Abstract
The Hedgehog (Hh) pathway plays conserved roles in regulating a diverse spectrum of developmental processes. In some developmental contexts, a gradient of Hh protein specifies multiple cell types in a dose-dependent fashion, thereby acting as a morphogen. Hh signaling ultimately acts on the transcriptional level through GLI proteins. In the presence of Hh signaling full length GLI proteins act as transcriptional activators of target genes. Conversely, in the absence of Hh, GLI proteins act as transcriptional repressors. This review will highlight mechanisms contributing to how graded Hh signaling might translate to differential GLI activity and be interpreted into distinct transcriptional responses.
Keywords: Gli, Hedgehog, Morphogen, Development
1. The Hedgehog Signaling Pathway Overview
The Hedgehog (Hh) signaling pathway regulates a large number of tissue patterning events during development, acting as a growth factor, survival agent, and inductive signal in a context-dependent fashion. Although aspects of the nature, timing and response to Hh ligands are all areas of active research, Hh ultimately acts through GLI transcription factors to elicit tissue-specific responses. Because the Hh ligand is secreted, it has the ability to evoke concentration-specific responses in some contexts, fitting the functional criteria for a morphogen [1–4].
The role of graded Hh signaling as a morphogen has been most intensively studied in the context of Drosophila wing imaginal disc patterning, as well as in the vertebrate limb bud and neural tube (Fig. 1). In all three systems, distinct transcriptional responses are associated with different doses of the Hh ligand. The underlying principles of Hh signaling and response are well conserved with the notable exception that the cilia functions as a processing center for Hh signal transduction and GLI processing in vertebrates [5]. Hh is secreted by a population of signaling cells into the surrounding tissue where it binds to the transmembrane receptor protein Patched (Ptch) on signal receiving cells. In the absence of Hh, Ptch inhibits the activity of another transmembrane protein, Smoothened (Smo). Upon Hh binding to Ptch, Smo is activated and subsequently results in the activation of GLI transcription factors which include GLI1–3 in vertebrates, and the homologous Cubitus interruptus (Ci) transcription factor in Drosophila [6–8]. All GLI proteins recognize and bind to the same binding motif (Gli binding motif or GBM) [9–12]. Transcriptional activation of Ptch by GLI transcription factors in response to Hh signaling provides negative feedback to restrict Hh signaling both spatially and temporally [13–15]. This negative feedback is integral to Hh signaling, as in the absence of Ptch, responsive tissues have constitutively high levels of Hh pathway activation [16, 17]. The transcriptional response to Hh signaling occurs solely through the activity of GLI family proteins [18, 19]. This review will highlight efforts towards understanding how GLI family proteins resolve graded Hh signaling and translate it into a discrete transcriptional output.
Figure 1. Model systems for hedgehog morphogens.
(L–R) Drosophila wing imaginal disc, vertebrate limb bud, vertebrate neural tube. The Hh secreting populations are shown in blue with the resulting protein gradient schematized in the triangle. The ventral progenitor domains are highlighted on the right side of the neural tube. Abbreviations: N, notochord; FP, floorplate.
2. Processing of GLI proteins into transcriptional repressors
In the absence of Hh signaling, GLI3 as well as the Drosophila Ci are subject to processing by the proteasome into their truncated, transcriptional repressor forms (GLI-R/Ci-R) (Fig. 1) [20–22]. GLI2 has the potential to be processed in a similar fashion but is primarily degraded in the absence of Hh signal [22]. The processing of GLIs is driven by a protein complex containing Suppressor of fused (Sufu) that results in Protein Kinase A (PKA) mediated phosphorylation [23–25]. Both GLI2 and GLI3 have a cluster of six conserved serine residues on the carboxy terminal side of their DNA binding domain (ser1–6). Phosphorylation of the first four serines (ser1–4) by PKA provokes a subsequent cascade of further phosphorylation by GSK3 and Casein Kinase 1 family proteins. The combined activity of these kinases on GLI2/3 and Ci ultimately leads to binding of E3 SCF ubiquitin ligase and processing of GLI2 and GLI3 to their truncated repressor forms [8, 22, 26–32]. The mechanisms by which GLI proteins repress target genes is poorly understood, but includes histone deacetylation [33].
3. Regulated activity of full length GLI proteins
Hh activation prevents processing of GLI2, GLI3, and Ci [19–21, 34, 35]. The resultant full length proteins then undergo additional processing steps that enable them to activate transcription (GLI-A/Ci-A) (Fig. 1). However, multiple mechanisms for modulating full length GLI activity have been described. These mechanisms include binding of proteins that either promote or antagonize the stability of full length GLI proteins [25, 36, 37], cytosolic sequestration [38–40], and differential post-translational modification events [28, 41–44].
4. Phosphorylation state can influence full length GLI activity
Phosphorylation events are implicated in tuning the activation state of full length GLI. For example, PKA phosphorylation on ser6 of the previously mentioned ser1–6 cluster propagates the binding of the 14-3-3 protein to GLI proteins. This interaction decreases the transcriptional activating potential of GLI proteins [45]. The phosphorylation of ser6 by PKA is sensitive to the state of Hh signaling, and therefore has the potential to serve as a method to tune GLI activity according to graded Hh exposure. A second mechanism for how phosphorylation influences the transition of GLI proteins into a fully activated state arose from the identification of a second conserved cluster of 5 phosphorylation sites (c–g) whose phosphorylation state correlates with GLI activity. Although sites c–g were identified as partial consensus sites for PKA, they are not subject to phosphorylation by PKA, and the kinase that phosphorylates these sites has not been identified (Fig. 2) [42]. In contrast to phosphorylation of ser1–6, the phosphorylation of sites c–g is countered by PKA activation and increases in a graded manner in response to graded activation of the Hh pathway [42]. The apparent graded responsiveness of GLI phosphorylation state to graded Hh input provides an attractive model for how incremental changes in Hh have the potential to directly translate into incremental increases in transcriptional activation. Although technically challenging, it will be very interesting to see how differential phosphorylation of GLI activators occurs in response to endogenous Hh gradients and how this phosphorylation changes over time. It is presently not known if analogous mechanisms govern the repressive potency of GLI-R.
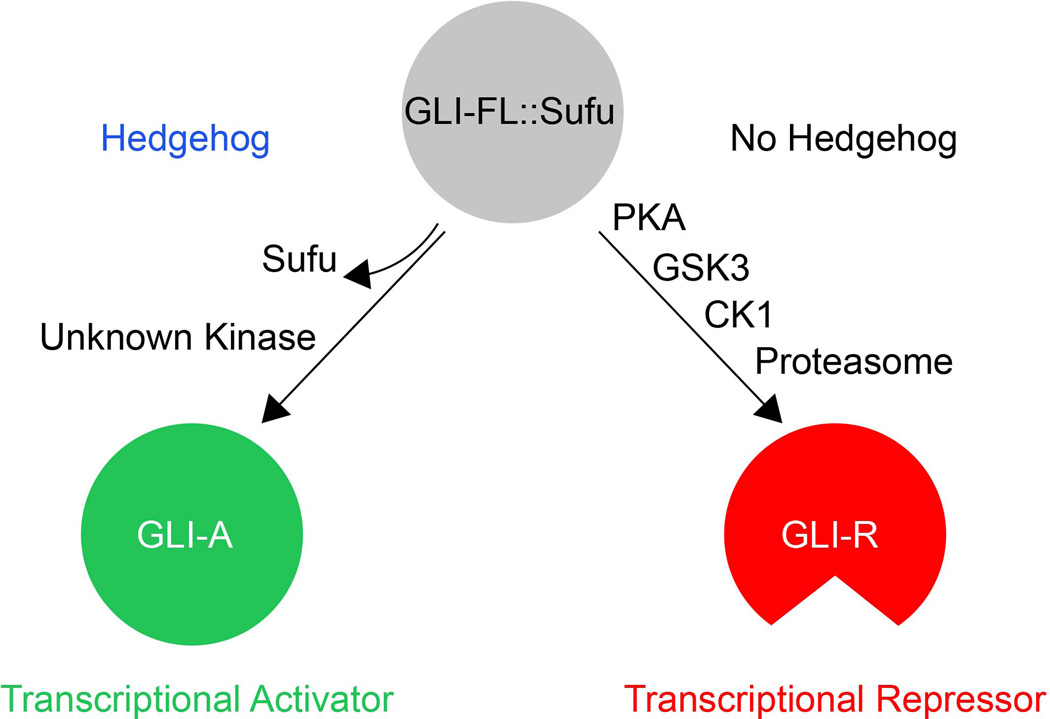
Figure 2. GLI proteins act as context-dependent transcriptional activators or repressors.
In presence of Hh, a full length form of GLI proteins undergoes phosphorylation into a transcriptional activator. In the absence of Hh, they undergo proteolytic cleavage into a truncated transcriptional repressor. Abbreviations: Sufu, suppressor of fused; CK2,; PKA, protein kinase A; GSK3, glycogen synthase kinase 3; CK1, casein kinase 1.
5. Hh driven patterning requires transcriptional activation and de-repression
Transcriptional targets of Hh signaling can broadly be placed in two categories. One set of Hh responsive genes requires transcriptional activation by GLI-A for expression while another set of targets are transcriptionally silenced by GLI-R and only require de-repression for their expression. This latter group is therefore activated by GLI-independent mechanisms, although GLI-A could still potentially regulate quantitative levels of response. While GLI1 and GLI2 serve primarily activating functions [46–49], the repressor function of GLI3 is indispensable to Hh driven patterning [21, 50–52].
6. The role of de-repression in patterning
Limb bud development is severely disrupted in embryos lacking Hh signaling, which have defects that include the loss of all digits except for a rudimentary thumb [53, 54]. These defects are significantly improved in embryos deficient in Hh activity and GLI3, which have largely normal skeletal patterns along with a polydactylous phenotype [54, 55]. Additionally, limb morphology is not disrupted in the absence of Gli1 or Gli2 [56, 57]. These studies indicate that de-repression is a major mechanism controlling limb bud development. Accordingly, expression of an obligate repressor form of GLI3 (GLI3Δ699) influenced both digit identity and number in a dose dependent manner [58]. Additionally, embryos containing an engineered locus that can only generate a repressor form of GLI3 fail to express GLI target genes in the limb bud, including Gremlin [59]. Consistent with this, the major patterning genes regulated by Hh signaling are expressed in the absence of GLI3 repression. These genes include Hand2, 5’HoxA and D family genes, and Gremlin [54, 55]. The symmetrical expression of these genes in embryos lacking both Hh and GLI3 activity suggests that one of the major roles for GLI activators and repressors is to impose asymmetry upon otherwise symmetrical gene expression patterns that ultimately give rise to five digits. Interestingly, embryos in which the activator specific Gli1 is driven by the Gli2 locus have alterations in skeletal identity of anterior digits, suggesting that GLI2 repressor may also play a role in sculpting this gradient [60]. The conversion of GLI3-R to GLI3-A requires minimal stimulation by Hh [22] and, in certain contexts, GLI3-A may serve a passive role in transcriptional activation by relieving the repressive input of GLI-R [50]. However, GLI3-A can also activate transcription directly [61–65] and is essential for Hh responsive transcriptional activation in Gli2 null embryos [60, 66].
7. The importance of transcriptional activation by GLI activator
The conversion of GLI2 to its activator form may require a higher concentration of Hh than GLI3 [22]. Additionally, GLI2 plays a more active role in initiating transcription [46, 47, 57, 67]. In the vertebrate neural tube, disruption of Gli2 disrupts the patterning of tissue types that are located closest to the origin of the Hh signal [46, 57, 67]. The expression patterns of Hh responsive genes that typify these proximal cell types are not disrupted upon perturbation of GLI3 [68, 69]. This is not surprising since GLI3 is primarily a transcriptional repressor and, at high Hh concentrations, only minimal amounts of GLI3 would be converted to its repressor form. Importantly, in both the limb bud and neural tube, the restoration of target genes in Hh deficient Gli3−/− mutants that are lost in Hh deficient mutants does not rescue the spatial expression of these genes, which are expressed in a scattered fashion throughout the neural tube or in a non-polarized fashion in the limb bud [52, 54, 55, 66, 69–71]. These studies suggest that GLI-A is also required for defining spatial patterning.
8. Relative levels of GLI proteins along the Hh gradient
The relative levels of different GLI proteins contribute to the magnitude of the Hh induced transcriptional response. As a testament to this, Gli1−/−;Gli2−/− double mutants have more severe defects than either single mutant. While Ci, GLI2 and GLI3 are present in the absence of Hh signaling, Gli1 is a direct transcriptional target of the Hh pathway [46, 54, 60, 66, 72]. Thus, the amount of GLI-A present in a cell exposed to the Hh signal will initially increase as a result of Hh stimulation, and then decrease with decreasing Hh signaling. The integration of negative feedback loops which dampen Hh signal transduction also antagonize Gli1 transcription [14]. Fluctuations in levels of GLI1 and 2 can influence transcriptional activation in opposition to GLI3 mediated transcriptional repression which can also be influenced by GLI3 protein levels. Importantly, Gli3 is negatively regulated by Hh signaling [15]. Therefore, the relative amount of these three GLI proteins in Hh responding cells has the potential to affect how efficiently those cells transduce the Hh signal. While the relative amounts of GLI 1, 2, and 3 in Hh responding cells has not been reported, concurrent changes in individual Gli mRNA levels have been demonstrated in response to Hh signaling. Analysis of real time transcription of Gli2 and Gli3 in response to Hh gradient over time showed that Gli3 transcription has an inverse relationship with the Hh gradient and that this pattern remains stable over time [9, 51]. In contrast, Gli2 mRNA levels fluctuate over time in response to Hh signal creating temporal variations in overall dosage of GLI-A proteins [9].
9. Deciphering the GLI activation gradient
The most common readouts for GLI activation states are either transcriptional induction of reporter constructs driven by GLI responsive enhancers or western blots showing the relative ratios of truncated (putative repressor) to full length (putative activator) forms of GLI3. Unfortunately, neither of these methods is likely to give an accurate measure of the GLI activation state. As will be discussed in detail below (section 16), enhancer driven reporter assays can only give a rough idea of relative GLI activity because they are taken out of the context of endogenous sequence and are often assayed in unrelated cell types. Western blots indicating the relative ratio of GLI-A to GLI-R are also likely to be a misleading method for determining GLI activity status. It has become increasingly evident that the presence of full length or truncated forms of GLI proteins does not necessarily translate to the amount of activating or repressive potential. It is yet unclear what fraction of the overall GLI pool is functionally inert due to sequestration in the cytosol, differential phosphorylation state, or perhaps other regulatory events. Without this information, it is impossible to know the true ratio of GLI transcriptional activator to repressor. With current approaches, defining the hyper-phosphorylation and/or acetylation state along with nuclear localization might provide better methods of quantifying GLI protein activity. Alternatively, techniques that measure de novo transcription have the potential to provide sharper resolution of temporal transcriptional status [9, 73].
10. Duration of Hh signaling contributes to tissue patterning
The temporal requirement for Hh signaling seems to be central for Hh morphogen patterning programs. A study of how Hh target gene expression patterns are dictated during the development of the Drosophila wing imaginal disc (Fig. 1) used mathematical modeling to demonstrate the need for temporal fluctuations in the Hh gradient [17]. Based on the relationship between a gradient of Hh signal and resultant differential transcriptional outputs, Nahmad et al [17] developed simulations in which the initial Hh gradient expands in a minimally restricted manner to serve as an “on” switch for transcription of Hh responsive targets. The Hh target, ptch, is then up-regulated to spatially temper the Hh signal. This results in three distinct populations of cells; those that never receive the Hh signal, those that are briefly exposed to Hh, and those that are close to the signal origin and experience continuous exposure to Hh [17]. Presumably, Hh targets that require activation by GLI-A for expression would require prolonged Hh signaling, while a different set of Hh targets which do not require GLI-A but are normally repressed by GLI-R might only need temporary de-repression to initiate expression. This differential exposure is hypothesized to be solely responsible for the induction of three unique expression domains [17]. In vivo analysis of differential expression in response to changes in the temporal dynamic of Hh gradient support this model for patterning [17]. Temporal dependence on Hh signaling has also been shown in vertebrate systems. Duration of exposure to Hh as a means for choreographing patterned expression is implicated in digit specification in the developing limb bud as well as the neural tube [74–77].
11. How prolonged Hh signaling translates to GLI activity during neural tube patterning
During ventral neural tube progenitor specification, Hh is produced and secreted from the notochord and forms a gradient along the ventral to dorsal axis (Fig. 1) [78]. As Hh driven patterning persists, progressively more ventral domains take progressively more time to emerge [77, 79]. This temporal pattern of domain specification was demonstrated to be a function of the duration of Hh signaling rather than of a continuing increase in relative concentration along the Hh gradient [76]. Dessaud and colleagues [76, 77] observed the dynamic transcriptional responses of the MN and V3 domains during neural tube development in order to elaborate on this relationship. GLI transcriptional activation potential was shown to be saturated in response to Hh concentrations that induce MN differentiation which occurs just dorsal to the V3 domain. This suggests that cells spanning what will become distinct progenitor domains initially experience maximal GLI activation in response to the Hh signal. Under these saturating conditions, cells that will ultimately contribute to distinct domains are unable to respond differentially to differences in Hh concentration. However, signal receiving cells become progressively insulated against the Hh signal by GLI-A mediated transcription of Ptch. In this de-sensitized state, longer exposure to high concentrations of Hh would be required to maintain maximal GLI transcriptional activation [77]. This requirement for prolonged activation of GLI has the potential to prime cells for a differential transcriptional response to a temporal gradient of Hh exposure. To support this model Dessaud [76] showed that without continued Hh stimulation, cells will revert to a more dorsal identity. The mechanism of this reversion could, in theory, hinge on either reduced GLI-A input or increased repression by GLI-R. In support of the latter, a recent study investigating the distinct contributions of GLI-A and GLI-R to Hh patterning in the neural tube indicate that an initial concentration gradient of Hh establishes a patterning response but that the persistence of this patterning response requires a sustained GLI-R gradient [80]. Presumably, reversion occurs through repressive input winning out over transcriptional activation. While these studies attest to the sensitivity of patterning responses to variations in Hh signal duration, it is not entirely clear how this mechanism of pattern maintenance is balanced with gene regulatory network feedback during patterning. Balaskas et al. [68] suggests that temporal Hh dynamics are integrated with the downstream gene regulatory network to robustly define expression domains. It will be interesting to see how additive repressive inputs that arise from the establishment of gene regulatory networks contribute to the temporal requirement for Hh signaling.
12. How graded GLI activation drives differential transcriptional responses
While the response of genes to high and low levels of Hh signaling appears to be relatively binary, the mechanisms underlying transcriptional response within gradients of Hh signaling are more complex (Fig. 3A). At a temporal level, the inception of Hh signaling will result in a cell that initially contains high levels of GLI-R. Over time the levels of Hh signaling will result in increasing amounts of GLI-A while GLI-R levels are reduced by degradation [74, 81]. Three basic models have been proposed to explain how Hh targets can respond to a gradient of GLI activation state. Ratio sensing models suggest that the relative ratio of GLI-A and GLI-R is integrated at a nuclear level, resulting in a graded transcriptional response to changes in ratio rather than concentration (Fig. 3B) [51, 82–84]. Limb buds from embryos containing both a GLI3-activator-specific allele as well as a GLI3-repressor-specific allele exhibit relatively normal expression domains for most GLI target genes [65]. This model argues against a strict ratio sensing mechanism. However, one caveat is that later studies indicate that the function of both the activator and repressor products are likely attenuated [42, 59]. Threshold activation models predict that threshold-specific concentrations of GLI activators activate target genes (Fig. 3C) [79, 83]. Oosterveen and colleagues found that electroporating an apparently inert GLI protein containing only the DNA binding domain into chick neural tubes resulted in the dorsal expansion of some GLI-A target genes rather than an overall decrease in expression that would indicate a block in direct activation by GLI-A. This experiment suggested that levels of GLI activator were permissive and that response was instead defined by a threshold repression model (Fig. 3D) [50]. It is important to note that the models are not mutually exclusive and different target genes could be regulated by different models in a context dependent fashion.
Figure 3. Mechanisms for transcriptional resolution of opposing GLI gradients.
(A) Graded Hh ligand results in the corresponding activation of graded Gli activator and an inverse gradient of Gli repressor. (B–D) Models by which competition between Gli activators and repressors could result in threshold dependent activation. (B) In a ratio sensing model, the relative ratio between Gli activator and repressor rather than the overall levels drive threshold-specific response. (C) In the threshold activation model, the presence or absence of threshold levels of activation by GLI-A defines a transcriptional response. (D) In the threshold repression model, levels of repression by GLI-R dictate threshold gene response boundaries with Gli activators providing permissive inputs.
The relative contribution of GLI activator versus repressor inputs for patterned expression of Hh target genes has also been described in Drosophila. Hh plays an integral role in defining expression domains during the development of wing imaginal discs. Each disc can be divided into a posterior, Hh secreting, region and an anterior, Hh signal receptive, region of cells (Fig. 1) [85]. Similar to vertebrate tissue patterning models, it is proposed that a gradient of Hh establishes a gradient of Ci-A:Ci-R to drive differential transcriptional responses. A subset of Hh responsive targets in the imaginal disc are ectopically expressed in the absence of all Ci activity, indicating that their expression is solely dependent on de-repression in the absence of Ci-R. Other targets require the presence of Ci for expression and therefore rely on activation by Ci-A, while the correct expression of a third set of targets is governed by a balance of de-repression with activation [21]. The disparate mechanisms governing these Hh target genes implies that they are not all equally responsive to either Ci-A or Ci-R. In support of this model, only 30% of regions bound by Ci-A are also bound by Ci-R [86]. Recent studies [83, 87, 88] hypothesize that binding site affinity along with co-operative Ci-R interactions are responsible for differences in regulatory mechanisms that govern the expression of different Hh responsive genes (discussed in sections 14 and 15).
13. Defining the tissue specific response to a generic GLI activation gradient
Hh signaling is known to drive disparate patterning events regulated by distinct sets of genes in a tissue specific manner. An understanding of how Hh is able to play such a multifaceted role during development is starting to emerge. Sox binding sites are enriched in GLI binding regions that are associated with neural Hh targets. Studies using cis-regulatory module (CRM) driven reporter constructs suggest that the expression of multiple Hh target genes in the ventral neural tube are completely silenced when Sox motifs are mutated, whereas abrogation of the associated GBMs elicits variable reductions in expression depending on the identity of the CRM [9, 50]. Together, these studies support a model where SoxB1 family proteins prime neural specific Hh responsive genes for activation and that the GLI activation gradient provides gated instructive input to direct spatially restricted SoxB1 induced expression domains. Accordingly, ChIP experiments using mouse embryonic limb bud tissue showed that GLI3 inertly binds neural specific GLI enhancers [89]. These neural specific GLI responsive genes can be activated when SoxB1 is expressed with concurrent activation of the Hh pathway [90]. As we explore the cis-regulatory inputs that co-operate with GLI transcription factors, it will be interesting to see if there are different cofactors in other Hh patterned tissues that produce tissue specific responses to Hh signaling.
14. GLI binding motif quality and distinct transcriptional responses
In the case of some morphogens, responsive genes that have high affinity binding motifs are able to respond to lower concentrations of morphogen and are expressed further from the morphogen source. In contrast, morphogen responsive genes with lower affinity binding motifs are only expressed in close proximity to the origin of the morphogen signal where morphogen concentration is at its highest [91–96]. Multiple studies have shown that GLI binding motif (GBM) affinities do not follow this model. Studies in Drosophila [83, 87], chick [50], and mouse [9] indicate that Hh responsive genes that have tightly restricted expression near the source of the Hh signal are associated with high affinity/quality GLI binding motifs while genes that are expressed at longer range tend to be associated with lower affinity GLI binding motifs. Furthermore, increasing the affinity of Ci/GLI binding motifs alters the expression pattern of transcriptional reporter constructs accordingly [9, 50, 87]. The importance of binding motif affinity to transcriptional response is evidenced by high conservation in the quality of non-consensus Ci binding sites among Drosophila species [87]. Analysis of GLI motif driven expression patterns in Drosophila indicate that repressor and activator forms of Ci/GLI have divergent binding motif preferences[87]. Consistent with this, all 3 vertebrate GLI family proteins have comparable binding affinity profiles [9, 10]. However, one study found that a GLI-R bound region acted as a silencer in the mouse limb, but lacked enhancer activity [89].
15. Additive inputs of multiple GLI binding motifs
Adding to the complexity of how variations in GLI binding motifs contribute to the resolution of the GLI activation landscape is the role of multiple GLI binding motifs within a single GLI binding region. In the Drosophila wing imaginal disc, the expression of two well characterized Hh responsive targets, Dpp and Ptch, is driven by Ci binding regions that contain multiple GLI motifs. The differential patterned expression of these Hh targets is dependent on both the relative quality as well as the number of their respective GLI motifs [83]. Thermodynamic modeling of how multiple GLI motifs could mediate the interpretation of a ratio of transcriptional activation and repression into distinct expression patterns suggests that co-operative interactions between Ci/GLI transcription factors play a role in Hh driven patterning [88]. Comparative analysis of expression patterns induced by single or multiple Ci binding motifs suggest that Ci-R but not Ci-A can co-operate to mediate transcriptional regulation [83]. This provides an attractive model for how CRMs respond differentially to Ci-R repressive inputs. The relevance of co-operative GLI input has not been well explored in vertebrate systems. In large scale GLI1 ChIP studies, only 25% of GLI binding regions (GBRs) identified in vertebrates contained multiple GLI motifs [9, 89]. However, these studies focused only on high affinity GLI motifs, likely overlooking lower consensus sites.
16. GLI binding regions are context dependent
While it is clear that both binding motif affinity and number play an integral role in the transcriptional response to Hh, most of the studies investigating how GLI binding motifs respond to Hh gradients rely on synthetic reporter constructs. These methods have the potential to confound a true understanding of how Hh responsive transcription is delegated within the context of the genome. As a testament to this issue, disruption of GLI motifs contained in different CRMs identified for the Hh responsive neural gene, Nkx6.1, have given variable results with respect to the importance of GLI binding for transcriptional activation. While most studies indicate that relieving repressive GLI input is sufficient for Nkx6.1 expression [22, 52, 70, 71], reporter construct assays driven from different putative GLI motif containing CRMs for Nkx6.1 have produced variable results that suggest anywhere from minimal dependence on GLI-A [9] to complete dependence on GLI-A [50]. These individual CRMs almost certainly do not act alone within the context of transcriptional regulation, and it is therefore not surprising that they behave quite differently when isolated. Peterson [9] demonstrated this phenomenon using two GLI binding regions associated with Nkx6.1. Each drove expression within the confines of the normal Nkx6.1 domain, however, they each displayed distinct patterns of expression within this domain. Similarly, the enhancer properties of two GLI binding regions associated with the Hh responsive target, Gremlin, have distinct properties [89, 97]. It is likely that a combination of GLI inputs from multiple enhancer regions coalesce into a defined transcriptional output. Recent studies of how Fgf8 and HoxD expression is regulated provide strong evidence for the long range co-operation of multiple CRMs that additively propagate a transcriptional response [98, 99]. Alternatively, multiple CRMs might act as semi-redundant “shadow elements” [100]. A recent study characterized a GBR associated with the Hh responsive target Gremlin. Besides minor changes to the expression pattern boundary, there was no phenotype when deleting the element by itself. When this element was removed along with one copy of Gli3, there was a loss of most anterior repression, suggesting that this element functions with other uncharacterized GBRs to robustly repress Gremlin [101]. The context within a given CRM may also play a role in contributing to the “enhanceosome”. The enhanceosome model describes the structural composition of an enhancer that results from DNA-protein complexing [102]. This structural composition is suggested to play an integral role in the regulatory behavior of a given enhancer. In support of this model, a minimal sequence contributing to the structure of the ZRS enhancer was recently demonstrated to play a significant role in the long range transcriptional control of Shh in the limb [103]. These studies speak to the limitations of simple CRM driven reporter assays for understanding the chromosomal interactions that motivate distinct transcriptional interpretations of the Hh gradient. In future studies it will be very interesting to determine if and how disparate GBRs interact to influence transcriptional responses.
17. Concluding remarks
In this review, we have highlighted some of the mechanisms by which GLI proteins translate graded Hedgehog signaling into distinct transcriptional responses. In the last few years, a great deal of progress has been made in understanding the mechanisms by which GLI proteins can provide graded transcriptional activity. One of the major current challenges lies in quantitative methods to detect real-time GLI activity within the embryo. The implementation of new approaches would provide mechanistic insight into how GLI proteins respond to signaling within a graded Hh environment. A second challenge lies in understanding the specificity of GLI proteins both in terms of recognizing tissue specific targets and in terms of understanding relative affinities of GLI activator and repressor forms for common binding regions. Increasingly sophisticated genomic studies have already provided some insight and should enable rapid progress in this area. Similar approaches should also shed light on the gene regulatory networks that seem likely to be hardwired into tissue-specific morphogen responses.
Highlights.
New insights into how GLI activity is shaped by a Hh gradient
The relative roles of transcriptional activation and repression by GLI proteins
Models for how GLI proteins interpret the duration of Hh signaling
Three mechanisms for transcriptional response to a GLI activation gradient
GLI binding motif characteristics contribute to transcriptional response
ACKNOWLEDGEMENTS
We would like to thank Dr. Kristin Patterson, Simone Giovanetti, and Paul Oliphint for their comments and suggestions during the preparation of this manuscript. This work was supported by the Cancer Prevention Research Institute of Texas #RP120343 (to S.A.V) and NIH R01HD073151 (to S.A.V)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, et al. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- 2.Gritli-Linde A, Lewis P, McMahon AP, Linde A. The whereabouts of a morphogen: direct evidence for short- and graded long-range activity of hedgehog signaling peptides. Developmental biology. 2001;236:364–386. doi: 10.1006/dbio.2001.0336. [DOI] [PubMed] [Google Scholar]
- 3.Hynes M, Ye W, Wang K, Stone D, Murone M, Sauvage F, et al. The seven-transmembrane receptor smoothened cell-autonomously induces multiple ventral cell types. Nature neuroscience. 2000;3:41–46. doi: 10.1038/71114. [DOI] [PubMed] [Google Scholar]
- 4.Lewis PM, Dunn MP, McMahon JA, Logan M, Martin JF, St-Jacques B, et al. Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell. 2001;105:599–612. doi: 10.1016/s0092-8674(01)00369-5. [DOI] [PubMed] [Google Scholar]
- 5.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nature reviews Genetics. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J, Kato M, Beachy PA. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21666–21671. doi: 10.1073/pnas.0912180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohlen VT, Lessing D, Nusse R, Hooper JE. Hedgehog signaling regulates transcription through Cubitus interruptus, a sequence-specific DNA binding protein. Proc Natl Acad Sci USA. 1997;94:2404–2409. doi: 10.1073/pnas.94.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wen X, Lai CK, Evangelista M, Hongo JA, de Sauvage FJ, Scales SJ. Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Molecular and cellular biology. 2010;30:1910–1922. doi: 10.1128/MCB.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson KA, Nishi Y, Ma W, Vedenko A, Shokri L, Zhang X, et al. Neural-specific Sox2 input and differential Gli-binding affinity provide context and positional information in Shh-directed neural patterning. Genes & development. 2012;26:2802–2816. doi: 10.1101/gad.207142.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, et al. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 11.Kinzler KW, Vogelstein B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Molecular and cellular biology. 1990;10:634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vortkamp A, Gessler M, Grzeschik KH. Identification of optimized target sequences for the GLI3 zinc finger protein. DNA and cell biology. 1995;14:629–634. doi: 10.1089/dna.1995.14.629. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Struhl G. Dual roles for patched in sequestering and transducing Hedgehog. Cell. 1996;87:553–563. doi: 10.1016/s0092-8674(00)81374-4. [DOI] [PubMed] [Google Scholar]
- 14.Goodrich LV, Jung D, Higgins KM, Scott MP. Overexpression of ptc1 inhibits induction of Shh target genes and prevents normal patterning in the neural tube. Developmental biology. 1999;211:323–334. doi: 10.1006/dbio.1999.9311. [DOI] [PubMed] [Google Scholar]
- 15.Marigo V, Tabin CJ. Regulation of patched by sonic hedgehog in the developing neural tube. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9346–9351. doi: 10.1073/pnas.93.18.9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong J, McMahon AP. Growth and pattern of the mammalian neural tube are governed by partially overlapping feedback activities of the hedgehog antagonists patched 1 and Hhip1. Development. 2005;132:143–154. doi: 10.1242/dev.01566. [DOI] [PubMed] [Google Scholar]
- 17.Nahmad M, Stathopoulos A. Dynamic interpretation of hedgehog signaling in the Drosophila wing disc. PLoS biology. 2009;7:e1000202. doi: 10.1371/journal.pbio.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hooper JE, Scott MP. Communicating with Hedgehogs. Nature reviews Molecular cell biology. 2005;6:306–317. doi: 10.1038/nrm1622. [DOI] [PubMed] [Google Scholar]
- 19.Methot N, Basler K. An absolute requirement for Cubitus interruptus in Hedgehog signaling. Development. 2001;128:733–742. doi: 10.1242/dev.128.5.733. [DOI] [PubMed] [Google Scholar]
- 20.Aza-Blanc P, Ramirez-Weber FA, Laget MP, Schwartz C, Kornberg TB. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- 21.Methot N, Basler K. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell. 1999;96:819–831. doi: 10.1016/s0092-8674(00)80592-9. [DOI] [PubMed] [Google Scholar]
- 22.Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Molecular and cellular biology. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes & development. 2010;24:670–682. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svard J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergstrom A, et al. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Developmental cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 25.Wang C, Pan Y, Wang B. Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development. 2010;137:2001–2009. doi: 10.1242/dev.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia J, Zhang L, Zhang Q, Tong C, Wang B, Hou F, et al. Phosphorylation by double-time/CKIepsilon and CKIalpha targets cubitus interruptus for Slimb/beta-TRCP-mediated proteolytic processing. Developmental cell. 2005;9:819–830. doi: 10.1016/j.devcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Smelkinson MG, Kalderon D. Processing of the Drosophila hedgehog signaling effector Ci-155 to the repressor Ci-75 is mediated by direct binding to the SCF component Slimb. Current biology : CB. 2006;16:110–116. doi: 10.1016/j.cub.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 28.Smelkinson MG, Zhou Q, Kalderon D. Regulation of Ci-SCFSlimb binding, Ci proteolysis, and hedgehog pathway activity by Ci phosphorylation. Developmental cell. 2007;13:481–495. doi: 10.1016/j.devcel.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tempe D, Casas M, Karaz S, Blanchet-Tournier MF, Concordet JP. Multisite protein kinase A and glycogen synthase kinase 3beta phosphorylation leads to Gli3 ubiquitination by SCFbetaTrCP. Molecular and cellular biology. 2006;26:4316–4326. doi: 10.1128/MCB.02183-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang J. Regulation of Hh/Gli signaling by dual ubiquitin pathways. Cell cycle. 2006;5:2457–2463. doi: 10.4161/cc.5.21.3406. [DOI] [PubMed] [Google Scholar]
- 31.Wang B, Li Y. Evidence for the direct involvement of {beta}TrCP in Gli3 protein processing. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:33–38. doi: 10.1073/pnas.0509927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang W, Zhao Y, Tong C, Wang G, Wang B, Jia J, et al. Hedgehog-regulated Costal2-kinase complexes control phosphorylation and proteolytic processing of Cubitus interruptus. Developmental cell. 2005;8:267–278. doi: 10.1016/j.devcel.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Feng J, Pan C, Lv X, Wu W, Zhou Z, et al. Atrophin-Rpd3 complex represses Hedgehog signaling by acting as a corepressor of CiR. The Journal of cell biology. 2013;203:575–583. doi: 10.1083/jcb.201306012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aza-Blanc P, Lin HY, Ruiz i, Altaba A, Kornberg TB. Expression of the vertebrate Gli proteins in Drosophila reveals a distribution of activator and repressor activities. Development. 2000;127:4293–4301. doi: 10.1242/dev.127.19.4293. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz i, Altaba A. Pattern formation in the vertebrate neural plate. Trends in neurosciences. 1994;17:233–243. doi: 10.1016/0166-2236(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 36.Seong KH, Akimaru H, Dai P, Nomura T, Okada M, Ishii S. Inhibition of the nuclear import of cubitus interruptus by roadkill in the presence of strong hedgehog signal. PloS one. 2010;5:e15365. doi: 10.1371/journal.pone.0015365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Developmental cell. 2006;10:719–729. doi: 10.1016/j.devcel.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 38.Cho HK, Kim SY, Kim KH, Kim HH, Cheong J. Tumor suppressor protein VHL inhibits Hedgehog-Gli activation through suppression of Gli1 nuclear localization. FEBS letters. 2013;587:826–832. doi: 10.1016/j.febslet.2013.01.050. [DOI] [PubMed] [Google Scholar]
- 39.Maurya AK, Ben J, Zhao Z, Lee RT, Niah W, Ng AS, et al. Positive and negative regulation of Gli activity by Kif7 in the zebrafish embryo. PLoS genetics. 2013;9:e1003955. doi: 10.1371/journal.pgen.1003955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monnier V, Ho KS, Sanial M, Scott MP, Plessis A. Hedgehog signal transduction proteins: contacts of the Fused kinase and Ci transcription factor with the kinesin-related protein Costal2. BMC developmental biology. 2002;2:4. doi: 10.1186/1471-213X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia H, Liu Y, Xia R, Tong C, Yue T, Jiang J, et al. Casein kinase 2 promotes Hedgehog signaling by regulating both smoothened and Cubitus interruptus. The Journal of biological chemistry. 2010;285:37218–37226. doi: 10.1074/jbc.M110.174565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niewiadomski P, Kong JH, Ahrends R, Ma Y, Humke EW, Khan S, et al. Gli protein activity is controlled by multisite phosphorylation in vertebrate hedgehog signaling. Cell reports. 2014;6:168–181. doi: 10.1016/j.celrep.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan Y, Wang C, Wang B. Phosphorylation of Gli2 by protein kinase A is required for Gli2 processing and degradation and the Sonic Hedgehog-regulated mouse development. Developmental biology. 2009;326:177–189. doi: 10.1016/j.ydbio.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuson M, He M, Anderson KV. Protein kinase A acts at the basal body of the primary cilium to prevent Gli2 activation and ventralization of the mouse neural tube. Development. 2011;138:4921–4930. doi: 10.1242/dev.070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asaoka Y, Kanai F, Ichimura T, Tateishi K, Tanaka Y, Ohta M, et al. Identification of a suppressive mechanism for Hedgehog signaling through a novel interaction of Gli with 14-3-3. The Journal of biological chemistry. 2010;285:4185–4194. doi: 10.1074/jbc.M109.038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding Q, Motoyama J, Gasca S, Mo R, Sasaki H, Rossant J, et al. Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development. 1998;125:2533–2543. doi: 10.1242/dev.125.14.2533. [DOI] [PubMed] [Google Scholar]
- 47.Matise MP, Epstein DJ, Park HL, Platt KA, Joyner AL. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development. 1998;125:2759–2770. doi: 10.1242/dev.125.15.2759. [DOI] [PubMed] [Google Scholar]
- 48.Ruiz i, Altaba A. Gli proteins encode context-dependent positive and negative functions: implications for development and disease. Development. 1999;126:3205–3216. doi: 10.1242/dev.126.14.3205. [DOI] [PubMed] [Google Scholar]
- 49.Bai CB, Joyner AL. Gli1 can rescue the in vivo function of Gli2. Development. 2001;128:5161–5172. doi: 10.1242/dev.128.24.5161. [DOI] [PubMed] [Google Scholar]
- 50.Oosterveen T, Kurdija S, Alekseenko Z, Uhde CW, Bergsland M, Sandberg M, et al. Mechanistic differences in the transcriptional interpretation of local and long-range Shh morphogen signaling. Developmental cell. 2012;23:1006–1019. doi: 10.1016/j.devcel.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 51.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 52.Litingtung Y, Chiang C. Specification of ventral neuron types is mediated by an antagonistic interaction between Shh and Gli3. Nature neuroscience. 2000;3:979–985. doi: 10.1038/79916. [DOI] [PubMed] [Google Scholar]
- 53.Chiang C, Litingtung Y, Harris MP, Simandl BK, Li Y, Beachy PA, et al. Manifestation of the limb prepattern: limb development in the absence of sonic hedgehog function. Developmental biology. 2001;236:421–435. doi: 10.1006/dbio.2001.0346. [DOI] [PubMed] [Google Scholar]
- 54.Litingtung Y, Dahn RD, Li Y, Fallon JF, Chiang C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 2002;418:979–983. doi: 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- 55.te Welscher P, Zuniga A, Kuijper S, Drenth T, Goedemans HJ, Meijlink F, et al. Progression of vertebrate limb development through SHH-mediated counteraction of GLI3. Science. 2002;298:827–830. doi: 10.1126/science.1075620. [DOI] [PubMed] [Google Scholar]
- 56.Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- 57.Park HL, Bai C, Platt KA, Matise MP, Beeghly A, Hui CC, et al. Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development. 2000;127:1593–1605. doi: 10.1242/dev.127.8.1593. [DOI] [PubMed] [Google Scholar]
- 58.Hill P, Gotz K, Ruther U. A SHH-independent regulation of Gli3 is a significant determinant of anteroposterior patterning of the limb bud. Developmental biology. 2009;328:506–516. doi: 10.1016/j.ydbio.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 59.Cao T, Wang C, Yang M, Wu C, Wang B. Mouse limbs expressing only the Gli3 repressor resemble those of Sonic hedgehog mutants. Developmental biology. 2013;379:221–228. doi: 10.1016/j.ydbio.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bowers M, Eng L, Lao Z, Turnbull RK, Bao X, Riedel E, et al. Limb anterior-posterior polarity integrates activator and repressor functions of GLI2 as well as GLI3. Developmental biology. 2012;370:110–124. doi: 10.1016/j.ydbio.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. The Journal of biological chemistry. 1999;274:8143–8152. doi: 10.1074/jbc.274.12.8143. [DOI] [PubMed] [Google Scholar]
- 62.Motoyama J, Milenkovic L, Iwama M, Shikata Y, Scott MP, Hui CC. Differential requirement for Gli2 and Gli3 in ventral neural cell fate specification. Developmental biology. 2003;259:150–161. doi: 10.1016/s0012-1606(03)00159-3. [DOI] [PubMed] [Google Scholar]
- 63.Shin SH, Kogerman P, Lindstrom E, Toftgard R, Biesecker LG. GLI3 mutations in human disorders mimic Drosophila cubitus interruptus protein functions and localization. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2880–2884. doi: 10.1073/pnas.96.6.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tyurina OV, Guner B, Popova E, Feng J, Schier AF, Kohtz JD, et al. Zebrafish Gli3 functions as both an activator and a repressor in Hedgehog signaling. Developmental biology. 2005;277:537–556. doi: 10.1016/j.ydbio.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 65.Wang C, Ruther U, Wang B. The Shh-independent activator function of the full-length Gli3 protein and its role in vertebrate limb digit patterning. Developmental biology. 2007;305:460–469. doi: 10.1016/j.ydbio.2007.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bai CB, Stephen D, Joyner AL. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Developmental cell. 2004;6:103–115. doi: 10.1016/s1534-5807(03)00394-0. [DOI] [PubMed] [Google Scholar]
- 67.Matise MP, Lustig M, Sakurai T, Grumet M, Joyner AL. Ventral midline cells are required for the local control of commissural axon guidance in the mouse spinal cord. Development. 1999;126:3649–3659. doi: 10.1242/dev.126.16.3649. [DOI] [PubMed] [Google Scholar]
- 68.Balaskas N, Ribeiro A, Panovska J, Dessaud E, Sasai N, Page KM, et al. Gene regulatory logic for reading the Sonic Hedgehog signaling gradient in the vertebrate neural tube. Cell. 2012;148:273–284. doi: 10.1016/j.cell.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Persson M, Stamataki D, te Welscher P, Andersson E, Bose J, Ruther U, et al. Dorsal-ventral patterning of the spinal cord requires Gli3 transcriptional repressor activity. Genes & development. 2002;16:2865–2878. doi: 10.1101/gad.243402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lei Q, Zelman AK, Kuang E, Li S, Matise MP. Transduction of graded Hedgehog signaling by a combination of Gli2 and Gli3 activator functions in the developing spinal cord. Development. 2004;131:3593–3604. doi: 10.1242/dev.01230. [DOI] [PubMed] [Google Scholar]
- 71.Wijgerde M, McMahon JA, Rule M, McMahon AP. A direct requirement for Hedgehog signaling for normal specification of all ventral progenitor domains in the presumptive mammalian spinal cord. Genes & development. 2002;16:2849–2864. doi: 10.1101/gad.1025702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Panman L, Galli A, Lagarde N, Michos O, Soete G, Zuniga A, et al. Differential regulation of gene expression in the digit forming area of the mouse limb bud by SHH and gremlin 1/FGF-mediated epithelial-mesenchymal signalling. Development. 2006;133:3419–3428. doi: 10.1242/dev.02529. [DOI] [PubMed] [Google Scholar]
- 73.Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Developmental cell. 2009;16:47–57. doi: 10.1016/j.devcel.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 74.Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118:517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 75.Yang Y, Drossopoulou G, Chuang PT, Duprez D, Marti E, Bumcrot D, et al. Relationship between dose, distance and time in Sonic Hedgehog-mediated regulation of anteroposterior polarity in the chick limb. Development. 1997;124:4393–4404. doi: 10.1242/dev.124.21.4393. [DOI] [PubMed] [Google Scholar]
- 76.Dessaud E, Ribes V, Balaskas N, Yang LL, Pierani A, Kicheva A, et al. Dynamic assignment and maintenance of positional identity in the ventral neural tube by the morphogen sonic hedgehog. PLoS biology. 2010;8:e1000382. doi: 10.1371/journal.pbio.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dessaud E, Yang LL, Hill K, Cox B, Ulloa F, Ribeiro A, et al. Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature. 2007;450:717–720. doi: 10.1038/nature06347. [DOI] [PubMed] [Google Scholar]
- 78.Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- 79.Stamataki D, Ulloa F, Tsoni SV, Mynett A, Briscoe J. A gradient of Gli activity mediates graded Sonic Hedgehog signaling in the neural tube. Genes & development. 2005;19:626–641. doi: 10.1101/gad.325905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Su CY, Bay SN, Mariani LE, Hillman MJ, Caspary T. Temporal deletion of Arl13b reveals that a mispatterned neural tube corrects cell fate over time. Development. 2012;139:4062–4071. doi: 10.1242/dev.082321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lum L, Beachy PA. The Hedgehog response network: sensors, switches, and routers. Science. 2004;304:1755–1759. doi: 10.1126/science.1098020. [DOI] [PubMed] [Google Scholar]
- 82.Jacob J, Briscoe J. Gli proteins and the control of spinal-cord patterning. EMBO reports. 2003;4:761–765. doi: 10.1038/sj.embor.embor896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parker DS, White MA, Ramos AI, Cohen BA, Barolo S. The cis-regulatory logic of Hedgehog gradient responses: key roles for gli binding affinity, competition, and cooperativity. Science signaling. 2011;4:ra38. doi: 10.1126/scisignal.2002077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruiz i, Altaba A. Catching a Gli-mpse of Hedgehog. Cell. 1997;90:193–196. doi: 10.1016/s0092-8674(00)80325-6. [DOI] [PubMed] [Google Scholar]
- 85.Lawrence PA, Struhl G. Morphogens, compartments, and pattern: lessons from drosophila? Cell. 1996;85:951–961. doi: 10.1016/s0092-8674(00)81297-0. [DOI] [PubMed] [Google Scholar]
- 86.Biehs B, Kechris K, Liu S, Kornberg TB. Hedgehog targets in the Drosophila embryo and the mechanisms that generate tissue-specific outputs of Hedgehog signaling. Development. 2010;137:3887–3898. doi: 10.1242/dev.055871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ramos AI, Barolo S. Low-affinity transcription factor binding sites shape morphogen responses and enhancer evolution. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2013;368:20130018. doi: 10.1098/rstb.2013.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.White MA, Parker DS, Barolo S, Cohen BA. A model of spatially restricted transcription in opposing gradients of activators and repressors. Molecular systems biology. 2012;8:614. doi: 10.1038/msb.2012.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vokes SA, Ji H, Wong WH, McMahon AP. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes & development. 2008;22:2651–2663. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Oosterveen T, Kurdija S, Enstero M, Uhde CW, Bergsland M, Sandberg M, et al. SoxB1-driven transcriptional network underlies neural-specific interpretation of morphogen signals. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7330–7335. doi: 10.1073/pnas.1220010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Driever W, Thoma G, Nusslein-Volhard C. Determination of spatial domains of zygotic gene expression in the Drosophila embryo by the affinity of binding sites for the bicoid morphogen. Nature. 1989;340:363–367. doi: 10.1038/340363a0. [DOI] [PubMed] [Google Scholar]
- 92.Jiang J, Kosman D, Ip YT, Levine M. The dorsal morphogen gradient regulates the mesoderm determinant twist in early Drosophila embryos. Genes & development. 1991;5:1881–1891. doi: 10.1101/gad.5.10.1881. [DOI] [PubMed] [Google Scholar]
- 93.Jiang J, Levine M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell. 1993;72:741–752. doi: 10.1016/0092-8674(93)90402-c. [DOI] [PubMed] [Google Scholar]
- 94.Papatsenko D, Levine M. Quantitative analysis of binding motifs mediating diverse spatial readouts of the Dorsal gradient in the Drosophila embryo. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4966–4971. doi: 10.1073/pnas.0409414102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thisse C, Perrin-Schmitt F, Stoetzel C, Thisse B. Sequence-specific transactivation of the Drosophila twist gene by the dorsal gene product. Cell. 1991;65:1191–1201. doi: 10.1016/0092-8674(91)90014-p. [DOI] [PubMed] [Google Scholar]
- 96.Wharton SJ, Basu SP, Ashe HL. Smad affinity can direct distinct readouts of the embryonic extracellular Dpp gradient in Drosophila. Current biology : CB. 2004;14:1550–1558. doi: 10.1016/j.cub.2004.08.053. [DOI] [PubMed] [Google Scholar]
- 97.Zuniga A, Laurent F, Lopez-Rios J, Klasen C, Matt N, Zeller R. Conserved cis-regulatory regions in a large genomic landscape control SHH and BMP-regulated Gremlin1 expression in mouse limb buds. BMC developmental biology. 2012;12:23. doi: 10.1186/1471-213X-12-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Marinic M, Aktas T, Ruf S, Spitz F. An integrated holo-enhancer unit defines tissue and gene specificity of the Fgf8 regulatory landscape. Developmental cell. 2013;24:530–542. doi: 10.1016/j.devcel.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 99.Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, et al. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 100.Frankel N. Multiple layers of complexity in cis-regulatory regions of developmental genes. Developmental dynamics : an official publication of the American Association of Anatomists. 2012;241:1857–1866. doi: 10.1002/dvdy.23871. [DOI] [PubMed] [Google Scholar]
- 101.Li Q, Lewandowski JP, Powell MB, Norrie JL, Cho SH, Vokes SA. A Gli silencer is required for robust repression of gremlin in the vertebrate limb bud. Development. 2014;141:1906–1914. doi: 10.1242/dev.104299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Panne D. The enhanceosome. Current opinion in structural biology. 2008;18:236–242. doi: 10.1016/j.sbi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 103.Lettice LA, Williamson I, Devenney PS, Kilanowski F, Dorin J, Hill RE. Development of five digits is controlled by a bipartite long-range cis-regulator. Development. 2014;141:1715–1725. doi: 10.1242/dev.095430. [DOI] [PMC free article] [PubMed] [Google Scholar]