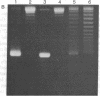
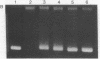
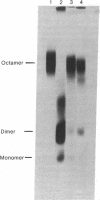
Abstract
We provide evidence that nucleosomes can assemble in vitro at physiological ionic strength (0.1-0.2 M NaCl/10 mM Tris . HCl, pH 8.0) in the absence of "assembly factors" and that poly(glutamic acid) greatly facilitates chromatin assembly under these conditions. We also show that in the presence of either poly(glutamic acid) or poly(aspartic acid), core histones assemble into octamers at physiological ionic strength. We suggest that it is a property of histones to assemble into octamers upon their interaction with macromolecules containing regions of high negative charge density, and we discuss several implications of this property.
Full text
PDF
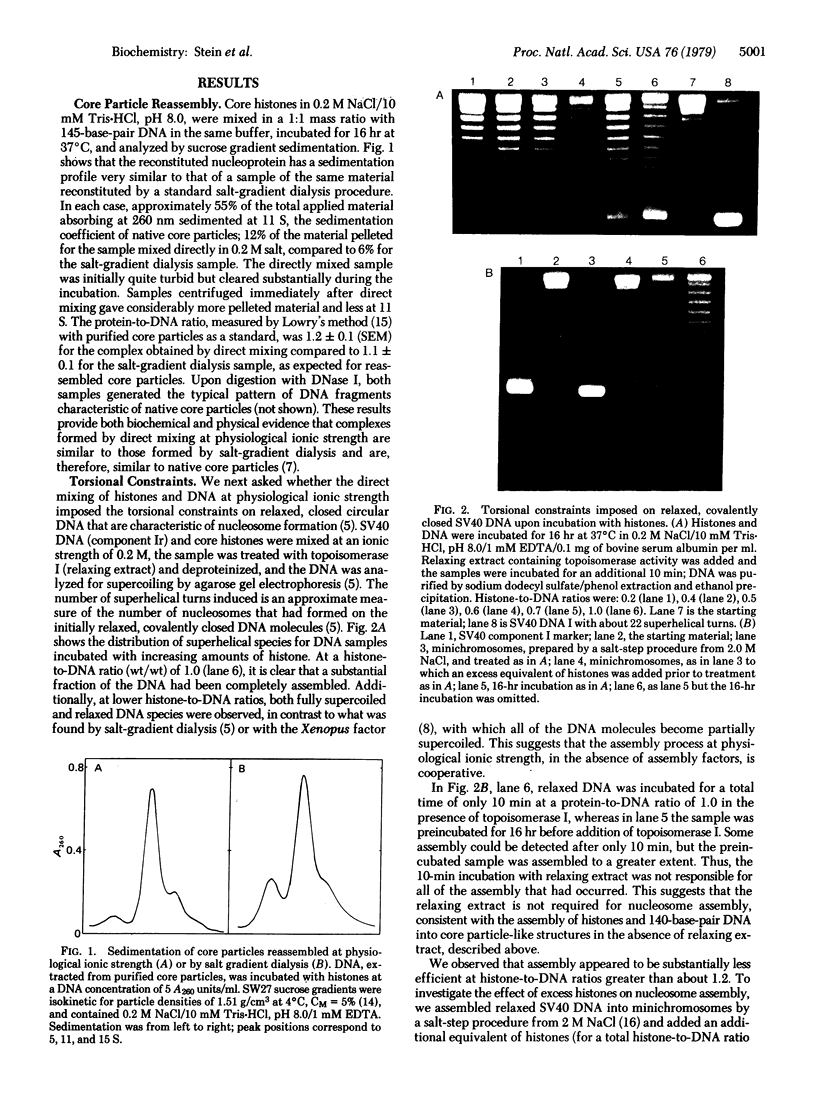



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bina-Stein M., Simpson R. T. Specific folding and contraction of DNA by histones H3 and H4. Cell. 1977 Jul;11(3):609–618. doi: 10.1016/0092-8674(77)90078-2. [DOI] [PubMed] [Google Scholar]
- Bina-Stein M., Singer M. F. The effect of H1 histone on the action of DNA-relaxing enzyme. Nucleic Acids Res. 1977 Jan;4(1):117–127. doi: 10.1093/nar/4.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Sollner-Webb B., Felsenfeld G. The organization of histones and DNA in chromatin: evidence for an arginine-rich histone kernel. Cell. 1976 Jul;8(3):333–347. doi: 10.1016/0092-8674(76)90145-8. [DOI] [PubMed] [Google Scholar]
- Eickbush T. H., Moudrianakis E. N. The histone core complex: an octamer assembled by two sets of protein-protein interactions. Biochemistry. 1978 Nov 14;17(23):4955–4964. doi: 10.1021/bi00616a016. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg R. D. Chromatin structure: a repeating unit of histones and DNA. Science. 1974 May 24;184(4139):868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laskey R. A., Honda B. M., Mills A. D., Finch J. T. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978 Oct 5;275(5679):416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D., Morris N. R. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977 Feb;10(2):237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- Levy W B., Wong N. C., Dixon G. H. Selective association of the trout-specific H6 protein with chromatin regions susceptible to DNase I and DNase II: possible location of HMG-T in the spacer region between core nucleosomes. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2810–2814. doi: 10.1073/pnas.74.7.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty K. S., Jr, Vollmer R. T., McCarty K. S. Improved computer program data for the resolution and fractionation of macromolecules by isokinetic sucrose density gradient sedimentation. Anal Biochem. 1974 Sep;61(1):165–183. doi: 10.1016/0003-2697(74)90343-1. [DOI] [PubMed] [Google Scholar]
- Nadeau P., Oliver D. R., Chalkley R. Effect of inhibition of DNA synthesis on histone synthesis and deposition. Biochemistry. 1978 Nov 14;17(23):4885–4893. doi: 10.1021/bi00616a005. [DOI] [PubMed] [Google Scholar]
- Olins A. L., Olins D. E. Spheroid chromatin units (v bodies). Science. 1974 Jan 25;183(4122):330–332. doi: 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- Oudet P., Germond J. E., Sures M., Gallwitz D., Bellard M., Chambon P. Nucleosome structure I: all four histones, H2A, H2B, H3, and H4, are required to form a nucleosome, but an H3-H4 subnucleosomal particle is formed with H3-H4 alone. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):287–300. doi: 10.1101/sqb.1978.042.01.031. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Stein A., Bina-Stein M., Simpson R. T. Crosslinked histone octamer as a model of the nucleosome core. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2780–2784. doi: 10.1073/pnas.74.7.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A. DNA folding by histones: the kinetics of chromatin core particle reassembly and the interaction of nucleosomes with histones. J Mol Biol. 1979 May 15;130(2):103–134. doi: 10.1016/0022-2836(79)90421-2. [DOI] [PubMed] [Google Scholar]
- Tatchell K., Van Holde K. E. Reconstitution of chromatin core particles. Biochemistry. 1977 Nov 29;16(24):5295–5303. doi: 10.1021/bi00643a021. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Butler P. J. The nucleosome core protein. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):119–125. doi: 10.1101/sqb.1978.042.01.013. [DOI] [PubMed] [Google Scholar]
- Van Holde K. E., Sahasrabuddhe C. G., Shaw B. R. A model for particulate structure in chromatin. Nucleic Acids Res. 1974 Nov;1(11):1579–1586. doi: 10.1093/nar/1.11.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordouw G., Eisenberg H. Binding of additional histones to chromatin core particles. Nature. 1978 Jun 8;273(5662):446–448. doi: 10.1038/273446a0. [DOI] [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]
- Weintraub H., Palter K., Van Lente F. Histones H2a, H2b, H3, and H4 form a tetrameric complex in solutions of high salt. Cell. 1975 Sep;6(1):85–110. doi: 10.1016/0092-8674(75)90077-x. [DOI] [PubMed] [Google Scholar]
- Weisbrod S., Weintraub H. Isolation of a subclass of nuclear proteins responsible for conferring a DNase I-sensitive structure on globin chromatin. Proc Natl Acad Sci U S A. 1979 Feb;76(2):630–634. doi: 10.1073/pnas.76.2.630. [DOI] [PMC free article] [PubMed] [Google Scholar]