Abstract
Endoplasmic reticulum-associated degradation (ERAD) is a mechanism during which native and misfolded proteins are recognized and retrotranslocated across the ER membrane to the cytosol for degradation by the ubiquitin-proteasome system. Like other cellular pathways, the factors required for ERAD have been analyzed using both conventional genetic and biochemical approaches. More recently, however, an integrated top-down approach has identified a functional network that underlies the ERAD system. In turn, bottom-up reconstitution has become increasingly sophisticated and elucidated the molecular mechanisms underlying substrate recognition, ubiquitylation, retrotranslocation, and degradation. In addition, a live cell imaging technique and a site-specific in vivo photo-crosslinking approach have further dissected specific steps during ERAD. These technical developments have revealed an unexpected dynamicity of the membrane-associated ERAD complex. In this article, we will discuss how these technical developments have improved our understanding of the ERAD pathway and have led to new questions.
Introduction
Approximately one-third of newly synthesized proteins are translocated into the endoplasmic reticulum (ER), where they must fold and assemble into their correct conformations. To accommodate these proteins, which exhibit diverse structures, oligomeric states, and folding rates, the ER harbors a high concentration of molecular chaperones that maintain polypeptide solubility, enzymes that posttranslationally modify proteins, and factors that directly assist in protein folding. However, the yield of correctly folded proteins is poor due to intracellular and external stresses, genetic mutations, or stochastic misfolding events [1–4].
Protein folding is monitored by the ER quality control machinery and terminally misfolded/unassembled proteins are recognized, retrotranslocated (i.e., transported) to the cytosol, and degraded by the ubiquitin-proteasome system. This process is referred to as ER associated degradation (ERAD) [5–12]. Since the discovery that soluble secretory and integral membrane proteins could be degraded in a pre-Golgi compartment, the components that constitute the ERAD pathway have been explored through the use of genetic and biochemical approaches and by examining a relatively limited number of model ERAD substrates. In recent years, considerable progress has been achieved by developing novel genetic and biochemical tools. For example, a top-down approach using mass spectrometry was introduced to systematically analyze physical interactions between components involved in ERAD. In addition, quantitative proteomics such as SILAC (stable isotope labeling using amino acids in cell culture) identified endogenous ERAD substrates. In parallel, bottom-up reconstitutions that recapitulate each step during the ERAD pathway have elucidated the mechanisms that lead to substrate recognition, ubiquitylation, retrotranslocation, and degradation. As a complementary approach to understand ERAD dynamics, a live cell imaging technique and a site-spedific in vivo photo-crosslinking technique were introduced.
Physical properties of the membrane-associated ERAD machineries
In the yeast, Saccharomyces cerevisiae, the Hrd1 E3 ubiquitin ligase is one of the central components of the membrane-associated ERAD machinery (Figure 1 and Table 1). Hrd1 has six transmembrane segments and harbors a cytosolically exposed RING domain at its C-terminal tail [13,14]. In yeast, the Hrd1 “core” complex comprises Hrd1, Hrd3, Usa1, and Der1. The molecular weight of the complex in digitonin-solubilized lysates was estimated by sucrose density gradient analysis to be >700 kD a [15,16]. In this complex, Hrd1 forms an oligomer and its formation is scaffolded by Usa1 [17–19], which spans the membrane twice. Usa1 also bridges the interaction between Hrd1 and Der1 [15]. A recent report indicated that Der1 forms an oligomer, and its formation is also scaffolded by Usa1 [20]. Another component, Hrd3, binds to and stabilizes Hrd1 and possesses a single transmembrane domain. Because robust ERAD activity was evident when yeast expressed a mutant form of Hrd3 lacking this domain, the lumenal domain of Hrd3 may bind to a Hrd1 lumenal loop [13]. In turn, the large lumenal domain of Hrd3 may directly recognize misfolded proteins [21–23] (Figure 1).
Figure 1.
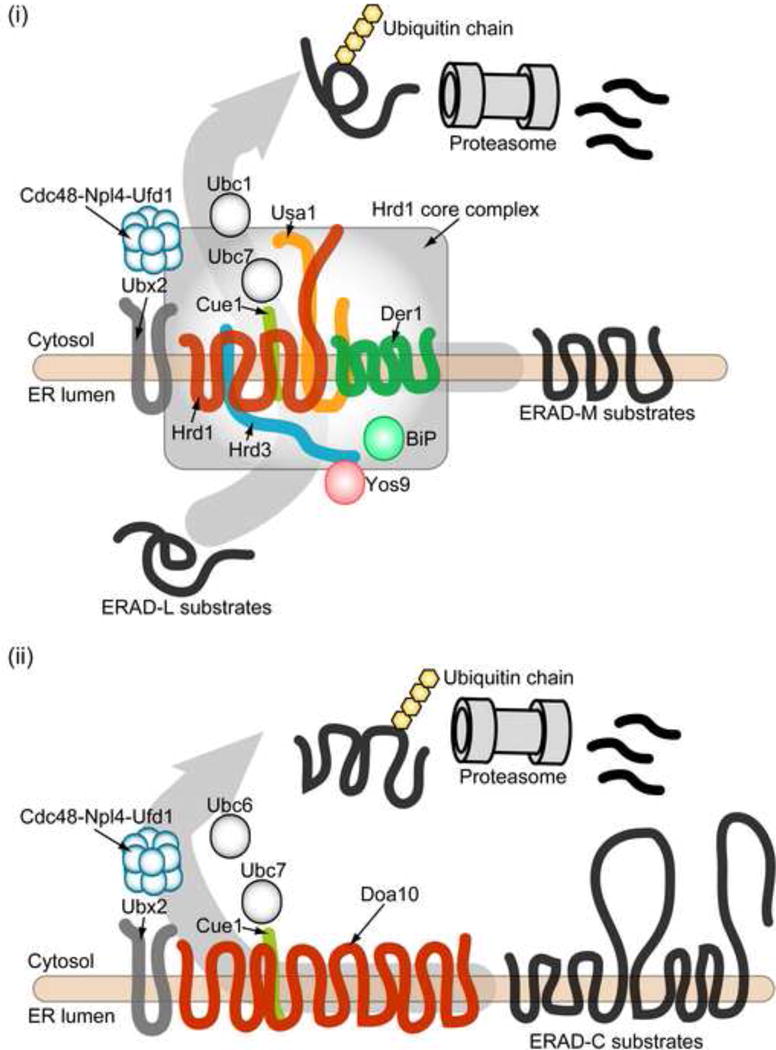
The membrane-associated E3 ligase complexes in yeast. (i) The Hrd1 “core” complex contains Hrd1, Hrd3, Usa1, and Der1. ERAD-L substrates are recognized by a lumenal Hsp70 (BiP; Kar2 in yeast), Yos9, and the lumenal domain of Hrd3. ERAD-M substrates may be directly recognized by the transmembrane domain of Hrd1. Recognized substrates are retrotranslocated to the cytosol and ubiquitylated by Ubc7 and Hrd1 before proteasomal degradation. (ii) Doa10 (TEB4 in mammals) ubiquitylates membrane substrates that have a misfolded domain facing the cytosol. Ubiquitylated membrane proteins may also be extracted to the cytosol and degraded by the proteasome.
Table 1.
Select, conserved components required for ubiquitylation and retrotranslocation during ERAD
| Yeast | Mammal | Function |
|---|---|---|
| Hrd1 complex | ||
|
| ||
| Hrd1 | HRD1, gp78 | E3 ubiquitin ligase |
| Hrd3 | SEL1 | Substrate recognition, Hrd1 stability |
| Usa1 | HERP | Hrd1 and Der1 oligomerization |
| Der1 | Derlin-1, -2, -3 | Substrate recognition, transfer of substrate to the Hrd1 complex |
| Yos9 | OS9, XTP3-B | Substrate recognition |
| Kar2 | BiP | Chaperone activity, substrate recognition |
| Doa10 complex | ||
|
| ||
| Doa10 | TEB4 (also known as MARCH-VI) | E3 ubiquitin ligase |
| Ubc6 | Ubc6, Ubc6e | E2 ubiquitin-conjugating enzyme |
| Common to the Hrd1 and Doa10 complexes | ||
|
| ||
| Ubc7 | UBE2G1 (also known as UBCH7 or UBC7), UBE2G2 | E2 ubiquitin-conjugating enzyme |
| Cue1 | Recruitment and activation of Ubc7 | |
| Ubx2 | UBXD8 | Membrane-recruiting factor for Cdc48 |
| Cdc48 | p97/VCP | Substrate retrotranslocation and membrane extraction |
| Npl4 | NPL4 | Cdc48 cofactor |
| Ufd1 | UFD1 | Cdc48 cofactor |
Components in the Hrd1 and Doa10 E3 ligase complexes shown in Figure 1 and their mammalian counterparts are listed. In both yeast and mammals, other ancillary factors including chaperones, E3 ubiquitin ligase enzymes, UBL-UBA domain-containing proteins, deglycosylating enzymes, and deubiquitylating enzymes facilitate ERAD.
The Hrd1 core complex also associates with a number of peripheral components in the cytoplasm and in the ER lumen. These components include Yos9 (a lumenal substrate recognition lectin) [24–28], Ubx2 (a UBX-UBA domain containing protein) [29,30], and Cdc48/p97-Npl4-Ufd1 complex (a AAA-ATPase complex, which drives the retrotranslocation of ubiquitylated substrates) [31–35]. The association of the peripheral components with the Hrd1 core complex seems to be weaker and/or transient because they do not co-migrate with the core complex in a density gradient but are co-immunoprecipitated with Hrd1 [15,36]. Together, the Hrd1 core complex links events that include substrate recognition, ubiquitylation, and retrotranslocation [16,36] (Figure 1).
Another E3 ligase involved in ERAD is Doa10, which contains fourteen transmembrane segments with both termini facing the cytoplasm [37] (Figure 1 and Table 1). Doa10 was initially found as an E3 ligase that functions in the degradation of a soluble substrate, the Matα2 transcriptional repressor [38,39]. It subsequently turned out that Doa10 is responsible for the ubiquitylation of ER membrane proteins with misfolded cytosolic domains (the “ERAD-C pathway”), whereas Hrd1 recognizes and ubiquitylates misfolded substrates containing intra-membrane or ER-lumenal lesions (the “ERAD-M” and “ERAD-L” pathways, respectively) [15,40,41]. For both Hrd1 and Doa10, two E2 ubiquitin conjugating enzymes, Ubc6 and Ubc7, are required for efficient substrate ubiquitylation [38,41–44]. Ubc6 is bound to the ER membrane via a C-terminal transmembrane anchor [42], whereas the soluble Ubc7 enzyme localizes to the ER by binding the transmembrane Cue1 receptor protein, which also allosterically activates the E2 enzyme [45–47] (Figure 1 and Table 1).
In mammals, as many as 16 E3 ligases have been implicated in ERAD [11], including Hrd1 [48], gp78 (also called AMFR or RNF45) [49] and TEB4 (which is a Doa10 homolog and also called MARCH-VI) [37,50]. Of these, Hrd1 complex assembly has been best documented. Constituents of the Hrd1 complex in mammals are similar to those in yeast. These include the adaptor protein SEL1L (which is similar to Hrd3) [51], the rhomboid pseudoprotease Derlin-1 (a homolog of Der1) [52–54], the scaffold protein HERP (which may be functionally related to Usa1) [15,55,56], and a series of membrane-embedded, lumenal and cytosolic peripheral factors [57]. The molecular weight of the Hrd1 core complex in CHAPS-solubilized lysate was estimated by sucrose density gradient analysis and was substantially larger than that found in yeast (>700 kDa, see above). The mammalian Hrd1 complex exhibited two peaks, and the larger one was >2000 kDa. This complex was retrotranslocation-competent and included HERP, SEL1L, and Derlin-1 [58] (see below).
The membrane-associated ERAD machinery is dynamic
In recent years, several lines of evidence suggested a dynamic behavior of the Hrd1 complex. Substrates in the ER lumen are targeted to the lumenal side of the Hrd1 complex and should re-emerge on the cytosolic face of the complex for ubiquitylation [13,43]. The potential role of the Hrd1 complex in facilitating the retrotranslocation of lumenal substrates is being investigated in many laboratories. However, because membrane protein complexes that facilitate the translocation of proteins into the ER, mitochondria, and chloroplast are known to be dynamic [59], it is not unreasonable to assume that the Hrd1 complex is also dynamic and its conformation may significantly change during substrate recognition and retrotranslocation. However, given the lack of an in vitro reconstitution system with purified membranous components and in the absence of structural data, definitive evidence for this model is lacking. Recently, however, an analysis of the Hrd1 complex’s catalytic cycle through the application of in vivo site-specific photocross-linking was reported. This approach provides protein-protein interaction data at amino acid resolution [60,61]. The details of this technique will be mentioned in a later section, but germane to this section we note that direct interactions between lumenal substrates and Hrd3, Hrd1, and Der1 were observed [62]. Within this complex, Der1 appears to facilitate the movement of misfolded lumenal proteins through the Hrd1 complex [20].
Recent studies have suggested that the assembly and disassembly of the Hrd1 complex is triggered by misfolded proteins. When retrotranslocation is blocked in yeast expressing a thermosensitive allele of CDC48, ubiquitylated substrates accumulate on the Hrd1 complex and nucleate the assembly of a supramolecular complex that includes the Hrd1 complex as well as Ubx2, Yos9, and the 19S proteasome [16]. These results suggest that the Hrd1 complex transiently forms a larger complex that links lumenal and cytosolic events during retrotranslocation [16]. Several lines of evidence hinted that the integrity of the Hrd1 complex in mammalian cells is also regulated by misfolded proteins. Hrd1 and HERP, which are normally short lived proteins [63–65], are stabilized when the concentration of misfolded proteins rises [58]. The stabilized Hrd1 complex is retrotranslocation competent and is >2000 kDa. These data imply that misfolded proteins may directly enhance the activity of the ERAD machinery, a concept referred to as “ERAD tuning” [58,66,67].
Components of the Hrd1 complex are transcriptionally upregulated by the unfolded protein response (UPR) [68], which is triggered in response to an increase in the concentration of non-native proteins in the ER [69,70]. Therefore, one might assume that the architecture of the Hrd1 complex is altered and its activity might be enhanced by the UPR. The response of the Hrd1 complex upon UPR activation could be different from and slower than ERAD tuning because the UPR requires transcriptional and translational upregulation of ERAD components [66]. One intriguing observation is that the members of the Hrd1 complex are induced to widely varying degrees (between 1.5- and 4-fold) [16]. Moreover, UPR signaling not only upregulates genes involved in the ERAD pathway but many other genes, including those encoding molecular chaperones and those required for protein translocation into the ER, glycosylation, vesicle transport, and lipid metabolism [68]. In addition, accumulation of misfolded proteins in the ER results in significant morphological changes of the ER membrane [71] and induces autophagy [72–78], which alleviate the ER stress. Given these many effects, it remains mysterious how the assembly of the Hrd1 complex is regulated by the UPR and how the rate-limiting step(s) during ERAD are modulated by ER stress.
Systematic approaches have expanded the genetic and physical linkages of the membrane-associated ERAD machineries
Until recently, the ERAD pathway and an analysis of how ERAD-requiring complexes interact have primarily been measured in yeast. In contrast, Kopito and colleagues performed a mass spectrometry analysis of proteins that co-precipitated with 25 “bait” proteins corresponding to different ERAD components in mammalian cells [57]. Over 170 unique interacting proteins were identified, including 71 proteins that were not previously associated with ERAD. Many of the genes encoding these proteins are upregulated by the UPR, suggesting a role in ER protein quality control. Consistent with previous observations [79–85], the lumenal recognition factors OS-9 (a homolog of yeast Yos9) and XTP3-B were associated with Hrd1 and SEL1L, whereas components involved in retrotranslocation such as Derlin and UBXD8 (a UBX domain containing protein) were connected to both Hrd1 and to gp78. Gp78 is another E3 ubiquitin ligase that plays a vital role in the modification and degradation of specific ERAD substrates [49]. The study also uncovered FAM8A1, which is a potential regulator of Hrd1; UBAC2, a ubiquitin binding protein associated with gp78; and the uncharacterized ER-membrane complex (EMC), which has been connected to various ERAD-related phenomena. For example, six out of 10 of the yeast EMC subunits were identified in a study that systematically identified genes contributing to ER protein folding [86].
Exploring the endogenous substrates highlights the importance of ERAD under normal physiological conditions
ERAD is often examined using artificial model substrates or disease relevant mutant proteins, which are usually overexpressed relative to their native levels in vivo. A notable exception is HMG CoA reductase-2 (Hmg2 in yeast), which catalyzes the rate-limiting step in sterol biosynthesis and whose stability is controlled by ERAD in a sterol intermediate-dependent manner [87–90]. Nevertheless, due to the presence of a small number of endogenous substrates, the importance of ERAD function under normal physiological conditions is poorly understood. Recently, however, the identities of endogenous ERAD substrates have been uncovered by proteomic methods and by taking advantage of specific cell lines. For example, squalene monooxygenase, which is encoded by ERG1 in yeast, was identified as a Doa10 substrate by SILAC and mass spectrometry [91]. The Doa10-mediated degradation of Erg1 is stimulated by lanosterol, which is a downstream product of this pathway. Depletion of Doa10 led to the accumulation of lanosterol and sterol esters, indicating feed-back regulation of Erg1 [91]. In mammalian cells, the Hedgehog precursor molecule was identified as an endogenous ERAD substrate [92]. This protein is processed in the ER by self-cleavage into the N-terminal Hedgehog ligand and the C-terminal fragment. The C-terminal fragment is a lumenal glycoprotein, which is constitutively degraded through the SEL1L-HRD1 pathway. Another endogenous substrate in mammalian cells is the core-glycosylated version of a protein, CD147, which was identified as a factor that co-precipitated with OS-9 and is degraded via the OS-9/SEL1L/Hrd1 pathway [93].
Finally, the integral membrane UPR transducer, ATF6, is degraded in a SEL1L-dependent manner, a result that was substantiated with stable knock-out SEL1L DT 40 cells [94]. Continued work in this area is sure to uncover other examples in which the ERAD pathway contributes to cellular homeostasis.
In vitro reconstitution of ERAD
Most cell free ERAD systems are substrate specific and require the addition of crude materials (e.g., microsome fractions and cytosol). Typically, substrates are translated in vitro, mixed with ER-derived microsomes, and then incubated with cytosol and/or purified components, which facilitate substrate ubiquitylation, retrotranslocation, and/or degradation. In some studies, especially when polytopic integral membrane proteins are examined, microsomes are prepared from cells expressing the substrate. In either case, retrotranslocation is typically assessed by detection of substrates in a cytosolic fraction after centrifugation. Alternatively, deglycosylation of N-glycosylated substrates, which occurs in the cytosol, is an indicator of retrotranslocation. Model substrates used for in vitro systems include ΔgpαF (an immature version of the yeast α-factor mating pheromone) [95–99], apolipoprotein B [100], cholera toxin [101,102], the cystic fibrosis transmembrane conductance regulator [103–106], Hmg2 (in yeast, see above) [107], HMGCoA-reductase (in mammalian cells) [108], Ste6* (a mutated form of a yeast plasma membrane transporter) [109], the cell surface CD4 receptor [110], and the MHC class I heavy chain [111,112].
In cell-free systems, the cytosol can be replaced with or added to purified components such as the proteasome or proteasome subparticles (i.e., the 19S “cap” and the 20S core) [98], the Cdc48 complex (see above; also known as p97 in mammalian cells), and/or other regulatory factors [31]. Alternatively, specific components can be immune-depleted from the cytosol to assess their function [106]. When ERAD is examined using components derived from yeast, membranes and cytosol from cells containing specific mutant alleles can be used [96,107,109]. However, replacement or just removal of lumenal and membrane-associated components presents a challenge when the goal is to reconstitute the ERAD pathway in mammalian cells. One way to replace lumenal components with a defined component is to use alkaline extraction. For example, Johnson and colleagues reconstituted rough mammalian microsomes with defined lumenal contents and measured the retrotranslocation and degradation of ΔgpαF, a model ERAD substrate in yeast that was used in the first reconstituted ERAD system. The ΔgpαF was covalently modified with a fluorescence dye, which permitted measurements of substrate retrotranslocation and degradation. In this assay, addition of antibodies against Derlin-1 blocked the ERAD of ΔgpαF but only after a short lag time, suggesting that Derlin-1 contributes to cycling the retrotranslocation complex for multiple rounds of retrotranslocation. This study also demonstrated that protein disulfide isomerase is necessary and sufficient for ΔgpαF retrotranslocation, at least in the mammalian ER [99]. A more recent study recapitulated the ubiquitylation of a single-spanning membrane protein embedded in a liposome with purified components. An HIV-encoded membrane protein, Vpu, bridges the interaction between the substrate, CD4, and the SCFβTrcP E3 ubiquitin ligase complex in the ER membrane. Surprisingly, the extent of ubiquitylation of wild-type CD4 (which is degraded) and a stable mutant (which is defective for Vpu binding) was similar. However, the addition of cytosol led to significantly greater levels of ubiquitin conjugation to the degraded versus the stable substrate. Ultimately, the activity of a deubiquitylating enzyme was found to mediate substrate discrimination. These data indicate that an ERAD substrate undergoes multiple rounds of ubiquitylation/deubiquitylation, an event that is required for the proteasome to engage only a bona fide substrate but not a wild type protein [110].
Fluorescent reporters of retrotranslocation
The ultimate goal of ERAD reconstitution is to recapitulate the entirety of the reaction with a minimal set of purified components and chemically defined lipids. Given the large number of potential ERAD regulators and parallel/redundant pathways that route a protein for ERAD, this goal has not been achieved. In contrast, the development of a live cell assay could significantly bolster our understanding of specific steps in the ERAD pathway, particularly with the development of powerful genetic tools that are available in yeast and increasingly in mammalian cells. One recent study took advantage of the split-GFP system [113], where the bulk of GFP (the N-terminal 10 β-strands, designated S1–10) was expressed in the cytosol and the other part (a C-terminal β -strand, designated as S11) was fused with model substrates. When S11-tagged ERAD substrates retrotranslocated back to the cytosol, S11 reassembled with S1–10, resulting in a fluorescent signal that was detected in the microscope [114]. Another study utilized variants of Venus fluorescent protein, which fluoresce after they are first glycosylated in the ER and subsequently deglycosylated by the cytosolic peptide-N-glycanase (PNGase) after retrotranslocation [115]. Both studies demonstrated the value of the split and/or mutant fluorescent protein system and supported the notion that Hrd1, HERP, Sel1L, and the p97/Cdc48 complex play a central role in ERAD. As evidenced in prior studies, this work also demonstrated that integral membrane proteins can be liberated into the cytosol and remain soluble before degradation [107,109]. It is important to bear in mind that the expression of GFP in the ER lumen can result in an unexpected post-translational modification and may be degraded by the proteasome, perhaps due to its slow rate of folding [116]. Nonetheless, the continued development of fluorescence-based live cell assays for ERAD will continue to be of great value [117].
In vivo site-specific crosslinking
Conventional biochemical methods to detect protein-protein interactions, such as co-immunoprecipitation, chemical crosslinking, and density gradient centrifugation, provide important information regarding component identities, the nature of their interactions, and the sizes of protein complexes. In principle, a systematic mapping of the direct interactions at the amino acid level between a substrate and an ERAD component with high resolution would provide a “snap-shot” of substrate retrotranslocation, a process that remains enigmatic. In the absence of x-ray cystallographic data, an alternate approach is to adopt in vivo site-specific photocrosslinking [60].
To assess the interactions between a lumenal substrate and the Hrd1 complex, the gene encoding CPY* (a model ERAD substrate) that contains an amber codon at several positions was expressed in yeast. The cells also expressed an orthogonal pair consisting of an amber suppressor tRNA and a cognate aminoacyl-tRNA synthetase specific for DL-2-amino-3-(p-benzoylphenyl)pentanoic acid (BPA), which is photoactive. Yeast cells were cultured in media containing BPA, which was incorporated into CPY* at the position specified by the amber codon, and UV irradiation resulted in the BPA-dependent crosslinking of CPY* to adjacent proteins. CPY* bound directly to Hrd1, Der1, and Hrd3, consistent with these proteins playing a central role during substrate recognition and retrotranslocation [62]. A more recent analysis employing the same method established the sequence of events during retrotranslocation: Misfolded lumenal substrates are first bound by Hrd3 and Yos9 and are then transferred to Der1, which initiates their insertion into the membrane. Next, the substrates are transported to the cytosolic face of Hrd1. Although the pore through which the substrate was threaded is still not completely clear, it may have been formed by the transmembrane regions of Der1 and Hrd1 [20]. An important next goal will be to trap a retrotranslocating substrate in the membrane in order to view this critical but undefined step in the ERAD pathway.
Conclusions
Significant progresses in our understanding of the ERAD pathway has been made by numerous technical developments, only some of which we were able to discuss here (Figure 2). These advances have uncovered key features of this pathway, but many questions remained answered. Specific events during retrotranslocation and the energetics that initially drive a protein across the membrane, before ubiquitylation and Cdc48/p97 engagement, remain unknown. It is also unknown which constituents are required to regulate the ERAD machinery in specific tissues and under conditions of stress or changes in nutrient availability. The continued development of techniques for in vitro reconstitution, for live cell imaging, for monitoring the interactions of components and substrates at the amino acid level, for the development of specific chemical inhibitors, and for structural analyses will provide important mechanistic insights relevant to multiple aspects of the ERAD pathway and to cellular function.
Figure 2.
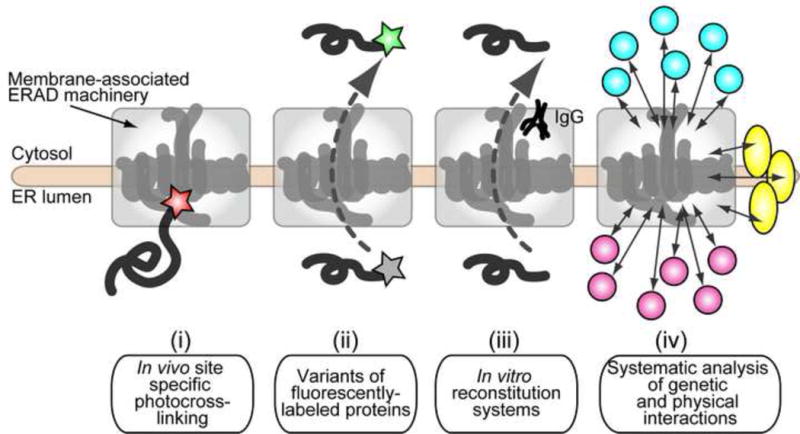
Select technologies that have improved our understanding of the ERAD pathway. (i) In vivo site specific photocross-linking revealed the direct interaction of substrates with components of the Hrd1 complex. (ii) Variants of fluorescent proteins were adopted to analyze retrotranslocation in vivo. (iii) Bottom-up reconstitutions that recapitulate each step during the ERAD pathway have elucidated the mechanisms that lead to substrate recognition, ubiquitylation, retrotranslocation, and degradation. (iv) Top-down and systematic genetic and biochemical approaches have identified novel genetic and physical interactions with membrane-associated ERAD machineries.
Acknowledgments
We would like to thank to S. D. Byrne for critical comments on the manuscript. This work was supported in part by a grant from the Ministry of Education, Science, Sports, and Culture of Japan to K. N and T. K., and by National Institutes of Health grant GM75061 and DK79307 to J. L. B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
• Of special interest
• • Of outstanding interest
- 1.Ellgaard L, Molinari M, Helenius A. Setting the standards: quality control in the secretory pathway. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- 2.Aridor M. Visiting the ER: the endoplasmic reticulum as a target for therapeutics in traffic related diseases. Adv Drug Deliv Rev. 2007;59:759–781. doi: 10.1016/j.addr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Hebert DN, Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 4.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romisch K. Endoplasmic reticulum-associated degradation. Annu Rev Cell Dev Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- 6.Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- 7.Araki K, Nagata K. Protein folding and quality control in the ER. Cold Spring Harb Perspect Biol. 2011;3:a007526. doi: 10.1101/cshperspect.a007526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hampton RY, Sommer T. Finding the will and the way of ERAD substrate retrotranslocation. Curr Opin Cell Biol. 2012;24:460–466. doi: 10.1016/j.ceb.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Thibault G, Ng DT. The endoplasmic reticulum-associated degradation pathways of budding yeast. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2012;151:1163–1167. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claessen JH, Kundrat L, Ploegh HL. Protein quality control in the ER: balancing the ubiquitin checkbook. Trends Cell Biol. 2012;22:22–32. doi: 10.1016/j.tcb.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sommer T, Wolf DH. The ubiquitin-proteasome-system. Biochim Biophys Acta. 2014;1843:1. doi: 10.1016/j.bbamcr.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, Kim C, Hampton RY. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deak PM, Wolf DH. Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. J Biol Chem. 2001;276:10663–10669. doi: 10.1074/jbc.M008608200. [DOI] [PubMed] [Google Scholar]
- 15.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 16•.Nakatsukasa K, Brodsky JL, Kamura T. A stalled retrotranslocation complex reveals physical linkage between substrate recognition and proteasomal degradation during ER-associated degradation. Mol Biol Cell. 2013;24:1765–1775. doi: 10.1091/mbc.E12-12-0907. This paper reported that the Hrd1 complex forms a large complex containing lumenal and cytosolic components when retrotranslocation is stalled by inactivating Cdc48/p97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horn SC, Hanna J, Hirsch C, Volkwein C, Schutz A, Heinemann U, Sommer T, Jarosch E. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol Cell. 2009;36:782–793. doi: 10.1016/j.molcel.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 18.Kim I, Li Y, Muniz P, Rao H. Usa1 protein facilitates substrate ubiquitylation through two separate domains. PLoS One. 2009;4:e7604. doi: 10.1371/journal.pone.0007604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carroll SM, Hampton RY. Usa1p is required for optimal function and regulation of the Hrd1p endoplasmic reticulum-associated degradation ubiquitin ligase. J Biol Chem. 2010;285:5146–5156. doi: 10.1074/jbc.M109.067876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20••.Mehnert M, Sommer T, Jarosch E. Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nat Cell Biol. 2014;16:77–86. doi: 10.1038/ncb2882. A synthetic photo-reactive amino acid was incorporated into lumenal substrates and Der1. This study provided a snap shot of lumenal substrates during retrotranslocation at amino acid level resolution. [DOI] [PubMed] [Google Scholar]
- 21.Gardner RG, Shearer AG, Hampton RY. In vivo action of the HRD ubiquitin ligase complex: mechanisms of endoplasmic reticulum quality control and sterol regulation. Mol Cell Biol. 2001;21:4276–4291. doi: 10.1128/MCB.21.13.4276-4291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 23.Stanley AM, Carvalho P, Rapoport T. Recognition of an ERAD-L substrate analyzed by site-specific in vivo photocrosslinking. FEBS Lett. 2011;585:1281–1286. doi: 10.1016/j.febslet.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buschhorn BA, Kostova Z, Medicherla B, Wolf DH. A genome-wide screen identifies Yos9p as essential for ER-associated degradation of glycoproteins. FEBS Lett. 2004;577:422–426. doi: 10.1016/j.febslet.2004.10.039. [DOI] [PubMed] [Google Scholar]
- 25.Bhamidipati A, Denic V, Quan EM, Weissman JS. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19:741–751. doi: 10.1016/j.molcel.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 26.Kim W, Spear ED, Ng DT. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol Cell. 2005;19:753–764. doi: 10.1016/j.molcel.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol Cell. 2005;19:765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 28.Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- 29.Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat Cell Biol. 2005;7:993–998. doi: 10.1038/ncb1298. [DOI] [PubMed] [Google Scholar]
- 30.Schuberth C, Buchberger A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat Cell Biol. 2005;7:999–1006. doi: 10.1038/ncb1299. [DOI] [PubMed] [Google Scholar]
- 31.Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 32.Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol Biol Cell. 2001;12:4114–4128. doi: 10.1091/mbc.12.12.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol. 2002;4:134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- 34.Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gauss R, Sommer T, Jarosch E. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J. 2006;25:1827–1835. doi: 10.1038/sj.emboj.7601088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kreft SG, Wang L, Hochstrasser M. Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI) J Biol Chem. 2006;281:4646–4653. doi: 10.1074/jbc.M512215200. [DOI] [PubMed] [Google Scholar]
- 38.Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006;25:533–543. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vashist S, Ng DT. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem. 2004;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- 42.Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- 43.Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 44.Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- 45.Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- 46•.Bagola K, von Delbruck M, Dittmar G, Scheffner M, Ziv I, Glickman MH, Ciechanover A, Sommer T. Ubiquitin binding by a CUE domain regulates ubiquitin chain formation by ERAD E3 ligases. Mol Cell. 2013;50:528–539. doi: 10.1016/j.molcel.2013.04.005. Structural and biochemical analyses elucidated the roles of Cue1-Ubc7 in the regulation of ubiquitin chain synthesis on the ERAD substrates. [DOI] [PubMed] [Google Scholar]
- 47•.Metzger MB, Liang YH, Das R, Mariano J, Li S, Li J, Kostova Z, Byrd RA, Ji X, Weissman AM. A structurally unique E2-binding domain activates ubiquitination by the ERAD E2, Ubc7p, through multiple mechanisms. Mol Cell. 2013;50:516–527. doi: 10.1016/j.molcel.2013.04.004. Structural and biochemical analyses elucidated the roles of Cue1-Ubc7 in the regulation of ubiquitin chain synthesis on the ERAD substrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kikkert M, Doolman R, Dai M, Avner R, Hassink G, van Voorden S, Thanedar S, Roitelman J, Chau V, Wiertz E. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J Biol Chem. 2004;279:3525–3534. doi: 10.1074/jbc.M307453200. [DOI] [PubMed] [Google Scholar]
- 49.Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hassink G, Kikkert M, van Voorden S, Lee SJ, Spaapen R, van Laar T, Coleman CS, Bartee E, Fruh K, Chau V, et al. TEB4 is a C4HC3 RING finger-containing ubiquitin ligase of the endoplasmic reticulum. Biochem J. 2005;388:647–655. doi: 10.1042/BJ20041241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mueller B, Lilley BN, Ploegh HL. SEL1L, the homologue of yeast Hrd3p, is involved in protein dislocation from the mammalian ER. J Cell Biol. 2006;175:261–270. doi: 10.1083/jcb.200605196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- 53.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 54.Greenblatt EJ, Olzmann JA, Kopito RR. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant alpha-1 antitrypsin from the endoplasmic reticulum. Nat Struct Mol Biol. 2011;18:1147–1152. doi: 10.1038/nsmb.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kokame K, Agarwala KL, Kato H, Miyata T. Herp, a new ubiquitin-like membrane protein induced by endoplasmic reticulum stress. J Biol Chem. 2000;275:32846–32853. doi: 10.1074/jbc.M002063200. [DOI] [PubMed] [Google Scholar]
- 56.Schulze A, Standera S, Buerger E, Kikkert M, van Voorden S, Wiertz E, Koning F, Kloetzel PM, Seeger M. The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum associated degradation pathway. J Mol Biol. 2005;354:1021–1027. doi: 10.1016/j.jmb.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 57.Christianson JC, Olzmann JA, Shaler TA, Sowa ME, Bennett EJ, Richter CM, Tyler RE, Greenblatt EJ, Harper JW, Kopito RR. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol. 2012;14:93–105. doi: 10.1038/ncb2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58••.Bernasconi R, Galli C, Kokame K, Molinari M. Autoadaptive ER-Associated Degradation Defines a Preemptive Unfolded Protein Response Pathway. Mol Cell. 2013;52:783–793. doi: 10.1016/j.molcel.2013.10.016. This paper reported that misfolded proteins accumulated in the ER stabilize Hrd1 and HERP, which are normally short-lived proteins. Stabilization of Hrd1 and HERP leads to the formation of a large, reactive retrotranslocation complex. [DOI] [PubMed] [Google Scholar]
- 59.Schnell DJ, Hebert DN. Protein translocons: multifunctional mediators of protein translocation across membranes. Cell. 2003;112:491–505. doi: 10.1016/s0092-8674(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 60.Chin JW, Cropp TA, Anderson JC, Mukherji M, Zhang Z, Schultz PG. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 61.Shiota T, Mabuchi H, Tanaka-Yamano S, Yamano K, Endo T. In vivo protein-interaction mapping of a mitochondrial translocator protein Tom22 at work. Proc Natl Acad Sci U S A. 2011;108:15179–15183. doi: 10.1073/pnas.1105921108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143:579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sai X, Kokame K, Shiraishi H, Kawamura Y, Miyata T, Yanagisawa K, Komano H. The ubiquitin-like domain of Herp is involved in Herp degradation, but not necessary for its enhancement of amyloid beta-protein generation. FEBS Lett. 2003;553:151–156. doi: 10.1016/s0014-5793(03)01009-3. [DOI] [PubMed] [Google Scholar]
- 64.Hori O, Ichinoda F, Yamaguchi A, Tamatani T, Taniguchi M, Koyama Y, Katayama T, Tohyama M, Stern DM, Ozawa K, et al. Role of Herp in the endoplasmic reticulum stress response. Genes Cells. 2004;9:457–469. doi: 10.1111/j.1356-9597.2004.00735.x. [DOI] [PubMed] [Google Scholar]
- 65.Miura H, Hashida K, Sudo H, Awa Y, Takarada-Iemata M, Kokame K, Takahashi T, Matsumoto M, Kitao Y, Hori O. Deletion of Herp facilitates degradation of cytosolic proteins. Genes Cells. 2010;15:843–853. doi: 10.1111/j.1365-2443.2010.01422.x. [DOI] [PubMed] [Google Scholar]
- 66.Merulla J, Fasana E, Solda T, Molinari M. Specificity and regulation of the endoplasmic reticulum-associated degradation machinery. Traffic. 2013;14:767–777. doi: 10.1111/tra.12068. [DOI] [PubMed] [Google Scholar]
- 67.Bernasconi R, Molinari M. ERAD and ERAD tuning: disposal of cargo and of ERAD regulators from the mammalian ER. Curr Opin Cell Biol. 2011;23:176–183. doi: 10.1016/j.ceb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 69.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 70.Mori K. Signalling pathways in the unfolded protein response: development from yeast to mammals. J Biochem. 2009;146:743–750. doi: 10.1093/jb/mvp166. [DOI] [PubMed] [Google Scholar]
- 71.Schuck S, Prinz WA, Thorn KS, Voss C, Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol. 2009;187:525–536. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yorimitsu T, Nair U, Yang Z, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kruse KB, Brodsky JL, McCracken AA. Characterization of an ERAD gene as VPS30/ATG6 reveals two alternative and functionally distinct protein quality control pathways: one for soluble Z variant of human alpha-1 proteinase inhibitor (A1PiZ) and another for aggregates of A1PiZ. Mol Biol Cell. 2006;17:203–212. doi: 10.1091/mbc.E04-09-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kamimoto T, Shoji S, Hidvegi T, Mizushima N, Umebayashi K, Perlmutter DH, Yoshimori T. Intracellular inclusions containing mutant alpha1-antitrypsin Z are propagated in the absence of autophagic activity. J Biol Chem. 2006;281:4467–4476. doi: 10.1074/jbc.M509409200. [DOI] [PubMed] [Google Scholar]
- 75.Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3:285–287. doi: 10.4161/auto.3930. [DOI] [PubMed] [Google Scholar]
- 77.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–239. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 78.Ding WX, Ni HM, Gao W, Hou YF, Melan MA, Chen X, Stolz DB, Shao ZM, Yin XM. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem. 2007;282:4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- 79.Okuda-Shimizu Y, Hendershot LM. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol Cell. 2007;28:544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mueller B, Klemm EJ, Spooner E, Claessen JH, Ploegh HL. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc Natl Acad Sci U S A. 2008;105:12325–12330. doi: 10.1073/pnas.0805371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hosokawa N, Wada I, Nagasawa K, Moriyama T, Okawa K, Nagata K. Human XTP3-B forms an endoplasmic reticulum quality control scaffold with the HRD1-SEL1L ubiquitin ligase complex and BiP. J Biol Chem. 2008;283:20914–20924. doi: 10.1074/jbc.M709336200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bernardi KM, Williams JM, Kikkert M, van Voorden S, Wiertz EJ, Ye Y, Tsai B. The E3 ubiquitin ligases Hrd1 and gp78 bind to and promote cholera toxin retro-translocation. Mol Biol Cell. 2010;21:140–151. doi: 10.1091/mbc.E09-07-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iida Y, Fujimori T, Okawa K, Nagata K, Wada I, Hosokawa N. SEL1L protein critically determines the stability of the HRD1-SEL1L endoplasmic reticulum-associated degradation (ERAD) complex to optimize the degradation kinetics of ERAD substrates. J Biol Chem. 2011;286:16929–16939. doi: 10.1074/jbc.M110.215871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Groisman B, Shenkman M, Ron E, Lederkremer GZ. Mannose trimming is required for delivery of a glycoprotein from EDEM1 to XTP3-B and to late endoplasmic reticulum-associated degradation steps. J Biol Chem. 2011;286:1292–1300. doi: 10.1074/jbc.M110.154849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brown MS, Goldstein JL. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res. 1980;21:505–517. [PubMed] [Google Scholar]
- 88.Hampton RY, Rine J. Regulated degradation of HMG-CoA reductase, an integral membrane protein of the endoplasmic reticulum, in yeast. J Cell Biol. 1994;125:299–312. doi: 10.1083/jcb.125.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hampton RY. Proteolysis and sterol regulation. Annu Rev Cell Dev Biol. 2002;18:345–378. doi: 10.1146/annurev.cellbio.18.032002.131219. [DOI] [PubMed] [Google Scholar]
- 90.Jo Y, Debose-Boyd RA. Control of cholesterol synthesis through regulated ER-associated degradation of HMG CoA reductase. Crit Rev Biochem Mol Biol. 2010;45:185–198. doi: 10.3109/10409238.2010.485605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91••.Foresti O, Ruggiano A, Hannibal-Bach HK, Ejsing CS, Carvalho P. Sterol homeostasis requires regulated degradation of squalene monooxygenase by the ubiquitin ligase Doa10/Teb4. Elife. 2013;2:e00953. doi: 10.7554/eLife.00953. This study identified an endogenous Doa10 substrate, Erg1, and elucidated the feed-back mechanism of Erg1 degradation in the sterol synthesis pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92•.Chen X, Tukachinsky H, Huang CH, Jao C, Chu YR, Tang HY, Mueller B, Schulman S, Rapoport TA, Salic A. Processing and turnover of the Hedgehog protein in the endoplasmic reticulum. J Cell Biol. 2011;192:825–838. doi: 10.1083/jcb.201008090. Reference 92 identified previously unknown ERAD substrates in mammalian cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93•.Tyler RE, Pearce MM, Shaler TA, Olzmann JA, Greenblatt EJ, Kopito RR. Unassembled CD147 is an endogenous endoplasmic reticulum-associated degradation substrate. Mol Biol Cell. 2012;23:4668–4678. doi: 10.1091/mbc.E12-06-0428. Reference 93 identified previously unknown ERAD substrates in mammalian cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94•.Horimoto S, Ninagawa S, Okada T, Koba H, Sugimoto T, Kamiya Y, Kato K, Takeda S, Mori K. The unfolded protein response transducer ATF6 represents a novel transmembrane-type endoplasmic reticulum-associated degradation substrate requiring both mannose trimming and SEL1L protein. J Biol Chem. 2013;288:31517–31527. doi: 10.1074/jbc.M113.476010. Reference 94 identified previously unknown ERAD substrates in mammalian cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McCracken AA, Brodsky JL. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Werner ED, Brodsky JL, McCracken AA. Proteasome-dependent endoplasmic reticulum-associated protein degradation: an unconventional route to a familiar fate. Proc Natl Acad Sci U S A. 1996;93:13797–13801. doi: 10.1073/pnas.93.24.13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pilon M, Schekman R, Romisch K. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 1997;16:4540–4548. doi: 10.1093/emboj/16.15.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee RJ, Liu CW, Harty C, McCracken AA, Latterich M, Romisch K, DeMartino GN, Thomas PJ, Brodsky JL. Uncoupling retro-translocation and degradation in the ER-associated degradation of a soluble protein. EMBO J. 2004;23:2206–2215. doi: 10.1038/sj.emboj.7600232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wahlman J, DeMartino GN, Skach WR, Bulleid NJ, Brodsky JL, Johnson AE. Real-time fluorescence detection of ERAD substrate retrotranslocation in a mammalian in vitro system. Cell. 2007;129:943–955. doi: 10.1016/j.cell.2007.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gusarova V, Caplan AJ, Brodsky JL, Fisher EA. Apoprotein B degradation is promoted by the molecular chaperones hsp90 and hsp70. J Biol Chem. 2001;276:24891–24900. doi: 10.1074/jbc.M100633200. [DOI] [PubMed] [Google Scholar]
- 101.Schmitz A, Herrgen H, Winkeler A, Herzog V. Cholera toxin is exported from microsomes by the Sec61p complex. J Cell Biol. 2000;148:1203–1212. doi: 10.1083/jcb.148.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moore P, He K, Tsai B. Establishment of an In Vitro Transport Assay That Reveals Mechanistic Differences in Cytosolic Events Controlling Cholera Toxin and T-Cell Receptor alpha Retro-Translocation. PLoS One. 2013;8:e75801. doi: 10.1371/journal.pone.0075801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sato S, Ward CL, Kopito RR. Cotranslational ubiquitination of cystic fibrosis transmembrane conductance regulator in vitro. J Biol Chem. 1998;273:7189–7192. doi: 10.1074/jbc.273.13.7189. [DOI] [PubMed] [Google Scholar]
- 104.Xiong X, Chong E, Skach WR. Evidence that endoplasmic reticulum (ER)-associated degradation of cystic fibrosis transmembrane conductance regulator is linked to retrograde translocation from the ER membrane. J Biol Chem. 1999;274:2616–2624. doi: 10.1074/jbc.274.5.2616. [DOI] [PubMed] [Google Scholar]
- 105.Oberdorf J, Carlson EJ, Skach WR. Uncoupling proteasome peptidase and ATPase activities results in cytosolic release of an ER polytopic protein. J Cell Sci. 2006;119:303–313. doi: 10.1242/jcs.02732. [DOI] [PubMed] [Google Scholar]
- 106.Carlson EJ, Pitonzo D, Skach WR. p97 functions as an auxiliary factor to facilitate TM domain extraction during CFTR ER-associated degradation. EMBO J. 2006;25:4557–4566. doi: 10.1038/sj.emboj.7601307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Garza RM, Sato BK, Hampton RY. In vitro analysis of Hrd1p-mediated retrotranslocation of its multispanning membrane substrate 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase. J Biol Chem. 2009;284:14710–14722. doi: 10.1074/jbc.M809607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108••.Elsabrouty R, Jo Y, Dinh TT, DeBose-Boyd RA. Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from membranes of permeabilized cells. Mol Biol Cell. 2013;24:3300–3308. doi: 10.1091/mbc.E13-03-0157. The sterol-induced membrane extraction and dislocation of ubiquitylated HMGCoA-Reductase from the membrane of permeabilized cells was reconstituted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110••.Zhang ZR, Bonifacino JS, Hegde RS. Deubiquitinases sharpen substrate discrimination during membrane protein degradation from the ER. Cell. 2013;154:609–622. doi: 10.1016/j.cell.2013.06.038. In this study the ubiquitylation of single transmembrane protein with defined components was recapitulated. It was shown that discrimination of ER-quality control substrate is mediated by deubiquitylating enzymes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shamu CE, Flierman D, Ploegh HL, Rapoport TA, Chau V. Polyubiquitination is required for US11-dependent movement of MHC class I heavy chain from endoplasmic reticulum into cytosol. Mol Biol Cell. 2001;12:2546–2555. doi: 10.1091/mbc.12.8.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Furman MH, Ploegh HL, Tortorella D. Membrane-specific, host-derived factors are required for US2- and US11-mediated degradation of major histocompatibility complex class I molecules. J Biol Chem. 2002;277:3258–3267. doi: 10.1074/jbc.M109765200. [DOI] [PubMed] [Google Scholar]
- 113.Cabantous S, Terwilliger TC, Waldo GS. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat Biotechnol. 2005;23:102–107. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- 114••.Zhong Y, Fang S. Live cell imaging of protein dislocation from the endoplasmic reticulum. J Biol Chem. 2012;287:28057–28066. doi: 10.1074/jbc.M112.381798. Reference 114 reported the first demonstrations of a live cell imaging technique to view the retrotranslocation reaction. The systems used either the split GFP system or engineered Venus fluorescent proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115••.Grotzke JE, Lu Q, Cresswell P. Deglycosylation-dependent fluorescent proteins provide unique tools for the study of ER-associated degradation. Proc Natl Acad Sci U S A. 2013;110:3393–3398. doi: 10.1073/pnas.1300328110. Reference 115 reported the first demonstrations of a live cell imaging technique to view the retrotranslocation reaction. The systems used either the split GFP system or engineered Venus fluorescent proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116•.Xu C, Wang S, Thibault G, Ng DT. Futile protein folding cycles in the ER are terminated by the unfolded protein O-mannosylation pathway. Science. 2013;340:978–981. doi: 10.1126/science.1234055. This study reported that unfolded protein O-mannosylation terminates failed folding attempts through the Pmt1/Pmt2 complex in yeast. [DOI] [PubMed] [Google Scholar]
- 117.Hipp MS, Bersuker K, Kopito RR. Live-cell imaging of ubiquitin-proteasome system function. Methods Mol Biol. 2012;832:463–472. doi: 10.1007/978-1-61779-474-2_33. [DOI] [PMC free article] [PubMed] [Google Scholar]


