Abstract
Two libraries of modestly reactive ureas containing either electron-deficient acyl anilines or acyl pyrazoles were prepared and are reported as screening libraries for candidate serine hydrolase inhibitors. Within each library is a small but powerful subset of compounds that serve as a chemotype fragment screening library capable of subsequent diversification. Elaboration of the pyrazole-based ureas provided remarkably potent irreversible structural inhibitors of fatty acid amide hydrolase (FAAH, apparent Ki = 100-200 pM) complementary to those previously disclosed enlisting electron-deficient aniline-based ureas.
The serine hydrolases constitute a superfamily of enzymes representing more than 1% of the predicted proteins in the human genome for which at least 40-50% are still uncharacterized, lacking known roles or identified endogenous substrates.1 Not only are they one of the largest and most diverse enzyme class in mammals, but they perform crucial roles in many biological processes.1,2 There are approximately 240 human serine hydrolases composed of a series of lipases, peptidases, esterases, thioesterases, and amidases that hydrolyze small molecules, signaling lipids, peptides or post-translational ester and thioester protein modifications. They share a conserved mechanism that enlists a key active site serine nucleophile within a catalytic triad that is activated by a proton relay often involving an acidic (aspartate/glutamate) and basic residue (histidine/lysine). More than half of the serine hydrolases (>120 enzymes) remain poorly annotated, with no described physiological function or identified substrates, and an even greater number of these enzymes (>80%) lack selective inhibitors to aid in their characterization. In cases where a comprehensive understanding has been established, this has not only improved our fundamental understanding of biology, but it has also led to invaluable new therapies to treat disease.1
Herein, we report two distinct efforts designed to assemble a powerful screening library targeting the serine hydrolases. As a complement to our early ad hoc synthesis of a trifluoromethyl ketone library (ca. 300 compounds),3 additional candidate inhibitors bearing activated electrophilic carbonyls,4 and the more recent synthesis of an extensive series of α-ketoheterocycles5 (ca. 1000 compounds) many of which target fatty acid amide hydrolase (FAAH),6 the efforts herein provide two libraries of modestly reactive urea derivatives capable of irreversible7 serine hydrolase inhibition by virtue of selective active site carbamoylation of the catalytic serine.8 Such ureas, typically unreactive unless activated for nucleophilic acylation within a serine hydrolase active site, can be profiled against all serine hydrolases in the proteome9 with confidence that they are not routinely reactive with enzymes or proteins outside the serine hydrolase family.9,10 Because of their irreversible mechanism of action, those that are sufficiently selective may serve as initial in vivo pharmacological probes to define the function of an uncharacterized serine hydrolase,11 used to confidently validate its potential as a therapeutic target, and optimized into drug candidates themselves.1,12-15 First identified as a class of inhibitors effective for selective and potent inhibition of FAAH,12-15 their utility for serine hydrolase inhibition requires a modestly reactive urea bearing an amine capable of behaving as an effective leaving group (e.g., electron-deficient aniline,13-15 tetrazole,16 triazole,17 or imidazole17), was found often to be dependent on the nature, size, and substitution (e.g., tertiary vs secondary) of the second attached urea amine,13-15 and can be exquisitely responsive to the recognition elements used to target the enzyme active site. Opportunities to create such comprehensive screening libraries for the serine hydrolases are enhanced with the availability and continued refinement of activity-based protein profiling (ABPP) proteomic screening technology9-11 that offer the advantage of testing enzymes in their native state and eliminate the need for their recombinant expression, purification, and the development of specific substrate assays. Because the inhibitors are screened against many enzymes in the proteome in parallel, both their potency and selectivity can be simultaneously evaluated.18,19 Such libraries, in combination with use of this technology to rapidly assess proteome-wide selectivity, provide a powerful paradigm for discovery of selective chemical probes of new targets, for optimization of an enzyme inhibitor selectivity concurrent with target affinity,19 and for the detection, identification, and characterization of new therapeutic targets.1,2
The first of the screening libraries detailed herein constitutes a 693-membered library of activated ureas that systematically vary the reactivity of an aniline leaving group (head group) as well as the structure and flexibility of the additional urea amine (linking unit) that bears a common resident functionalization site amenable to a final divergent introduction of a variable unit capable of modulating active site recognition (Figure 1). In addition to providing a template that may be rapidly expanded following lead identification using a solution-phase synthetic protocol for the parallel synthesis of individual compounds where each intermediate and final product is isolated and purified by a simple acid/base liquid/liquid extraction,20 the library immediately informs on the influence of the urea reactivity, the importance of the linker and its rigidity or orientation, and the nature of the active site recognition features.
Figure 1.
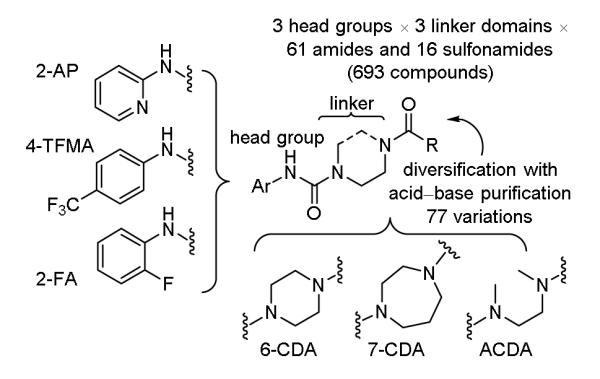
Aniline-based urea library of candidate serine hydrolase inhibitors.
Three leaving group amines were selected for incorporation into the library: 2-aminopyridine (2-AP), 4-trifluoromethylaniline (4-TFMA), and 2-fluoroaniline (2-FA). Both the former 2-AP13d and the latter 2-FA14a,d have been examined in series irreversibly targeting FAAH, whereas 4-TFMA appears unique to the studies herein (Figure 1, head group). A set of three linker domains were incorporated as mono Boc protected derivatives of three symmetrical diamines: a six-membered cyclic diamine (6-CDA, piperazine) widely used in past efforts,13-15 a seven-membered cyclic diamine (7-CDA, 1,4-diazepine or homopiperazine), and an acyclic diamine (ACDA, N,N’-dimethyl-1,2-diaminoethane), each of which bear a protected amine spatially proximal within the series for subsequent divergent functionalization (Figure 1, linker group). The nine individual combinations of the three head groups and three linkers were assembled using one of two protocols (Scheme 1). The first (method A) illustrated with 1, entailed treatment of 4-Boc-piperazine with carbonyldiimidazole (CDI, THF, 25 °C, 14 h) to provide the activated urea (93%) followed by MeI treatment (4 equiv MeI, CH3CN, 25 °C, 24 h) to provide the N-methyl imidazolium salt (quant.) activated for subsequent displacement.21 Its treatment with the lithium salt of 2-aminopyridine generated in situ (n-BuLi, THF) provided 1 (55%). Alternatively and in a procedure that proved more general and effective for all nine intermediates (method B), treatment of 2-AP (58%), 4-TFMA (97%), or 2-FA (88%) with 2-propenyl chloroformate (N-methylmorpholine, THF)22 provided the corresponding activated 2-propenyl carbamates that undergo smooth urea formation upon reaction with the mono Boc protected diamines (N-methylpyrrolidine, THF, reflux, 57-70%).
Scheme 1.
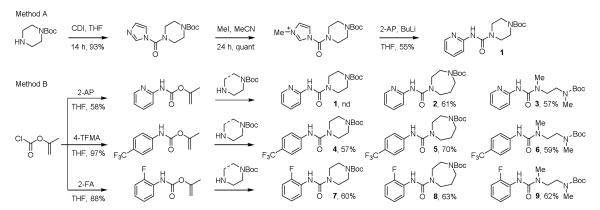
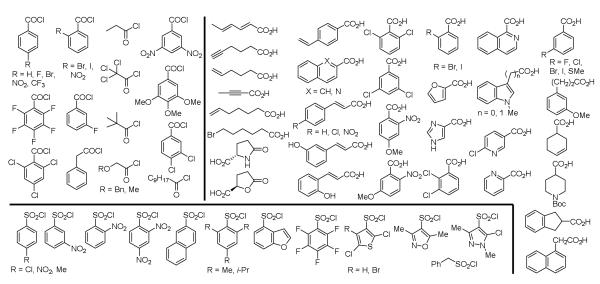
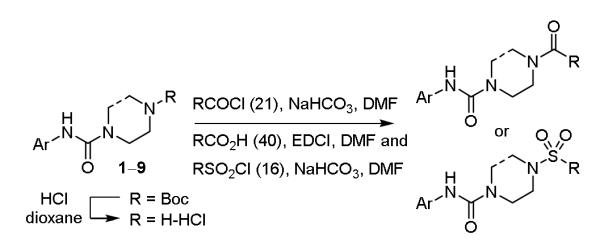
Each of the nine core structures (1-9) was Boc deprotected by treatment with anhydrous HCl in dioxane (25 °C) and each crude amine hydrochloride salt was subjected to coupling (30 mg/reaction) with the identical series (Figure 2) of 21 acid chlorides (2 equiv, 10 equiv NaHCO3, 0.2 M in DMF, 0 to 25 °C, 12 h), 40 carboxylic acids (1 equiv, 1.2 equiv EDCI, 1 equiv HOAt, 10 equiv NaHCO3, 0.2 M in DMF, 25 °C, 24 h), and 16 arylsulfonyl chlorides (2 equiv, 10 equiv NaHCO3, 0.2-0.4 M in DMF, 25 °C, 12 h) to provide nine sublibraries, each composed of 77 compounds (61 amides and 16 sulfonamides) (Scheme 2). Isolation and purification of the final products was conducted by simple acid/base extraction of the crude reaction mixture following aqueous workup for removal of excess reagents, their byproducts, and unreacted starting materials. Assessment of the 693-membered library by LCMS revealed that nearly all expected products were generated (ca. random 5-10% failure rate in each sublibrary) and a subsequent chromatographic purification of the individual compounds provided the individual library members in purified amounts ranging from 1-33 mg (>95% purity).
Figure 2.
The 21 acid chlorides, 40 carboxylic acids, and 16 sulfonyl chlorides used in the library.
Scheme 2.
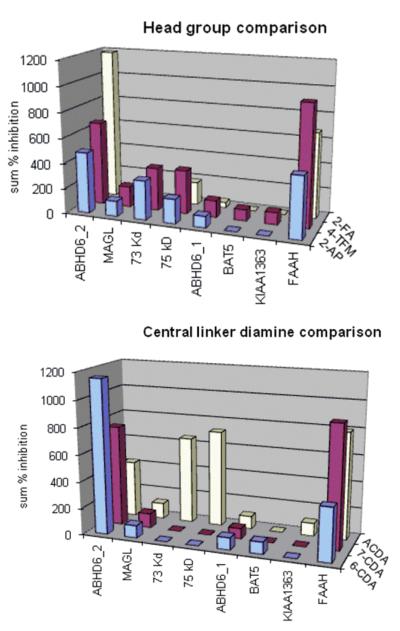
In principle, the systematic variations in the candidate inhibitors provide an initial SAR immediately following its screening that informs the subsequent optimization of screening hits. Illustrating this feature of the library, a small screening campaign with the library (screened at 5 μM) was conducted against a modestly sized serine hydrolase library to establish its utility. In addition to providing a distinct lead compound for inhibiting the individual serine hydrolases, the informative and digested screening data (sum % inhibition) for the library against the eight serine hydrolases is summarized in Figure 3. Using as an example the serine hydrolase ABHD6-2 that is upregulated in metastatic tumors, both the leaving group aniline (2-FA > 4-TFMA > 2-AP) and the linker domain (6-CDA > 7-CDA > ACDA) follow well-defined trends that can be exploited immediately in ongoing second generation optimization libraries directly following a screening campaign.
Figure 3.
Digested data (summed % inhibition) from the urea compound library screening (at 5 μM) against a panel of eight serine hydrolases in the proteome.
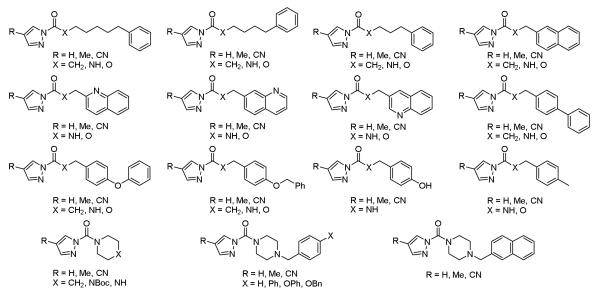
The second library evolved in a different manner and ultimately resulted in a library designed to screen for minimal, more promiscuous serine hydrolase inhibitors capable of subsequent immediate lead optimization. In the course of efforts targeting FAAH,23,24 we examined a series of N-acyl pyrazole amides, carbamates and ureas as candidate inhibitors of the enzyme. Precedent for their potential utility was based on the reported behavior of related azoles (tetrazoles,16 triazoles,17 imidazoles17, and indazoles17), although we were not aware of detailed accounts of N-acyl pyrazole ureas or carbamates reacting with serine hydrolases.25 The candidate acyl pyrazoles display an intrinsic modest reactivity toward nucleophilc attack and represent an activating group capable of formation of a metal-chelated stable tetrahedral intermediate. Additionally, the pyrazoles represent a series in which its acylating reactivity and leaving group propensity may be tuned not only through the nature of the second acyl site (amide, carbamate, urea), but also through symmetrical substitution of the pyrazole with electron-withdrawing or electron-donating substituents. The importance and remarkable impact of these features were examined and defined with a series of 111 acyl pyrazoles as inhibitors of FAAH (Figure 4), a collection in itself that represents a useful serine hydrolase inhibitor screening library.
Figure 4.
N-Acyl pyrazole amides, carbamates, and ureas examined against FAAH.
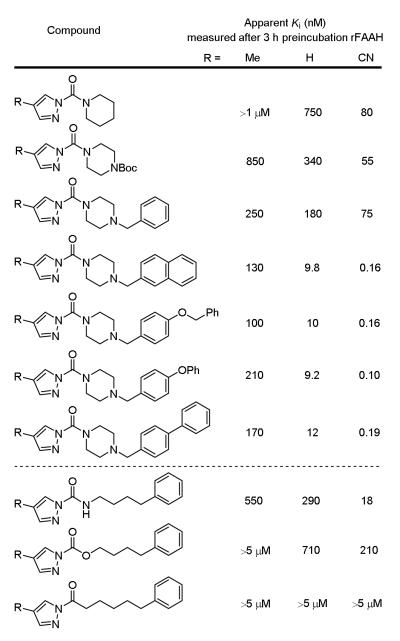
Several key structural features emerged from examination of the small library where the impact on FAAH inhibitory activity displayed clear pronounced effects, improving substantially as one alters both the nature of the reacting acyl group (urea > carbamate > amide) and the pyrazole C4 substituent (CN > H > Me). This is illustrated in Figure 5 with a representative series of increasingly potent FAAH inhibitors, the most potent of which display apparent Ki values of 100-200 pM when substituted with acyl chains known to be especially effective for targeting FAAH.5 Two of these potent series, the three pyrazole ureas bearing the naphthyl tail as well as the three containing the benzyloxyphenyl tail, were each confirmed to be irreversible FAAH inhibitors (dialysis dilution, not shown). Interestingly, it was the least reactive chemotype (urea > carbamate > amide) that proved to provide the more potent inhibitors, suggesting a unique FAAH active site activation for carbamoylation.
Figure 5.
Recombinant rFAAH inhibition, apparent Ki (3 h preincubation with enzyme).
Especially interesting in the series was the unusually large and remarkable impact of the pyrazole C4 substituent. Although this effect could reflect both an electronic and steric impact of the substituent on the activity, FAAH possess a large cytosolic port adjacent to the active site capable of accommodating large substituted heterocycles,26 suggesting the impact is largely or solely due to the electronic properties of the pyrazole C4 substituent. In order to establish whether this truly reflects the electronic impact of the substituent, an expanded series of substituted pyrazoles was examined in which the electron-withdrawing properties of the C4 substituent was systematically varied without significantly altering the substituent size or polarity (Figure 6). Beautifully, the series exhibited pronounced and incremental increases in potency as the electron-withdrawing properties of the C4 substituent increases (CN > CO2Me > Br > H > Me), where the change in apparent Ki values across the series spans a stunning 103-fold.
Figure 6.
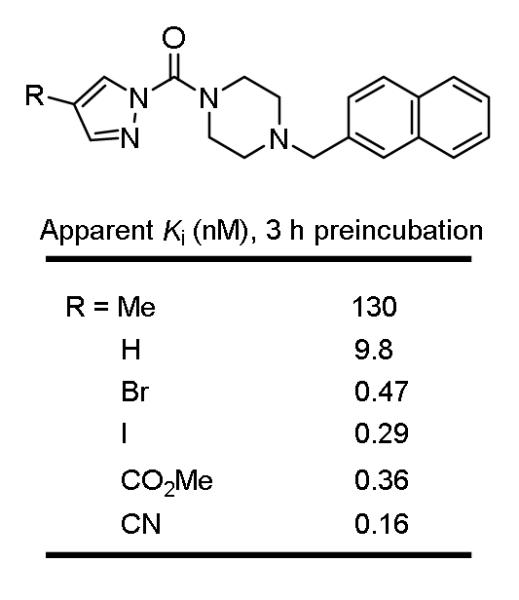
C4-Substituent impact on pyrazole urea inhibition of FAAH.
Notable in this work, the pyrazole ureas 10-12 that contain only a simple piperidine displayed robust inhibitory activity against FAAH even though they do not contain a recognition group mimicking the substrate27,28 fatty acid amide long chain acyl groups (Figure 5). Rather, they and a small series of pyrazole ureas including those containing pendant protected secondary amines in the acyl chain may serve as effective screening compounds used to identify chemotypes selective for inhibition of a subset of serine hydrolases, being more promiscuous across the enzyme class (10-21, Figure 7).17a Combined with 1-9, 10-21 represent a small, but powerful serine hydrolase screening library. Following detection of a targeted serine hydrolase inhibition, the informative screening leads, which define trends in acyl urea reactivity (head group) and the importance of the second attached amine (linker), may be divergently modified to improve active site recognition features, simultaneously improving potency and selectivity for a targeted serine hydrolase.
Figure 7.
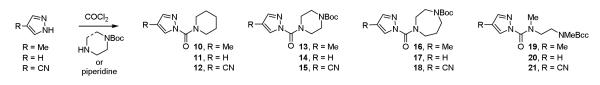
Key members of a serine hydrolase screening library.
Representative of such screening results and highlighted in Figure 8, the comparison of 10 and 12 screened at 20 μM against all proteins in the HeLa cell membrane fraction indicate the more selective nature of the least reactive pyrazole urea 10 and the more sensitive nature of the most reactive pyrazole urea 12. Combined their concurrent screening allows the identification of serine hydrolases sensitive to inhibition by the pyrazole urea chemotype, provide insights into how reactivity can impact selectivity among the distinguished candidate serine hydrolases, define candidate off target enzymes in the class whose activity should be monitored when addressing optimization against a single subsequently chosen enzyme, and provide superb starting fragments that can be further elaborated to tailor the potency and selectivity of the inhibitor class to a targeted serine hydrolase. Most notably and by using the nitrile substituted pyrazole urea 12, serine hydrolases sensitive to pyrazole urea inhibition are detected that would be overlooked in such fragment screening if only the unsubstituted pyrazole 10 was employed.
Figure 8.
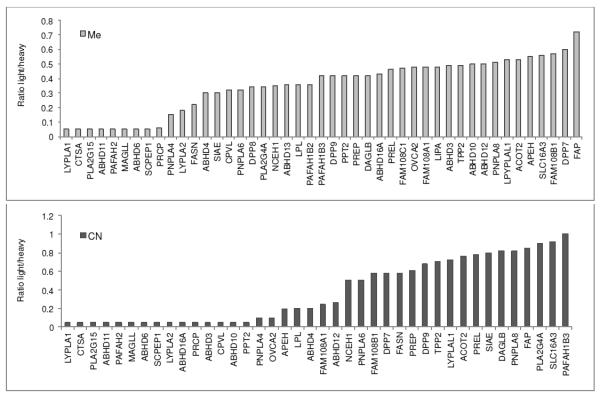
Identification of serine hydrolase targets for 10 (top) and 12 (bottom) in the HeLa cell membrane protein fraction established by ABPP-SILAC.17a Cells were cultured with each inhibitor at 20 μM (or DMSO controls) for 4 h before isolation of the membrane proteins and analysis by ABPP-SILAC (two independent replicates, FP-biotin probe9b,17a). Ratio light/heavy = 1, no inhibition; ratio = 0, complete inhibition. Only those serine hydrolases displaying some inhibition are shown.
Complementary to our screening libraries comprehensively targeting protein-protein29-31 or protein-DNA32 interactions, the urea-based serine hydrolase libraries disclosed herein and the related libraries of candidate inhibitors bearing electrophilic carbonyls3,4 represent our first of several screening libraries assembled to effectively target the major enzyme classes. The results of these and related studies, as well as their utility will be reported in due course.
Supplementary Material
Acknowledgement
This work was supported by the National Institutes of Health (DA015648). We thank Landon Whitby for digestion of the screening results obtained with the aniline urea library and William Parsons and Professor B. F. Cravatt for assistance with the ABPP-SILAC.
Footnotes
Supporting Information. Supplementary data associated with this article include full details of the synthesis, characterization and purity of the screening library and can be found in the online version at xxxxx.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Bachovchin DA, Cravatt BF. Nat. Drug Disc. Rev. 2012;11:52. doi: 10.1038/nrd3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long JZ, Cravatt BF. Chem, Rev. 2011;111:6022. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boger DL, Sato H, Lerner AE, Austin BJ, Patterson JE, Patricelli MP, Cravatt BF. Bioorg. Med. Chem. Lett. 1999;9:265. doi: 10.1016/s0960-894x(98)00734-3. [DOI] [PubMed] [Google Scholar]
- 4 (a).Patterson JE, Ollmann IR, Cravatt BF, Boger DL, Wong C-H, Lerner RA. J. Am. Chem. Soc. 1996;118:5938. [Google Scholar]; (b) Patricelli MP, Patterson JP, Boger DL, Cravatt BF. Bioorg. Med. Chem. Lett. 1998;8:613. doi: 10.1016/s0960-894x(98)00073-0. [DOI] [PubMed] [Google Scholar]
- 5 (a).Boger DL, Sato H, Lerner AE, Hedrick MP, Fecik RA, Miyauchi H, Wilkie GD, Austin BJ, Patricelli MP, Cravatt BF. Proc. Natl. Acad. Sci. USA. 2000;97:5044. doi: 10.1073/pnas.97.10.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Boger DL, Miyauchi H, Hedrick MP. Bioorg. Med. Chem. Lett. 2001;11:1517. doi: 10.1016/s0960-894x(01)00211-6. [DOI] [PubMed] [Google Scholar]; (c) Boger DL, Miyauchi H, Du W, Hardouin C, Fecik RA, Cheng H, Hwang I, Hedrick MP, Leung D, Acevedo O, Guimarães CRW, Jorgensen WL, Cravatt BF. J. Med. Chem. 2005;48:1849. doi: 10.1021/jm049614v. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Romero FA, Hwang I, Boger DL. J. Am. Chem. Soc. 2006;68:14004. doi: 10.1021/ja064522b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Boger DL, Miyauchi H, Hedrick MP, Romero FA, Du W, Hwang I, Rayl TJ, Kimball FS, Leung D, Hoover HS, Apodaca RL, Breitenbucher JG, Cravatt BF, Boger DL. J. Med. Chem. 2007;50:1058. doi: 10.1021/jm0611509. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Hardouin C, Kelso MJ, Romero FA, Rayl TJ, Leung D, Hwang I, Cravatt BF, Boger DL. J. Med. Chem. 2007;50:3359. doi: 10.1021/jm061414r. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Kimball FS, Romero FA, Ezzili C, Garfunkle J, Rayl TJ, Hochstatter DG, Hwang I, Boger DL. J. Med. Chem. 2008;51:937. doi: 10.1021/jm701210y. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Garfunkle J, Ezzili C, Rayl TJ, Hochstatter DG, Hwang I, Boger DL. J. Med. Chem. 2008;51:4392. doi: 10.1021/jm800136b. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) DeMartino JK, Garfunkle J, Hochstatter DG, Cravatt BF, Boger DL. Bioorg. Med. Chem. Lett. 2008;18:5842. doi: 10.1016/j.bmcl.2008.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]; (j) Ezzili C, Mileni M, McGlinckey N, Long JZ, Kinsey SG, Hochstatter DG, Stevens RC, Lichtman AH, Cravatt BF, Bilsky EJ, Boger DL. J. Med. Chem. 2011;54:2805. doi: 10.1021/jm101597x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) Otrubova K, Brown M, McCormick MS, Han GW, O’Neal ST, Cravatt BF, Stevens RC, Lichtman AH, Boger DL. J. Am. Chem. Soc. 2013;135:6289. doi: 10.1021/ja4014997. [DOI] [PMC free article] [PubMed] [Google Scholar]; (l) Otrubova K, Cravatt BF, Boger DL. J. Med. Chem. 2014;57:1079. doi: 10.1021/jm401820q. [DOI] [PMC free article] [PubMed] [Google Scholar]; (m) Duncan KK, Otrubova K. Bioorg. Med. Chem. 2014;22:2763. doi: 10.1016/j.bmc.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otrubova K, Boger DL. ACS Chem. Neurosci. 2012;3:340. doi: 10.1021/cn2001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powers JC, Asgian JL, Ekici OD, James KE. Chem. Rev. 2002;102:4639. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 8 (a).Alexander JP, Cravatt BF. J. Am. Chem. Soc. 2006;128:9699. doi: 10.1021/ja062999h. [DOI] [PubMed] [Google Scholar]; (b) Alexander J, Cravatt BF. Chem. Biol. 2005;12:1179. doi: 10.1016/j.chembiol.2005.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9 (a).Liu Y, Patricelli MP, Cravatt BF. Proc. Natl. Acad. Sci. USA. 1999;96:14694. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Patricelli MP, Cravatt BF. Biochemistry. 1999;38:14125. doi: 10.1021/bi991876p. [DOI] [PubMed] [Google Scholar]
- 10 (a).Barglow KT, Cravatt BF. Nat. Methods. 2007;4:822. doi: 10.1038/nmeth1092. [DOI] [PubMed] [Google Scholar]; (b) Evans MJ, Cravatt BF. Chem. Rev. 2006;106:3279. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- 11 (a).Li W, Blankman JL, Cravatt BF. J. Am. Chem. Soc. 2007;129:9594. doi: 10.1021/ja073650c. [DOI] [PubMed] [Google Scholar]; (b) Chiang KP, Niessen S, Saghatelian A, Cravatt BF. Chem. Biol. 2006;13:1041. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Otrubova K, Ezzili C, Boger DL. Bioorg. Med. Chem. Lett. 2011;21:4674. doi: 10.1016/j.bmcl.2011.06.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13 (a).Ahn K, Johnson DS, Fitzgerald LR, Liimatta M, Arendse A, Stevenson T, Lund ET, Nugent RA, Normanbhoy T, Alexander JP, Cravatt BF. Biochemistry. 2007;46:13019. doi: 10.1021/bi701378g. [DOI] [PubMed] [Google Scholar]; (b) Johnson DS, Ahn K, Kesten S, Lazerwith SE, Song Y, Morris M, Fay L, Gregory T, Stiff C, Dunbar JB, Jr., Liimatta M, Beidler D, Smtih S, Nomanbhoy TK, Cravatt BF. Bioorg. Med. Chem. Lett. 2009;19:2865. doi: 10.1016/j.bmcl.2009.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ahn K, Johnson DS, Mileni M, Beidler D, Long JZ, McKinney MK, Weerapana E, Sadagopan N, Liimatta M, Smith SE, Lazerwith S, Stiff C, Kamtekar S, Bhattacharya K, Zhang Y, Swaney S, Van Becelaere K, Stevens RC, Cravatt BF. Chem. Biol. 2009;16:411. doi: 10.1016/j.chembiol.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Johnson DS, Stiff C, Lazerwith SE, Kesten SR, Fay LK, Morris M, Beidler D, Liimatta MB, Smith SE, Dudley DT, Sadagopan N, Bhattachar SN, Kesten SJ, Nomanbhoy TK, Cravatt BF, Ahn K. ACS Med. Chem. Lett. 2011;2:91. doi: 10.1021/ml100190t. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Meyers JM, Long AS, Pelc JM, Wang LJ, Bowen JS, Walker CM, Schweitzer AB, Madsen MH, Tenbrink ER, McDonald J, Smith ES, Foltin S, Beidler F, Thorarensen A. Bioorg. Med. Chem. Lett. 2011;21:6538. doi: 10.1016/j.bmcl.2011.08.055. [DOI] [PubMed] [Google Scholar]; (f) Meyers JM, Long AS, Pelc JM, Wang LJ, Bowen JS, Schweitzer AB, Wilcox CM, McDonald J, Smith ES, Foltin S, Rumsey J, Yang SY, Walker CM, Kamtekar S, Beidler F, Thorarensen A. Bioorg. Med. Chem. Lett. 2011;21:6545. doi: 10.1016/j.bmcl.2011.08.048. [DOI] [PubMed] [Google Scholar]
- 14 (a).Keith JM, Apocada R, Xiao W, Seierstad M, Pattabiraman K, Wu J, Webb M, Karbarz MJ, Brown S, Wilson S, Scott B, Tham C-S, Luo L, Palmer J, Wennerholm M, Chaplan S, Breitenbucher JG. Bioorg. Med. Chem. Lett. 2008;18:4838. doi: 10.1016/j.bmcl.2008.07.081. [DOI] [PubMed] [Google Scholar]; (b) Karbarz MJ, Luo L, Chang L, Tham C-S, Palmer JA, Wilson SJ, Wennerholm ML, Brown SM, Scott BP, Apocada RL, Keith JM, Wu J, Breitenbucher JG, Chaplan SR, Webb M. Anesth. Analg. 2009;108:316. doi: 10.1213/ane.0b013e31818c7cbd. [DOI] [PubMed] [Google Scholar]; (c) Keith JM, Apodaca R, Tichenor M, Xiao W, Jones W, Pierce J, Seierstad M, Palmer J, Webb M, Karbarz M, Scott B, Wilson S, Luo L, Wennerholm M, Chang L, Brown S, Rizzolio M, Rynberg R, Chaplan S, Breitenbucher JG. ACS Med. Chem. Lett. 2012;3:823. doi: 10.1021/ml300186g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Tichenor MS, Keith JM, Jones WM, Pierce JM, Merit J, Hawryluk N, Seierstad M, Palmer JA, Webb M, Karbarz MJ, Wilson SJ, Wennerholm ML, Woestenborghs F, Beerens D, Luo L, Brown SM, De Boeck M, Chaplan SR, Breitenbucher JG. Bioorg. Med. Chem. Lett. 2012;22:7357. doi: 10.1016/j.bmcl.2012.10.076. [DOI] [PubMed] [Google Scholar]
- 15 (A).Kono M, Matsumoto T, Kawamura T, Nishimura A, Kiyota Y, Oki H, Miyazaki J, Igaki S, Behnke CA, Shimojo M, Kori M. Bioorg. Med. Chem. 2013;21:28. doi: 10.1016/j.bmc.2012.11.006. [DOI] [PubMed] [Google Scholar]; (b) Kono M, Matsumoto T, Imaeda T, Kawamura T, Fujimoto S, Kosugi Y, Odani O, Shimizu Y, Matsui H, Shimojo M, Kori M. Bioorg. Med. Chem. 2014;22:1468. doi: 10.1016/j.bmc.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 16.Moore SA, Nomikos GG, Dickason–Chesterfield AK, Sohober DA, Schaus JM, Ying BP, Xu YC, Phebus L, Simmons RM, Li D, Iyengar S, Felder CC. Proc. Natl. Acad. Sci. USA. 2005;102:17852. doi: 10.1073/pnas.0507470102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17 (a).Adibekian A, Martin BR, Wang C, Hsu KL, Bachovchin DA, Niessen S, Hoover H, Cravatt BF. Nat. Chem. Biol. 2011;7:469. doi: 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Morera L, Labar G, Ortar G, Lambert DM. Bioorg. Med. Chem. 2012;20:6260. doi: 10.1016/j.bmc.2012.09.011. [DOI] [PubMed] [Google Scholar]; (c) Aaltonen N, Savinainen JR, Ribas CR, Ronkko J, Kuusisto A, Korhonen J, Navia-Paldanius D, Hayrinen J, Takabe P, Kasnanen H, Pantsar T, Laitinen T, Lehtonen M, Pasonen-Seppanen S, Poso A, Nevalainen T, Laitinen JT. Chem. Biol. 2013;20:379. doi: 10.1016/j.chembiol.2013.01.012. [DOI] [PubMed] [Google Scholar]; (d) Ebdrup S, Sorensen LG, Olsen OH, Jacobsen P. J. Med. Chem. 2004;47:400. doi: 10.1021/jm031004s. [DOI] [PubMed] [Google Scholar]; (e) Crocetti L, Schepetkin IA, Cilibrizzi A, Graziano A, Vergelli C, Giomi D, Khlebnikov AI, Quinn MT, Giovannoni MP. J. Med. Chem. 2013;56:6259. doi: 10.1021/jm400742j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung D, Hardouin C, Boger DL, Cravatt BF. Nat. Biotechnol. 2003;21:687. doi: 10.1038/nbt826. [DOI] [PubMed] [Google Scholar]
- 19.Leung D, Du W, Hardouin C, Cheng H, Hwang I, Cravatt BF, Boger DL. Bioorg. Med. Chem. Lett. 2005;15:1423. doi: 10.1016/j.bmcl.2004.12.085. [DOI] [PubMed] [Google Scholar]
- 20 (a).Cheng S, Comer DD, Williams JP, Boger DL. J. Am. Chem. Soc. 1996;118:2567. [Google Scholar]; (b) Boger DL, Tarby CM, Caporale LH. J. Am. Chem. Soc. 1996;118:2109. [Google Scholar]; (c) Cheng S, Tarby CM, Comer DD, Williams JP, Caporale LH, Boger DL. Bioorg. Med. Chem. 1996;4:727. doi: 10.1016/0968-0896(96)00069-7. [DOI] [PubMed] [Google Scholar]
- 21.Saha AK, Schultz P, Rapoport H. J. Am. Chem. Soc. 1989;111:4856. [Google Scholar]
- 22.Jouin P, Castro B, Zeggaf A, Pantaloni A, Senet JP, Lecolier S, Sennyey G. Tetrahedron Lett. 1987;28:1661. [Google Scholar]
- 23 (a).Cravatt BF, Lichtman AH. Curr. Opin. Chem. Biol. 2003;7:469. doi: 10.1016/s1367-5931(03)00079-6. [DOI] [PubMed] [Google Scholar]; (b) Ahn K, Johnson DS, Cravatt BF. Exp. Opin. Drug Discov. 2009;4:763–784. doi: 10.1517/17460440903018857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24 (a).Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB. Nature. 1996;384:83. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]; (b) Giang DK, Cravatt BF. Proc. Natl. Acad. Sci. USA. 1997;94:2238. doi: 10.1073/pnas.94.6.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Cravatt BF, Prospero–Garcia O, Suizdak G, Gilula NB, Henriksen SJ, Boger DL, Lerner RA. Science. 1995;268:1506. doi: 10.1126/science.7770779. [DOI] [PubMed] [Google Scholar]
- 25 (a).Lone AM, Bachovchin DA, Westwood DB, Speers AE, Spicer TP, Fernandez-Vega V, Chase P, Hodder PS, Rosen H, Cravatt BF, Saghatelian A. J. Am. Chem. Soc. 2011;133:11665. doi: 10.1021/ja2036095. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chang JW, Cognetta AB, III, Niphakis MJ, Cravatt BF. ACS Chem. Biol. 2013;8:1590. doi: 10.1021/cb400261h. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Blankman JL, Cravatt BF. Pharmacol. Rev. 2013;65:849. doi: 10.1124/pr.112.006387. [DOI] [PMC free article] [PubMed] [Google Scholar]; For N-acyl pyrazole amides, see: Schepetkin IA, Khlebnikov AI, Quinn MT. J. Med. Chem. 2007;50:4928. doi: 10.1021/jm070600+.
- 26 (a).Bracey MH, Hanson MA, Masuda KR, Stevens RC, Cravatt BF. Science. 2002;298:1793. doi: 10.1126/science.1076535. [DOI] [PubMed] [Google Scholar]; (b) Mileni M, Johnson DS, Wang Z, Everdeen DS, Liimatta M, Pabst B, Bhattacharya K, Nugent RA, Kamtekar S, Cravatt BF, Ahn K, Stevens RC. Proc. Natl. Acad. Sci. USA. 2008;105:12820. doi: 10.1073/pnas.0806121105. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Mileni M, Garfunkle J, DeMartino JK, Cravatt BF, Boger DL, Stevens RC. J. Am. Chem. Soc. 2009;131:10497. doi: 10.1021/ja902694n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Mileni M, Garfunkle J, Kimball FS, Cravatt BF, Stevens RC, Boger DL. J. Med. Chem. 2010;53:230. doi: 10.1021/jm9012196. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Mileni M, Garfunkle J, Ezzili C, Cravatt BF, Stevens RC, Boger DL. J. Am. Chem. Soc. 2011;133:4092. doi: 10.1021/ja110877y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27 (a).Boger DL, Henriksen SJ, Cravatt BF. Curr. Pharm. Des. 1998;4:303. [PubMed] [Google Scholar]; (b) Cravatt BF, Lerner RA, Boger DL. J. Am. Chem. Soc. 1996;118:580. [Google Scholar]; (c) Lerner RA, Siuzdak G, Prospero–Garcia O, Henriksen SJ, Boger DL, Cravatt BF. Proc. Natl. Acad. Sci. USA. 1994;91:9505. doi: 10.1073/pnas.91.20.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, Gibson D, Mandelbaum A, Etinger A, Mechoulam R. Science. 1992;258:1946–1949. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- 28 (a).Boger DL, Fecik RA, Patterson JE, Miyauchi H, Patricelli MP, Cravatt BF. Bioorg. Med. Chem. Lett. 2000;10:2613. doi: 10.1016/s0960-894x(00)00528-x. [DOI] [PubMed] [Google Scholar]; (b) Ezzili C, Otrubova K, Boger DL. Bioorg. Med. Chem. Lett. 2010;20:5959. doi: 10.1016/j.bmcl.2010.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29 (a).Boger DL, Desharnais J, Capps K. Angew. Chem., Int. Ed. 2003;42:4138. doi: 10.1002/anie.200300574. [DOI] [PubMed] [Google Scholar]; (b) Whitby LR, Boger DL. Acc. Chem. Res. 2012;45:1698. doi: 10.1021/ar300025n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30 (a).Whitby LR, Ando Y, Setola V, Vogt PK, Roth BL, Boger DL. J. Am. Chem. Soc. 2011;133:10184. doi: 10.1021/ja201878v. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Shaginian A, Whitby LR, Hong S, Hwang I, Farooqi B, Searcey M, Chen JC, Vogt PK, Boger DL. J. Am. Chem. Soc. 2009;131:5564. doi: 10.1021/ja810025g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31 (a).Whitby LR, Boyle KE, Cai L, Yu X, Gochin M, Boger DL. Bioorg. Med. Chem. Lett. 2012;22:2861. doi: 10.1016/j.bmcl.2012.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ambrus G, Whitby LR, Singer EL, Trott O, Choi E, Olson AJ, Boger DL, Gerace L. Bioorg. Med. Chem. 2010;18:7611. doi: 10.1016/j.bmc.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Chang MW, Giffin MJ, Muller R, Savage J, Lin YC, Hong S, Jin W, Whitby LR, Elder JH, Boger DL, Torbett BE. Biochem. J. 2010;429:527. doi: 10.1042/BJ20091645. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Lee AM, Rojek JM, Spiropoulou CF, Gundersen AT, Jin W, Shaginian A, York J, Nunberg JH, Boger DL, Oldstone MBA, Kunz S. J. Biol. Chem. 2008;283:18734. doi: 10.1074/jbc.M802089200. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Whitby LR, Lee AM, Kunz S, Oldstone MBA, Boger DL. Bioorg. Med. Chem. Lett. 2009;19:3771. doi: 10.1016/j.bmcl.2009.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Thomas CJ, Casquilho HE, York J, DeCamp DL, Dai D, Petrilli EB, Boger DL, Slayden RA, Amberg SM, Sprang SR, Nunberg JH. J. Biol. Chem. 2011;286:6192. doi: 10.1074/jbc.M110.196428. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Eubanks LM, Hixon MS, Jin W, Hong S, Clancy CM, Tepp WH, Malizio CJ, Goodnough MC, Barbieri JT, Johnson EA, Boger DL, Dickerson TJ, Janda KD. Proc. Natl. Acad. Sci. USA. 2007;104:2602. doi: 10.1073/pnas.0611213104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Capps KJ, Humiston J, Dominique R, Hwang I, Boger DL. Bioorg. Med. Chem. Lett. 2005;15:2840. doi: 10.1016/j.bmcl.2005.03.094. [DOI] [PubMed] [Google Scholar]; (i) Ambroise Y, Yuspan B, Ginsberg MH, Boger DL. Chem. Biol. 2002;9:1219. doi: 10.1016/s1074-5521(02)00246-6. [DOI] [PubMed] [Google Scholar]; (j) Goldberg J, Jin Q, Ambroise Y, Satoh S, Desharnais J, Capps K, Boger DL. J. Am. Chem. Soc. 2002;124:544. doi: 10.1021/ja0118789. [DOI] [PubMed] [Google Scholar]; (h) Berg T, Cohen SB, Desharnais J, Sonderegger C, Maslyar DJ, Goldberg J, Boger DL, Vogt PK. Proc. Natl. Acad. Sci. USA. 2002;99:3830. doi: 10.1073/pnas.062036999. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) Shi J, Stover JS, Whitby LR, Vogt PK, Boger DL. Bioorg. Med. Chem. Lett. 2009;19:6038. doi: 10.1016/j.bmcl.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]; (l) Silletti S, Kessler T, Goldberg J, Boger DL, Cheresh DA. Proc. Natl. Acad. Sci. USA. 2001;98:119. doi: 10.1073/pnas.011343298. [DOI] [PMC free article] [PubMed] [Google Scholar]; (m) Boger DL, Goldberg J, Silletti S, Kessler T, Cheresh DA. J. Am. Chem. Soc. 2001;123:1280. doi: 10.1021/ja003579+. [DOI] [PubMed] [Google Scholar]; (n) Boger DL, Goldberg J, Satoh S, Ambroise Y, Cohen SB, Vogt PK. Helv. Chim. Acta. 2000;83:1825. [Google Scholar]
- 32 (a).Stover JS, Shi J, Jin W, Vogt PK, Boger DL. J. Am. Chem. Soc. 2009;131:3342. doi: 10.1021/ja809083d. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Boger DL, Fink BE, Hedrick MP. J. Am. Chem. Soc. 2000;122:6382. [Google Scholar]; (c) Boger DL, Fink BF, Brunette SR, Tse W, Hedrick MP. J. Am. Chem. Soc. 2001;123:5878. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]; (d) Tse W, Boger DL. Acc. Chem. Res. 2004;37:61. doi: 10.1021/ar030113y. [DOI] [PubMed] [Google Scholar]; (e) Hauschild KE, Stover JS, Boger DL, Ansari AZ. Bioorg. Med. Chem. Lett. 2009;19:3779. doi: 10.1016/j.bmcl.2009.04.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.