic potential (Figures 3e-g). This indicates that the MTOR-dependent up regulation of CDKN1A contributes, at least in part, to the loss of proliferative potential of PIP4K2A KD AML cells.
Figure 3.
Apoptosis and induction of CDKN1A after PIP4K2A KD is dependent on MTOR. Human THP1 AML cells were infected with a lentiviral vector targeting PIP4K2A for KD, or a non-targeting control (NTC), with puromycin as the selectable marker. Cells were treated with puromycin for 48 hours to kill untransduced cells. Control and KD cells demonstrated similar viabilities at this time point, indicating equivalent transduction rates (data not shown). (a) Bar chart shows mean±SEM percentage of apoptotic cells seven days following lentiviral infection for the indicated conditions, as determined by annexin V binding (n=6). * indicates p<0.001 using one-way ANOVA followed by Fisher’s least significant difference post hoc test. (b) & (c) Western blots show expression of the indicated proteins, and in the indicated conditions, in THP1 AML cells 72 hours following lentiviral infection. Data from two separate experiments are shown (lanes 1 & 2 for each condition). (d-g) THP1 cells were double infected with lentiviral vectors targeting PIP4K2A or CDKN1A for KD, or a non-targeting control, with GFP and puromycin as selectable markers. Puromycin was added 24 hours later and viable GFP+ cells were FACS purified 48 hours following spinfection. Bar charts show (d) mean±SEM expression of PIP4K2A and CDKN1A relative to control cells, as determined by quantitative PCR, in FACS purified cells 72 hours following lentiviral infection (n=6); (e) mean±SEM fold change in resorufin (alamarBlue) signal of puromycin-resistant, GFP+ cells cultured for 72 hours following FACS purification (n=3); and (f) mean±SEM colony forming cell (CFC) frequencies (n=3) for the indicated control and KD conditions (n=3). For (e) and (f), * indicates p<0.01 for the indicated comparisons, as determined by an unpaired t-test. (g) Representative images from (f).
Selective requirement for PIP4K2A in primary human AML cells
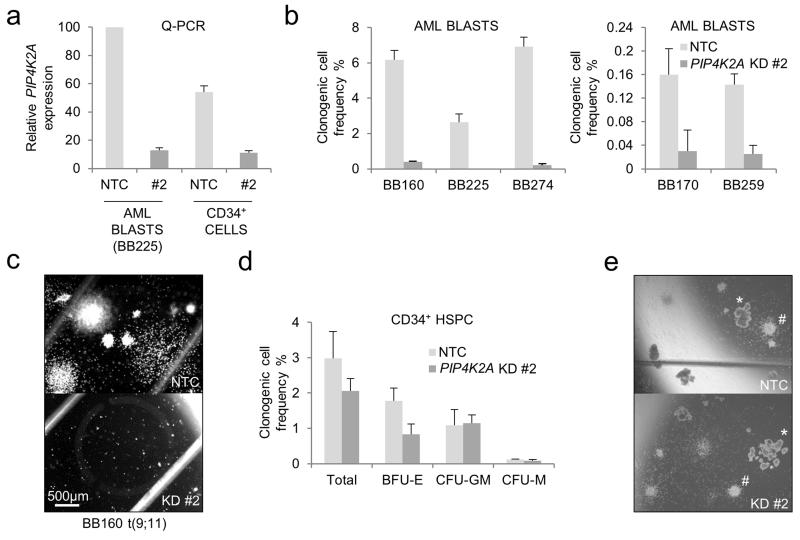
To determine whether primary human AML cells are also dependent upon PIP4K2A, we performed KD experiments (Figure 4a) using samples from five separate patients with different molecular subtypes of disease, including one with an MLL gene rearrangement and two from patients with disease relapse (Supplementary Table 2). As expected, cells from different patients exhibited different baseline clonogenic frequencies in semi-solid culture. However, in every case PIP4K2A KD reduced the frequency of clonogenic cells to between 0 and 15% of control levels (Figures 4b, c). In complete contrast, PIP4K2A KD in normal CD34+ HSPC from three separate donors had no significant effect on either clonogenic cell frequency or multilineage differentiation potential (Figures 4d, e). In our analyses, expression levels of PIP4K2A relative to ACTB were 1.8-fold higher in primary AML cells versus normal HSPC, but the proportional extent of KD was broadly similar (Figure 4a). Of note, analysis of published microarray data demonstrates no significant difference between PIP4K2A expression in human immunophenotypic LSC by comparison with normal HSPC (data not shown).32,33
Figure 4.
Selective requirement for PIP4K2A in primary human AML cells. Primary human normal CD34+ HSPC or AML cells were infected with lentiviral vectors targeting PIP4K2A for KD, or a non-targeting control (NTC), with eGFP as the selectable marker. GFP+ cells were FACS purified 48 hours later. Typical transduction efficiencies for both cell types were 20-50%. (a) Bar chart shows mean±SEM (of triplicate analyses) expression of PIP4K2A relative to control AML blasts for the indicated normal and leukemic populations, as determined by quantitative PCR, 48 hours following initiation of KD. (b) Bar charts shows mean clonogenic cell frequencies of primary AML blasts from five separate patients (see Supplementary Table 2) infected with control or PIP4K2A KD lentiviral vectors. Error bars refer to SEM of triplicate analyses. BB numbers refer to the Biobank identifier. Data are shown in two graphs with different y-axes due to the inherent variability in clonogenic cell frequencies between samples from different patients. (c) Representative image from (b). Scale bar applies to images shown in (c) and (e). (d) Bar chart shows mean±SEM frequencies of the indicated colony forming cells (n= 3, separate donors) (BFU-E – erythroid burst-forming unit; CFU-GM – granulocyte/macrophage colony forming unit; CFU-M – macrophage colony forming unit). (e) Representative image from (d). # indicates CFU-GM; * indicates BFU-E.
Selective requirement for Pip4k2a in murine AML cells
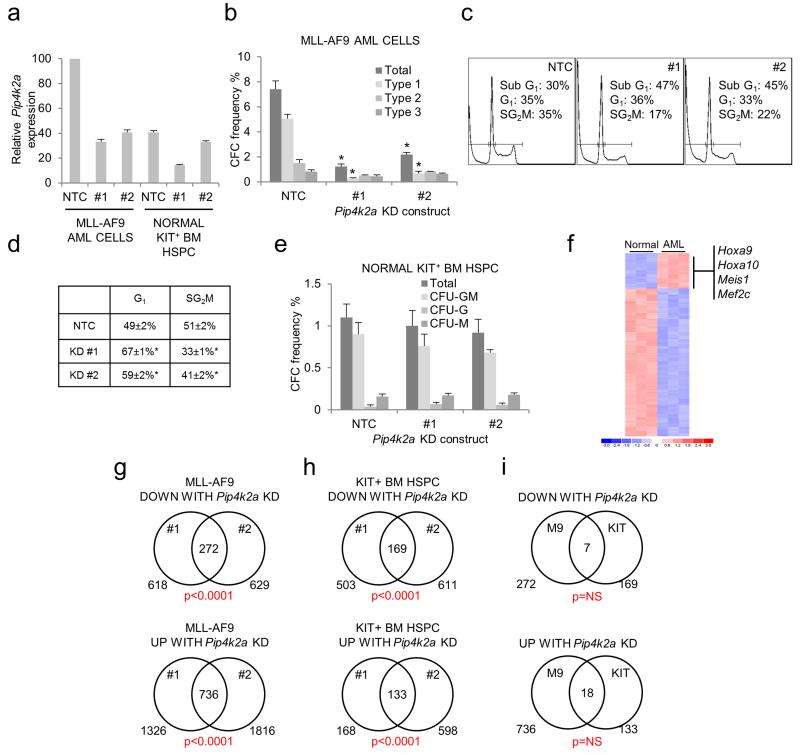
We next investigated whether a similar selective dependency of AML cells on Pip4k2a was observed in murine AML. Depletion of Pip4k2a KD in MLL-AF9 AML cells obtained from a retroviral transduction and transplantation mouse model 34-36 led to a significant reduction in clonogenic potential (Figures 5a, b). This was due to a lower frequency of Type 1 colonies, which contain poorly differentiated myeloblasts, rather than a reduction in the frequency of Type 2 colonies, which contain a mixed population of blasts and differentiating myeloid cells, or Type 3 colonies which contain terminally differentiated macrophages.34 Given that the formation of blast-like (Type 1) colonies in semi-solid culture by murine MLL-AF9 AML-CFCs directly correlates with LSC potential,34 these observations provide additional support for a critical role for Pip4k2a in leukemia-initiating cells. Loss of clonogenic potential due to Pip4k2a KD was again due to G1 cell cycle arrest and induction of apoptosis (in this case shown by the accumulation of a sub-G1 population in Pip4k2a KD cells) (Figures 5c, d). Forced expression of PIP4K2A in murine MLL-AF9 AML cells did not affect clonogenic potential (data not shown).
Figure 5.
Selective requirement for Pip4k2a in murine MLL-AF9 AML cells. Murine normal KIT+ or MLL-AF9 AML cells were infected with lentiviral vectors targeting Pip4k2a for KD, or a non-targeting control (NTC), with eGFP as the selectable marker. GFP+ cells were FACS purified 48 hours following lentiviral infection and initiation of KD. (a) Bar chart shows mean±SEM (of triplicate analyses) expression of Pip4k2a relative to control MLL-AF9 AML cells for the indicated normal and leukemic populations, as determined by quantitative PCR, 48 hours following initiation of KD. (b) Bar chart shows mean±SEM colony forming cell (CFC) frequencies (n=3) of control and Pip4k2a KD MLL-AF9 AML cells. * indicates p<0.05 by comparison with control cells, as determined by one way ANOVA followed by Fisher’s least significant difference post hoc test. (c) Representative cell cycle profiles show the proportion of MLL-AF9 AML cells in each stage of the cell cycle, or sub-G1 apoptotic cells, in control or Pip4k2a KD cells 96 hours following initiation of KD. (d) Excluding sub-G1 cells and related to (c), table indicates mean±SEM percentage of murine MLL-AF9 AML cells in the indicated phase of the cell cycle (n=3 triplicate analyses). * indicates p<0.05 by comparison with control cells, as determined by one way ANOVA followed by Fisher’s least significant difference post hoc test. (e) Bar chart shows mean±SEM colony forming cell (CFC) frequencies (n=3) of control and Pip4k2a KD KIT+ normal HSPC. (f) Heat map represents 421 protein coding genes significantly up or down regulated (p<0.001 (unpaired t-test) and more than two-fold change in expression) in MLL-AF9 AML cells by comparison with KIT+ HSPC, with key genes highlighted. Color scale indicates normalized expression values. Venn diagrams indicate numbers of protein coding genes up or down regulated (defined as those whose expression increased or decreased by 25% compared with control samples) following Pip4k2a KD for each of the tested KD constructs (#1 and #2) in (g) MLL-AF9 AML cells and (h) KIT+ BM HSPC. The number of co-regulated genes is shown in the intersect, together with the significance of the overlap (Chi-square test). (i) Venn diagrams indicate extent of overlap of genes up or down regulated by Pip4k2a KD in MLL-AF9 AML cells (M9) or KIT+ BM HSPC (KIT).
In keeping with our findings in normal human HSPC, depletion of Pip4k2a transcripts in murine HSPC did not significantly alter either colony forming or multilineage differentiation potential (Figure 5e). This was not because Pip4k2a was expressed in one cell setting and not the other, nor due to different KD efficiencies (Figure 5a). Analysis of published microarray data demonstrates no significant difference between Pip4k2a expression in murine MLL-mutated LSC populations versus normal myeloblasts with a similar immunophenotype (data not shown).35 Our attempts to compare PIP4K2A expression levels in murine normal and leukemic hematopoietic cells by western blotting were unsuccessful using the antibodies available which did not detect bands of the predicted mass. There was no significant difference in whole cell levels of PtdIns5P in murine KIT+ normal HSPC versus MLL-AF9 AML cells following their recovery from normal or leukemic animals respectively and culture for five days in liquid conditions with growth factors supporting myeloid differentiation (data not shown).
Given the differential dependency of normal versus leukemic AML cells on Pip4k2a we next performed microarray analyses to investigate the transcriptional programs regulated by Pip4k2a in each cell setting. To enhance confidence in the analysis, and the resulting set of Pip4k2a-regulated genes, array data from two different KD constructs (#1 and #2) were analysed. As expected,34-36 MLL-AF9 AML cells exhibited significantly higher expression of genes such as Hoxa9 and Meis1 (Figure 5f) by comparison with normal BM HSPC. There was very substantial overlap in the genes up or down regulated by the separate Pip4k2a KD constructs in each cell setting (Figures 5g, h and Supplementary Table 3). However, when the Pip4k2a-regulated transcriptional programs in normal HSPC and AML cells were compared, there was no significant overlap (Figure 5i). Thus, in keeping with the different functional consequences following its knockdown, Pip4k2a regulates different transcriptional programs in normal HSPC by comparison with AML cells. Of note, genes down regulated following Pip4k2a-KD in MLL-AF9 AML cells did not include those such as Hoxa9, Meis1, Myb, Cbx5 and Hmgb3 which have previously been shown to regulate MLL LSC potential.34,36 Furthermore, there was no significant overlap in the Pip4k2a-regulated transcriptional programs we report and a recently reported PIP4K2A/PIP4K2B-regulated program in TP53-mutated breast cancer cells (data not shown).21 Gene ontology analysis of Pip4K2a-regulated gene sets (Supplementary Tables 3, 4) demonstrated significant enrichments for terms relating to the membrane, secretion, receptor signaling and extracellular matrix among those down regulated in AML cells, and nuclear chromatin among those up regulated in AML cells. Interestingly, in normal KIT+ BM HSPC there was significant enrichment for terms relating to platelets and cell adhesion among those down regulated and those relating to leukocyte activation and cytokine activation among those up regulated; the former likely reflects the role of Pip4k2a in megakaryocyte differentiation.29
DISCUSSION
Reflecting the importance of lipid signaling in general, and disordered lipid signaling in cancer in particular, our use of a targeted but unbiased KD screen has identified novel candidate regulators of myeloid leukemia cell proliferation and survival, in particular PIP4K2A. While MLL-AF9 expressing human and murine AML cells were used as exemplar lines in our studies, it is of note that all human AML cell lines and primary cells tested in the study, including relapse samples, demonstrated sensitivity to PIP4K2A depletion to variable extent (Figure 4b and Supplementary Figure 4). Coupled with the dependency of KRAS mutated colorectal cancer cells on PIP4K2A,16 the dependency of TP53 null breast cancer cells on PIP4K2A/PIP4K2B21 and the observation that PIP4K2A mice appear normal,21 our observations add to the case for the development and evaluation of PIP4K2A inhibitors in cancer in general, and leukemia in particular. The case is further enhanced given the high level expression of PIP4K2A in poor risk AML and in chemoresistant pediatric acute lymphoblastic leukemia (ALL), and the unmet need for novel therapies in these areas.24-26,37 Interestingly, susceptibility loci for ALL have been reported in both intronic and exonic sequences of PIP4K2A, although the causative mechanism remains unclear.38,39 While no PIP4K2A inhibitors are commercially available as yet, an in vitro screen of a library of pharmacologically active compounds identified several tyrphostins as having inhibitory activity,40 although compound stability, cell permeability and functional activity in cells remain to be determined.
While the cellular consequences of PIP4K2A depletion in AML cells are likely pleiotropic, we observed an accumulation of the cyclin-dependent kinase inhibitor CDKN1A (p21) with concomitant apoptosis and both were MTOR dependent. CDKN1A is regulated transcriptionally, translationally and by protein degradation and our data reinforce previous work implicating MTOR activation in the regulation of CDKN1A/p21 accumulation.41 PIP4K2A might regulate MTOR through altered levels of cellular PIs and we observed significant increases in PtdIns5P following PIP4K2A depletion as has been demonstrated to occur following depletion of other isoforms of PIP4K.42-45 PtdIns5P has been linked to the regulation of gene transcription,46,47 acetylation and activation of TP53, activation of AKT48 and the accumulation of cellular reactive oxygen species.43 PtdIns5P levels increase in response to various cellular stressors and its accumulation in AML cells might mimic the induction of cell stress thereby inducing cell cycle arrest and apoptosis, or perhaps increase susceptibility to other stresses such as DNA damage. The reason why normal HSPC and AML cells differ so markedly in their transcriptional and functional response to PIP4K2A depletion remains unclear, but our observations are nevertheless in keeping with other recently reported examples of the differential functional dependency of AML cells versus their normal progenitor counterparts (on, for example, KDM1A/LSD1 or EPC1).34,49 Furthermore, such differential functional dependencies represent strong candidate targets for future therapeutic intervention.
MATERIALS AND METHODS
Cell lines and tissue culture
Cell lines and culture conditions were as descibed.34,49 Murine AML cells were from a cohort of C57BL/6 mice with MLL-AF9 AML, initiated using a retroviral transduction and transplantation protocol as described.34 Normal HSPC were immunomagnetically selected using anti-CD117 or anti-CD34 microbeads (Miltenyi Biotec, Bisley, UK) for murine or human cells respectively, and an automated cell separator (AutoMACS Pro, Miltenyi Biotec). Semisolid colony assays and liquid culture conditions of human and murine cells were performed as described.34,49
Flow cytometry, apoptosis assays, cell cycle assays, quantitative PCR, western blotting, lipid assays and inhibitors
For experiments involving purification of cell populations by flow sorting, an Influx or a FACSAria II was used (BD Biosciences, Oxford, UK). Apoptosis and cell cycle analyses were performed using an LSR Model II flow cytometer (BD Biosciences). Apoptosis was measured using an Annexin V-APC kit (BD Pharmingen, Oxford UK) according to the manufacturer’s instructions. Cell cycle analyses were performed as described.50 Quantitative PCR was performed as described 34 using Universal Probe Library System (Roche, UK) designed primers and probes (Supplementary Table 5). Antibodies for western blotting and pharmacological inhibitors are shown in Supplementary Tables 6 and 7 respectively. Lipid assays were performed as detailed elsewhere.51,52
Lentiviral knockdown library, viral supernatant production and transduction of target cells
pLKO-puro shRNA KD constructs were from Sigma-Aldrich (St. Louis, MO) (Supplementary Table 1). Plasmid DNA was prepared with a NucleoSpin® Plasmid kit (Macherey-Nagel, Dueran, Germany). Purities were confirmed, and DNA concentrations measured, by spectrophotometry and adjusted to 50ng/μL. Lentiviral supernatants were generated in a 96-well plate format. To each well of HEK293FT cells at 80-90% confluence in 100μL culture medium was added 50μL of OptiMEM containing 0.8μL of Lipofectamine2000 and 350ng of plasmid DNA (pLKO, pCMVΔR8.91 and pMD2.G plasmids, 10:10:1 ratio). Next day, medium was replaced with 170μL DMEM supplemented with 1% fetal bovine serum and 5mM L-glutamine. Viral supernatants were harvested 48 and 72 hours post-transfection, pooled and frozen in aliquots of 60μL per well in 96-well plates.
AML cells were infected with viral particles in flat bottomed 384-well plates. For a single library screen, four plates were required. Each test well contained 2000 AML cells in 50μL of medium supplemented with 8μg/mL polybrene (Millipore, Billerica, MA). Viral supernatant plates were thawed and mixed, and each supernatant was divided between a pair of test wells side-by-side on the plate, with 25μL viral supernatant per well. Plates were centrifuged at 900xg at 37°C for 30 minutes prior to overnight incubation at 37°C and 5% CO2. After 16 hours, cells were selected for viral integration for 48 hours by addition of 20 μL of medium containing 2.5μg/mL puromycin (Sigma-Aldrich). Each plate additionally contained (i) 24 pairs of control wells containing cells infected with a pLKO-puro lentiviral vector expressing a non-targeting control shRNA (SH002, Sigma); (ii) one or more additional control wells containing cells infected with pLKO-eGFP30 used to confirm that puromycin drug treatment killed all untransduced cells within 48 hours; and (iii) twelve pairs of wells containing medium only to provide background fluorescence values.
Additional lentiviral vectors targeting PIP4K2A, Pip4k2a or CDKN1A for KD were purchased from Sigma (PIP4K2A: TRCN0000010988, TRCN0000356495, TRCN0000194659; Pip4k2a: TRCN0000025577, TRCN0000025578; CKDN1A: TRCN0000054901). For some constructs, the PGK-puromycin cassette was exchanged for an SFFV-eGFP cassette.34 Larger scale viral production and transduction of normal and leukemic target cells was as previously described.34 For lentiviral infection of primary human CD34+ cells or AML blasts, cells were thawed and incubated overnight in StemSpan (Stem Cell Technologies) with a 9-growth factor mix 34 at a density of 50-250 × 103/ml. Next day, medium was replaced with lentiviral supernatant containing polybrene 8μg/ml and 9-factor mix. Twenty-four hours later cells were returned to StemSpan with 9-factor mix. GFP+ cells were FACS sorted 24 hours later.
Cellular biomass estimation
To estimate cell biomass at the initial and final time points, 5μL alamarBlue (Invitrogen) was added to each well. Plates were incubated for three hours at 37°C before measuring resorufin fluorescence on a POLARstar Omega microplate reader (BMG Labtech) (excitation 544nm, emission 590nm). Fluorescence signal was background corrected by subtraction of the median fluorescence signal of blank wells (containing medium and alamarBlue alone) for each of the two reading points.
Murine experiments
Experiments were approved by the Cancer Research UK Manchester Institute’s Animal Ethics Committee and performed under a UK Home Office project license. NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ mice were purchased from Jackson Laboratories (Bar Harbor, ME) and bred in-house. For xenograft experiments, human THP1 AML cells were infected with lentiviruses targeting PIP4K2A for KD, or a non-targeting control vector, with puromycin as the selectable marker. Next day, cells were treated with puromycin 2.5μg/mL and then 48 hours later 104 viable cells, as determined by trypan blue dye exclusion, were transplanted in to sub-lethally irradiated (200cGy) mice by tail vein injection.
Human tissue
Use of human tissue was in compliance with the Human Tissue Act, 2004. Normal human CD34+ mobilized HSPC surplus to requirements were from patients undergoing chemotherapy and autologous transplantation for lymphoma and myeloma. Their use was authorized by the Salford and Trafford Research Ethics Committee and, where possible, following the written informed consent of donors. Primary human AML cells were obtained from Manchester Cancer Research Centre’s Tissue Biobank, instituted with the approval of the South Manchester Research Ethics Committee. Use of AML cells was authorized following ethical review by the Tissue Biobank’s scientific sub-committee and with the written informed consent of the patient.
Microarray analysis
Microarray CEL files and detailed methods are available at the Gene Expression Omnibus with accession number GSE54309.
Supplementary Material
Acknowledgements
We thank Morgan Blaylock, Jeff Barry, Michael Hughes, Gail Bruder, Angela Cooke and Yaoyong Li for technical support. This work was supported by Cancer Research UK grant number C5759/A12328.
Funding source: Cancer Research UK grant number C5759/A12328
Footnotes
Supplementary Information Supplementary Information accompanies the paper on the Oncogene website.
Conflict of interest The authors declare no conflict of interest.
References
- 1.Bunney TD, Katan M. Phosphoinositide signalling in cancer: beyond PI3K and PTEN. Nat Rev Cancer. 2010 May;10(5):342–352. doi: 10.1038/nrc2842. [DOI] [PubMed] [Google Scholar]
- 2.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006 Oct 12;443(7112):651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 3.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008 Feb;9(2):99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 4.Martelli AM, Nyakern M, Tabellini G, Bortul R, Tazzari PL, Evangelisti C, et al. Phosphoinositide 3-kinase/Akt signaling pathway and its therapeutical implications for human acute myeloid leukemia. Leukemia. 2006 Jun;20(6):911–928. doi: 10.1038/sj.leu.2404245. [DOI] [PubMed] [Google Scholar]
- 5.Polak R, Buitenhuis M. The PI3K/PKB signaling module as key regulator of hematopoiesis: implications for therapeutic strategies in leukemia. Blood. 2012 Jan 26;119(4):911–923. doi: 10.1182/blood-2011-07-366203. [DOI] [PubMed] [Google Scholar]
- 6.Vergez F, Recher C, Payrastre B. Class I phosphoinositide 3-kinases in normal and pathologic hematopoietic cells. Curr Top Microbiol Immunol. 2012;362:163–184. doi: 10.1007/978-94-007-5025-8_8. [DOI] [PubMed] [Google Scholar]
- 7.Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, Wu H, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006 May 25;441(7092):475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Grindley JC, Yin T, Jayasinghe S, He XC, Ross JT, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006 May 25;441(7092):518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- 9.Helgason CD, Damen JE, Rosten P, Grewal R, Sorensen P, Chappel SM, et al. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Dev. 1998 Jun 1;12(11):1610–1620. doi: 10.1101/gad.12.11.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Q, Simpson SE, Scialla TJ, Bagg A, Carroll M. Survival of acute myeloid leukemia cells requires PI3 kinase activation. Blood. 2003 Aug 1;102(3):972–980. doi: 10.1182/blood-2002-11-3429. [DOI] [PubMed] [Google Scholar]
- 11.Min YH, Eom JI, Cheong JW, Maeng HO, Kim JY, Jeung HK, et al. Constitutive phosphorylation of Akt/PKB protein in acute myeloid leukemia: its significance as a prognostic variable. Leukemia. 2003 May;17(5):995–997. doi: 10.1038/sj.leu.2402874. [DOI] [PubMed] [Google Scholar]
- 12.Kubota Y, Ohnishi H, Kitanaka A, Ishida T, Tanaka T. Constitutive activation of PI3K is involved in the spontaneous proliferation of primary acute myeloid leukemia cells: direct evidence of PI3K activation. Leukemia. 2004 Aug;18(8):1438–1440. doi: 10.1038/sj.leu.2403402. [DOI] [PubMed] [Google Scholar]
- 13.Zhao S, Konopleva M, Cabreira-Hansen M, Xie Z, Hu W, Milella M, et al. Inhibition of phosphatidylinositol 3-kinase dephosphorylates BAD and promotes apoptosis in myeloid leukemias. Leukemia. 2004 Feb;18(2):267–275. doi: 10.1038/sj.leu.2403220. [DOI] [PubMed] [Google Scholar]
- 14.Sykes SM, Lane SW, Bullinger L, Kalaitzidis D, Yusuf R, Saez B, et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 2011 Sep 2;146(5):697–708. doi: 10.1016/j.cell.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myhre S, Mohammed H, Tramm T, Alsner J, Finak G, Park M, et al. In silico ascription of gene expression differences to tumor and stromal cells in a model to study impact on breast cancer outcome. PLoS One. 2010;5(11):e14002. doi: 10.1371/journal.pone.0014002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009 May 29;137(5):835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimwade D, Hills RK, Moorman AV, Walker H, Chatters S, Goldstone AH, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patientstreated in the United Kingdom Medical Research Council trials. Blood. 2010 Jul 22;116(3):354–365. doi: 10.1182/blood-2009-11-254441. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, Huang J, Yang N, Greshock J, Liang S, Hasegawa K, et al. Integrative genomic analysis of phosphatidylinositol 3′-kinase family identifies PIK3R3 as a potential therapeutic target in epithelial ovarian cancer. Clin Cancer Res. 2007 Sep 15;13(18 Pt 1):5314–5321. doi: 10.1158/1078-0432.CCR-06-2660. [DOI] [PubMed] [Google Scholar]
- 19.Bertagnolo V, Benedusi M, Querzoli P, Pedriali M, Magri E, Brugnoli F, et al. PLC-beta2 is highly expressed in breast cancer and is associated with a poor outcome: a study on tissue microarrays. Int J Oncol. 2006 Apr;28(4):863–872. [PubMed] [Google Scholar]
- 20.Bertagnolo V, Benedusi M, Brugnoli F, Lanuti P, Marchisio M, Querzoli P, et al. Phospholipase C-beta 2 promotes mitosis and migration of human breast cancer-derived cells. Carcinogenesis. 2007 Aug;28(8):1638–1645. doi: 10.1093/carcin/bgm078. [DOI] [PubMed] [Google Scholar]
- 21.Emerling BM, Hurov JB, Poulogiannis G, Tsukazawa KS, Choo-Wing R, Wulf GM, et al. Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell. 2013 Nov 7;155(4):844–857. doi: 10.1016/j.cell.2013.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive mode of anticancer drug sensitivity. Nature. 2012 Mar 29;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011 Jan;39(Database issue):D945–950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007 Feb;9(2):166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wouters BJ, Louwers I, Valk PJ, Lowenberg B, Delwel R. A recurrent in-frame insertion in a CEBPA transactivation domain is a polymorphism rather than a mutation that does not affect gene expression profiling-based clustering of AML. Blood. 2007 Jan 1;109(1):389–390. doi: 10.1182/blood-2006-08-042325. [DOI] [PubMed] [Google Scholar]
- 26.Szczepanek J, Jarzab M, Oczko-Wojciechowska M, Kowalska M, Tretyn A, Haus O, et al. Gene expression signatures and ex vivo drug sensitivity profiles in children with acute lymphoblastic leukemia. J Appl Genet. 2012 Feb;53(1):83–91. doi: 10.1007/s13353-011-0073-x. [DOI] [PubMed] [Google Scholar]
- 27.van den Bout I, Divecha N. PIP5K-driven PtdIns(4,5)P2 synthesis: regulation and cellular functions. J Cell Sci. 2009 Nov 1;122(Pt 21):3837–3850. doi: 10.1242/jcs.056127. [DOI] [PubMed] [Google Scholar]
- 28.Ling LE, Schulz JT, Cantley LC. Characterization and purification of membrane-associated phosphatidylinositol-4-phosphate kinase from human red blood cells. J Biol Chem. 1989 Mar 25;264(9):5080–5088. [PubMed] [Google Scholar]
- 29.Schulze H, Korpal M, Hurov J, Kim SW, Zhang J, Cantley LC, et al. Characterization of the megakaryocyte demarcation membrane system and its role in thrombopoiesis. Blood. 2006 May 15;107(10):3868–3875. doi: 10.1182/blood-2005-07-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones DR, Foulger R, Keune WJ, Bultsma Y, Divecha N. PtdIns5P is an oxidative stress-induced second messenger that regulated PKB activation. FASEB J. 2013 Apr 27;27(4):1644–1656. doi: 10.1096/fj.12-218842. [DOI] [PubMed] [Google Scholar]
- 31.Viale A, De Franco F, Orleth A, Cambiaghi V, Giuliani V, Bossi D, et al. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature. 2009 Jan 1;457(7225):51–56. doi: 10.1038/nature07618. [DOI] [PubMed] [Google Scholar]
- 32.Saito Y, Uchida N, Tanaka S, Suzuki N, Tomizawa-Murasawa M, Sone A, et al. Induction of cell cycle entry eliminates human leukemia stem cells in a mouse model of AML. Nat Biotechnol. 2010 Mar;28(3):275–280. doi: 10.1038/nbt.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goardon N, Marchi E, Atzberger A, Quek L, Schuh A, Soneji S, et al. Coexistence of LMPP-like and GMP-like leukemia stem cells in acute myeloid leukemia. Cancer Cell. 2011 Jan 18;19(1):138–152. doi: 10.1016/j.ccr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 34.Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012 Apr 17;21(4):473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 35.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006 Oct;10(4):257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 36.Somervaille TC, Matheny CJ, Spencer GJ, Iwasaki M, Rinn JL, Witten DM, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009 Feb 6;4(2):129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiseman D, Greystoke BF, Somervaille TC. The variety of leukemic stem cells in myeloid malignancy. Oncogene. 2013 doi: 10.1038/onc.2013.269. [DOI] [PubMed] [Google Scholar]
- 38.Migliorini G, Fiege B, Hosking FJ, Ma Y, Kumar R, Sherborne AL, et al. Variation at 10p12.2 and 10p14 influences risk of childhood B-cell acute lymphoblastic leukemia and phenotype. Blood. 2013;122(19):3298–3307. doi: 10.1182/blood-2013-03-491316. [DOI] [PubMed] [Google Scholar]
- 39.Xu H, Yang W, Perez-Andreu V, Devidas M, Fan Y, Cheng S, et al. Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J Natl Cancer Inst. 2013;105(10):733–742. doi: 10.1093/jnci/djt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis MI, Sasaki AT, Shen M, Emerling BM, Thorne N, Michael S, et al. A homogeneous, high-throughput assay for phosphatidylinositol 5-phosphate 4-kinase with a novel, rapid substrate preparation. PLoS One. 2013;8(1):e54127. doi: 10.1371/journal.pone.0054127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beuvink I, Boulay B, Fumagalli S, Zilbermann F, Ruetz S, O’Reilly T, et al. The mTOR inhibitor RAD001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell. 2005;120(6):747–759. doi: 10.1016/j.cell.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 42.Wilcox A, Hinchliffe KA. Regulation of extranuclear PtdIns5P production by phosphatidylinositol phosphate 4-kinase 2alpha. FEBS Lett. 2008 Apr 16;582(9):1391–1394. doi: 10.1016/j.febslet.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 43.Keune WJ, Jones DR, Bultsma Y, Sommer L, Zhou XZ, Lu KP, et al. Regulation of phosphatidylinositol-5-phosphate signaling by Pin1 determines sensitivity to oxidative stress. Sci Signal. 2012 Nov 27;5(252):ra86. doi: 10.1126/scisignal.2003223. [DOI] [PubMed] [Google Scholar]
- 44.Sarkes D, Rameh LE. A novel HPLC-based approach makes possible the spatial characterization of cellular PtdIns5P and other phosphoinositides. Biochem J. 2010 Jun 15;428(3):375–384. doi: 10.1042/BJ20100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones DR, Bultsma Y, Keune WJ, Halstead JR, Elouarrat D, Mohammed S, et al. Nuclear PtdIns5P as a transducer of stress signaling: an in vivo role for PIP4Kbeta. Mol Cell. 2006 Sep 1;23(5):685–695. doi: 10.1016/j.molcel.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 46.Ndamukong I, Jones DR, Lapko H, Divecha N, Avramova Z. Phosphatidylinositol 5-phosphate links dehydration stress to the activity of ARABIDOPSIS TRITHORAX-LIKE factor ATX1. PLoS One. 2010;5(10):e13396. doi: 10.1371/journal.pone.0013396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keune WJ, Sims AH, Jones DR, Bultsma Y, Lynch JT, Jirstrom K, et al. Low PIP4K2B expression in human breast tumors correlates with reduced patient survival: a role for PIP4K2B in the regulation of E-cadherin expression. Cancer Res. 2013;73(23):6913–6925. doi: 10.1158/0008-5472.CAN-13-0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones DR, Foulger R, Keune WJ, Bultsma Y, Divecha N. PtdIns5P is an oxidative stress-induced second messenger that regulates PKB activation. FASEB J. 2012 Dec 14; doi: 10.1096/fj.12-218842. [DOI] [PubMed] [Google Scholar]
- 49.Huang X, Spencer GJ, Lynch JT, Ciceri F, Somerville TD, Somervaille TC. Enhancers of Polycomb EPC1 and EPC2 sustain the oncogenic potential of MLL leukemia stem cells. Leukemia. 2013 doi: 10.1038/leu.2013.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Somervaille TC, Linch DC, Khwaja A. Different levels of p38 MAP kinase activity mediate distinct biological effects in primary human erythroid progenitors. Br J Haematol. 2003 Mar;120(5):876–886. doi: 10.1046/j.1365-2141.2003.04204.x. [DOI] [PubMed] [Google Scholar]
- 51.Jones DR, Ramirez IB, Lowe M, Divecha N. Measurement of phosphoinositides in the zebrafish Danio rerio. Nat Protoc. 2013;8(6):1058–72. doi: 10.1038/nprot.2013.040. [DOI] [PubMed] [Google Scholar]
- 52.Halstead JR, Roefs M, Ellson CD, D’Andrea S, Chen C, D’Santos CS, et al. A novel pathway of cellular phosphatidylinositol(3,4,5)-trisphosphate synthesis is regulated by oxidative stress. Curr Biol. 2001;11(6):386–95. doi: 10.1016/s0960-9822(01)00121-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.