Abstract
Maintenance of cellular protein quality – by restoring misfolded proteins to their native state and by targeting terminally misfolded or damaged proteins for degradation – is a critical function of all cells. To ensure protein quality, cells have evolved various organelle-specific quality control mechanisms responsible recognizing and responding to misfolded proteins at different subcellular locations of the cell. Recently, several publications have begun to elucidate mechanisms of quality control that operate at the plasma membrane (PM), recognizing misfolded PM proteins and targeting their endocytic trafficking and lysosomal degradation. Here, I discuss these recent developments in our understanding of PM quality control mechanisms and how they relate to global protein quality control strategies in the cell.
Introduction
Proteins are constantly exposed to different cellular microenvironments that exert various stresses – including oxidative stress, thermal stress, physical stress, and chemical stress – all of which threaten the native fold of proteins and generally contribute to protein damage and misfolding. Therefore, it is not surprising that eukaryotic cells have evolved multiple elaborate and interconnected mechanisms dedicated to maintaining protein quality in specific organelles and subcellular compartments. In general, these quality control mechanisms exhibit several shared features, including: (i) the ability to distinguish between native and non-native substrate, or “client”, proteins, (ii) the potential to interact with a broad array of misfolded clients, and (iii) the ability to protect cells from the toxic effects protein misfolding in both physiological conditions and during cellular stress. Over the course of the past decade, many specialized quality control mechanisms that fit these criteria have been characterized in the cytosol [1,2], the endoplasmic reticulum (ER) [3-7], the nucleus [8,9], and the mitochondria [10,11]. From this research, it is clear that the specific chemistry and context of some organelles necessitates dedicated quality control mechanisms, but it is also clear that many different quality control mechanisms are overlapping and interwoven to provide robust protein quality control throughout the cell. Importantly, lessons from different quality control mechanisms are leading to the emergence of common themes and design patterns that will guide us in our efforts to understand how protein quality is maintained at different compartments in the cell.
Until recently, very little was known about quality control mechanisms that operate at the plasma membrane (PM). This is surprising, given the variety and importance of physiological processes that occur at the PM, including sensing of environmental cues, transduction of signals across the PM bilayer, uptake of nutrients, ion flux, and adhesion to other cells and surfaces. However, recent studies have highlighted the critical role of ubiquitination pathways both as sensors of PM protein misfolding and as mediators of plasma membrane quality control (PMQC). Specifically, ubiquitin modification of misfolded integral membrane proteins in the PM targets endocytosis and subsequent trafficking to the lysosome, resulting in protein degradation which generates free amino acids that can either be stored or transported to the cytosol and recycled [12,13]. Here, we review recent findings that have expanded our understanding of PMQC, comparing these systems with other cellular quality control pathways and highlighting the most important unresolved issues that need to be addressed in future studies.
PMQC: Unique Challenges and High Stakes at the Cell Surface
Maintaining high PM protein quality control and preventing the accumulation of misfolded integral membrane proteins at the cell surface is critical, not only to ensure proper physiological responses to and interactions with the environment, but also to maintain essential ion and chemical gradients between the cytosol and the extracellular space that are vital for life. Indeed, each channel and transporter at the surface is a potential liability which could threaten the integrity of the cell if misfolded variants persist at the PM. Along with such high stakes come substantial challenges. For example, changes to the extracellular microenvironment, mechanical stresses, or extrinsic factors that affect membrane fluidity could all promote misfolding of integral membrane proteins at the PM. Furthermore, PMQC has unique limitations compared to QC at other locations in the cell. For example, ER quality control can potentially detect protein misfolding of an integral membrane protein on its cytosolic domains (ERAD-C), its membrane spanning domains (ERAD-M), or domains accessible to the lumen of the ER (ERAD-L), and these distinct pathways contribute to robust quality control in the ER [14]. In contrast, it is unclear how misfolding of extracellular domains in PM proteins would be detected and, once detected, how such a signal would be transduced to ubiquitination machinery in the cytosol. Also in contrast to other quality control mechanisms such as ERAD, where substrate ubiquitination results in rapid retrotransolocation and proteasomal degradation, it is clear that misfolded PM proteins targeted for degradation along the endocytic pathway have multiple opportunities for ubiquitin modifications to be reversed by deubiquitylating enzymes to allow for recycling [15]. This important feature – the chance for multiple quality checkpoints along the endocytic trafficking pathway – suggests that the overall quality of PM proteins is determined by multiple sequential quality control mechanisms, and not just the ability to recognize misfolding of proteins at the PM.
CHIPing away at Garbage: Chaperone-mediated PMQC
While the first evidence of PMQC came from reports over a decade ago [16,17], it is only in recent years that the first mechanisms of PMQC have been reported. Two important studies demonstrated that conditional misfolding of integral membrane proteins at the PM – either mutant CFTR (ΔF506) or chimera constructs containing domains known to misfold at restrictive temperature – triggers ubiquitin-dependent endocytosis and lysosomal trafficking for degradation [18,19]. Both studies demonstrate that the detection of misfolded PM proteins is mediated by the CHIP (C- terminus of Hsp70 interacting protein) ubiquitin ligase, which catalyzes the ubiquitination of misfolded substrates and is thus required for subsequent endocytosis and lysosomal trafficking events (Figure 1 and Table 1). CHIP has a domain architecture that includes a C-terminal U-box domain responsible for recruitment of E2 ubiquitin conjugating enzymes and an N-terminal tetratricopeptide (TPR) domain that interacts with Hsc70 and Hsp90. These interactions allow these chaperones to function as adaptors for the CHIP ligase, effectively promoting the ubiquitin-mediated degradation of chronic Hsc70 and Hsp90 clients. In between the TPR and U-box domains, CHIP contains a dimerization domain, but structural studies have demonstrated that CHIP homodimers are asymmetrical and only one E2 conjugating enzyme can be recruited per complex [20].
Figure 1.
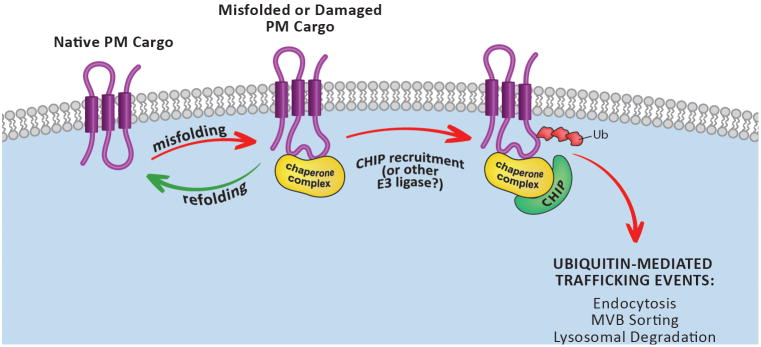
Model of chaperone-mediated PMQC via the CHIP E3 ubiquitin ligase.
Table 1. Examples of PMQC in Yeast and Mammalian Cells.
| PM Protein | Misfolding Condition | Recognition Mechanism | Ubiquitin Ligase | Reference |
|---|---|---|---|---|
| Yeast | ||||
| Ste2 | ste2-3 allele (ts) | unknown | Rsp5 | [16,17] |
| Can1 | can1-ts allele | unknown | Rsp5 | [17] |
| Pma1 | pma1-10 allele | unknown | Rsp5 | [57] |
| Gap1 | low [sphingolipids] | unknown | Rsp5 | [45] |
| Lyp1 | heat stress | Art1 and Art2 | Rsp5 | [30] |
| Fur4 | heat stress | Rsp5 | Rsp5 | [31] |
| Mammalian | ||||
| CFTR | ΔF508 CFTR | Hsc70, Hsp90 | CHIP | [18] |
| hERG | mutation, altered [K+] | CHIP | [22] | |
| Kir2.1 | unknown | unknown | unknown | [39] |
More recently, several reports have linked CHIP to the endocytic trafficking and lysosomal degradation of additional PM proteins. For example, CHIP in conjunction with Ubc13 was shown to be required for the endocytic downregulation of growth hormone receptor (GHR) expressed in different mammalian cell lines[21], although it is not entirely clear if CHIP is recognizing misfolded GHR as a quality control mechanism. Another recent study demonstrated that misfolding of the human ether-a-go-go-related gene (hERG), the α-subunit of the Kv11.1 K+ channel critical for repolarization of cardiac cells, resulted in CHIP-mediated ubiquitination, endocytosis, and lysosomal degradation [22] (Table 1). Thus, there is an emerging consensus that CHIP is a major factor of PM quality control in mammalian cells.
Importantly, CHIP has also been shown to function in cytosolic quality control and in ER quality control [23-25], and can catalyze the addition of both K48-linked and K63-linked polyubiquitin chains. This versatility suggests that CHIP contributes to overall cellular protein quality control, capable of being recruited to multiple subcellular locations to ubiquitinate chaperone clients and target their proteasomal or lysosomal degradation. Perhaps not surprisingly, a recent report found a link between mutations in CHIP and autosomal recessive cerebellar ataxias, a group of inherited neurodegenerative disorders [26]. Interestingly, this study demonstrated that, in contrast to its wildtype counterpart, mutant CHIP was associated with accumulation of N-methyl-D-aspartate receptors (NMDARs) at a cellular level [26]. Future analysis of these disease-linked CHIP mutants will need to address if the defect is associated with accumulation of receptors at the PM, in the ER, or both.
PMQC and the ART-Rsp5 network in Yeast
In yeast, endocytosis of specific cargo typically requires ubiquitination by the Rsp5 E3 ubiquitin ligase (the only member of the Nedd4 family of ubiquitin ligases encoded in the yeast genome), which can either target substrates directly or indirectly via a family of adaptor proteins known as ARTs (arrestin-related trafficking adaptors) [12,27-29]. Recent studies have implicated the ART-Rsp5 network in the targeted ubiquitination and endocytosis of misfolded proteins at the PM (Figure 2 and Table 1) [27,30]. ART family adaptors are absolutely required for the endocytosis and degradation of many different PM proteins following exposure to misfolding stress, and cells with defects in the specific nodes of the ART-Rsp5 network are extremely sensitive to these stresses [30]. This sensitivity correlates with the accumulation of proteins at the surface, and is also observed for other endocytosis mutants but not for later trafficking events like endosomal sorting by the ESCRT pathway or vacuolar fusion [30]. In most cases, blocking endocytosis of misfolded proteins required deletion of two or more ART family adaptors, which is indicative of a highly redundant and robust system [30]. Importantly, the ART-Rsp5 network exhibited synthetic genetic interactions with other membrane protein quality control pathways functioning at the ER and Golgi, indicating that these sequential quality control mechanisms operate in series to maintain membrane protein quality control throughout the cell [30]. Thus, the ART-Rsp5 network exhibits critical features of many QC systems, including (i) the ability to specifically recognize misfolded PM proteins for targeted ubiquitination, (ii) broad substrate recognition potential, either via direct interaction with Rsp5 or its adaptor network, and (iii) the ability to protect cells during conditions of proteotoxic stress [30].
Figure 2.
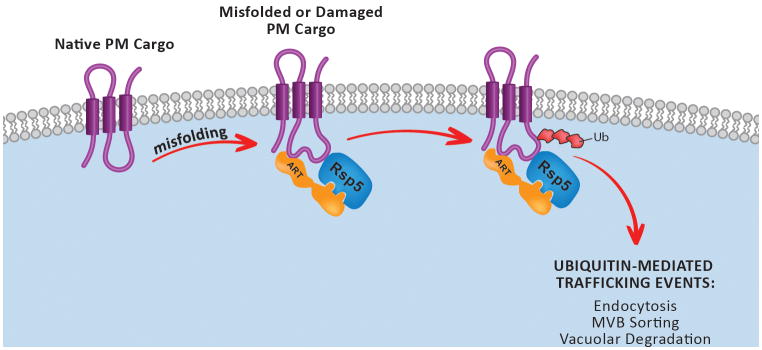
Model of ART-Rsp5-mediated PMQC in yeast.
While it is clear that Rsp5 and specific ARTs function in PMQC, the precise mechanism of recognition and the basis for distinction between native and non-native conformations of the same protein is not well understood. One recent investigation of the yeast uracil transporter Fur4 elucidated a key mechanism for how cell surface transporters may be recognized and targeted for removal [31]. This study proposes that, in the absence of uracil, the peptide sequence in Fur4 that is recognized by Rsp5 is apposed to the cytosolic leaflet of the plasma membrane and inaccessible for binding to Rsp5 [31] (Table 1). However, in the course of the uracil transport cycle, conformational changes in the transporter would destabilize this peptide, making it more accessible to Rsp5. While additional biochemical analysis will be necessary to confirm these ideas, it is clear that many PM transporters transit through a state (or states) of conformational instability during the transport process [32-34]. Thus, it is tempting to speculate that intermediates of substrate transport cycles may expose peptide domains that are recognized by the ubiquitination machinery in the same way that misfolded or damaged transporters would be recognized and targeted for degradation.
Another possible model for how misfolded PM proteins could be recognized and targeted for degradation involves the detection of misfolded proteins by intrinsically disordered domains present in the quality control machinery. Indeed, several recent studies have provided evidence that intrinsically disordered domains can function in the recognition of misfolded proteins. The yeast protein San1, a nuclear-localized RING ubiquitin ligase, is predicted to be >50% disordered, and these disordered domains were shown to be critical for the recognition, ubiquitination, and proteasomal degradation of misfolded substrates in the nucleus [35,36]. In a similar manner, the chaperone Hsp33 was shown to undergo a redox-dependent transition from an ordered conformation to a disordered conformation, with the regions of disorder critical for recognition of misfolded clients [37,38]. Given these and other examples, there is an emerging consensus that intrinsically disordered domains have the potential to recognize misfolded proteins. It is worth noting that many ART family proteins, and particularly the ones critical for PMQC [30], are predicted to have high degrees of intrinsic disorder, with three ART family members >50% disordered and all but two >40% disordered. It is not yet known if these disordered segments of ART family proteins are important for recognition of misfolded cargo, but this property of ART family proteins might explain some key features of the ART network such as its broad cargo recognition capability and the redundancy which makes the system so robust for PMQC.
Exploring New Mechanisms of PMQC
As mechanisms of PMQC are only now coming into focus, it is very likely that novel PMQC mechanisms and pathways will emerge from diverse strategies and types of analysis. One recent study investigated quality control pathways for the human Kir2.1 potassium channel with an elegant screen in yeast. The authors were able to express Kir2.1 in strains lacking the endogenous yeast potassium uptake system, which complemented growth in low potassium conditions [39]. This allowed the authors to conduct a screen for mutants that suppressed or enhanced this complementation, and the authors found that mutations that prevented lysosomal degradation (particularly mutations in the ESCRT pathway) enhanced the ability of Kir2.1 to complement the growth phenotype in low potassium [39]. Importantly, the authors then analyzed the degradation of Kir2.1 in human cells and found that degradation was mediated primarily by lysosomal degradation and, to a lesser extent, proteasomal degradation [39]. Although the exact mechanism of detection and ubiquitination remains unclear, these results suggested a role for PMQC in determining the stability of Kir2.1 at the PM (Table 1). This study also illustrates how synthetic genetic analysis in yeast can be used to screen for quality control pathways that recognized misfolded PM proteins and are highly conserved across evolution.
Fundamentally, protein quality control is about how cells distinguish between native and non-native conformations of a given protein, and this is a particularly challenging problem for integral membrane proteins, which must be monitored for proper folding of cytosolic domains, extracellular domains, and transmembrane domains. Using an in vitro system that reconstitutes ubiquitination events that occur at the ER and target substrates for ERAD, one recent study demonstrated that association with deubiquitylating enzymes (DUBs) enhances discrimination potential of ubiquitin ligases [40]. Using in vitro measurements and computational modeling, the authors demonstrate that DUB association with an E3 ligase can prevent polyubiquitin accumulation on “weak” substrates without dramatically affecting polyubiquitin accumulation on “strong” substrates, thus enhancing discrimination between native and misfolded substrates during ubiquitination [40]. While this study implicated this mechanism for integral membrane proteins in the ER, it is possible that such a mechanism also operates at the PM. Indeed, DUBs have been shown to associate with Rsp5 in yeast [41-43], but whether or not DUBs associate with CHIP in mammalian cells remains to be determined.
As discussed above in the case of CHIP-mediated PMQC, a key strategy for recognizing misfolded proteins throughout the cell is to use molecular chaperones as adaptors that recruit the CHIP E3 ubiquitin ligase, but these mechanisms are limited to detection of the misfolding of large cytosolic domains. Although decades of research have elucidated many fundamental mechanisms for a handful of highly-conserved molecular chaperones, new chaperones are still being discovered and it is clear that many remain to be discovered [44]. This is certainly the case for chaperones of integral membrane proteins at the PM. While canonical chaperones of soluble proteins, like Hsp70 or Hsp90, promote the folding and maintenance of the soluble cytosolic domains of PM proteins, chaperones that promote the folding and stability of transmembrane domains are not well established. Certainly lipids can function to promote the stability of integral membrane proteins, as evidenced from experiments showing that sphingolipids at the cell surface help stabilize the yeast general amino acid transporter, Gap1 [45]. It is also likely that carbohydrates play a role in promoting general PM protein stability, and in particular molecules like trehalose, a disaccharide, seem to function as molecular chaperones that prevent protein aggregation and promote membrane integrity and cell survival in conditions of proteotoxic stress [46-48]. Clues for potential chaperones that act within membrane lipid bilayers may come from studies that profiled yeast transcriptional responses to various different proteotoxic stresses, which reveal that many small (∼100 amino acids in length or less), hydrophobic ORFs are transcriptionally induced in response to proteotoxic stress conditions [49-54]. One such candidate chaperone is Hsp12, a small (109 amino acids in length) unstructured protein that acquires secondary structure in a membrane environment [55]. Hsp12 is a potential candidate membrane protein chaperone, as it is required for yeast survival to proteotoxic stresses (heat, osmotic, and oxidative stresses) and it is clearly important for maintenance of PM integrity under these conditions [55], but further studies will need to address if Hsp12 functions as a chaperone for integral membrane proteins at the PM. In addition to Hsp12, many other small, hydrophobic ORFs have been shown to be required for yeast survival during proteotoxic stress [56], suggesting that various small hydrophobic peptides play a crucial role in resistance to protein misfolding. Whether or not any of these stress-induced hydrophobic peptides function as chaperones for PM proteins remains to be determined.
Conclusions and Perspectives
Although research over the past two decades has provided evidence for cellular maintenance of PM protein quality control, it is only in recent years that specific mechanisms for targeted removal of misfolded PM proteins have been elucidated. These mechanisms involve chaperone-mediated targeting of ubiquitin ligases as well as mechanisms that recognize misfolded proteins via networks of ubiquitin ligase adaptors. In terms of human disease, PMQC pathways appear to be a double-edged sword. Stringent PMQC may facilitate human disease by recognizing mutant alleles of transporters or ion channels (such recognition of misfolded CFTR by CHIP) and mediating their ubiquitination and endocytic downregulation, making these pathways potential therapeutic targets for various channelopathies. On the other hand, relaxing PMQC has the potential to promote hyperproliferation by tolerating (or sustaining) signals from non-native, oncogenic variants of signaling receptors. Only through investigation of these pathways and analysis of the cellular consequences of modulating PMQC activity can we fully appreciate the role of PM quality control pathways in health and disease.
Acknowledgments
J.A.M is grateful to Scott Emr for helpful comments and discussion. J.A.M. is supported by an NIH Pathway to Independence Award (1R00 GM101077-01).
Glossary Box
- ERAD
Endoplasmic Reticulum Associated Degradation
- DUB
deubiquitinating enzyme
- CFTR
cystic fibrosis transmembrane conductance regulator
- ART
arrestin-related trafficking adaptor
- MVB
multivesicular body
- Channelopathies
human diseases caused by mutations that affect ion channel function
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Theodoraki MA, Nillegoda NB, Saini J, Caplan AJ. A network of ubiquitin ligases is important for the dynamics of misfolded protein aggregates in yeast. J Biol Chem. 2012;287:23911–23922. doi: 10.1074/jbc.M112.341164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nillegoda NB, Theodoraki MA, Mandal AK, Mayo KJ, Ren HY, Sultana R, Wu K, Johnson J, Cyr DM, Caplan AJ. Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol Biol Cell. 2010;21:2102–2116. doi: 10.1091/mbc.E10-02-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 4.Gardner BM, Pincus D, Gotthardt K, Gallagher CM, Walter P. Endoplasmic reticulum stress sensing in the unfolded protein response. Cold Spring Harb Perspect Biol. 2013;5:a013169. doi: 10.1101/cshperspect.a013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2012;151:1163–1167. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thibault G, Ng DT. The endoplasmic reticulum-associated degradation pathways of budding yeast. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a013193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hampton RY, Sommer T. Finding the will and the way of ERAD substrate retrotranslocation. Curr Opin Cell Biol. 2012;24:460–466. doi: 10.1016/j.ceb.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Gardner RG, Nelson ZW, Gottschling DE. Degradation-mediated protein quality control in the nucleus. Cell. 2005;120:803–815. doi: 10.1016/j.cell.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 9.Rosenbaum JC, Gardner RG. How a disordered ubiquitin ligase maintains order in nuclear protein homeostasis. Nucleus. 2011;2:264–270. doi: 10.4161/nucl.2.4.16118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker BM, Haynes CM. Mitochondrial protein quality control during biogenesis and aging. Trends Biochem Sci. 2011;36:254–261. doi: 10.1016/j.tibs.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Haynes CM, Ron D. The mitochondrial UPR - protecting organelle protein homeostasis. J Cell Sci. 2010;123:3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 12.Macgurn JA, Hsu PC, Emr SD. Ubiquitin and membrane protein turnover: from cradle to grave. Annu Rev Biochem. 2012;81:231–259. doi: 10.1146/annurev-biochem-060210-093619. [DOI] [PubMed] [Google Scholar]
- 13.Henne WM, Buchkovich NJ, Emr SD. The ESCRT Pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 14.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 15.Sharma M, Pampinella F, Nemes C, Benharouga M, So J, Du K, Bache KG, Papsin B, Zerangue N, Stenmark H, et al. Misfolding diverts CFTR from recycling to degradation: quality control at early endosomes. J Cell Biol. 2004;164:923–933. doi: 10.1083/jcb.200312018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenness DD, Li Y, Tipper C, Spatrick P. Elimination of defective alpha-factor pheromone receptors. Mol Cell Biol. 1997;17:6236–6245. doi: 10.1128/mcb.17.11.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Kane T, Tipper C, Spatrick P, Jenness DD. Yeast mutants affecting possible quality control of plasma membrane proteins. Mol Cell Biol. 1999;19:3588–3599. doi: 10.1128/mcb.19.5.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okiyoneda T, Barrière H, Bagdány M, Rabeh W, Du K, Höhfeld J, Young J, Lukacs G. Peripheral Protein Quality Control Removes Unfolded CFTR from the Plasma Membrane. Science. 2010;329:805–810. doi: 10.1126/science.1191542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Apaja PM, Xu H, Lukacs GL. Quality control for unfolded proteins at the plasma membrane. J Cell Biol. 2010;191:553–570. doi: 10.1083/jcb.201006012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 21.Slotman JA, da Silva Almeida AC, Hassink GC, van de Ven RH, van Kerkhof P, Kuiken HJ, Strous GJ. Ubc13 and COOH terminus of Hsp70-interacting protein (CHIP) are required for growth hormone receptor endocytosis. J Biol Chem. 2012;287:15533–15543. doi: 10.1074/jbc.M111.302521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22**.Apaja PM, Foo B, Okiyoneda T, Valinsky WC, Barriere H, Atanasiu R, Ficker E, Lukacs GL, Shrier A. Ubiquitination-dependent quality control of hERG K+ channel with acquired and inherited conformational defect at the plasma membrane. Mol Biol Cell. 2013;24:3787–3804. doi: 10.1091/mbc.E13-07-0417. Here the authors demonstrate CHIP-mediated PMQC for the human potassium channel hERG, which is mutated or otherwise destabilized in long QT type 2 syndrome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsumura Y, Sakai J, Skach WR. Endoplasmic reticulum protein quality control is determined by cooperative interactions between Hsp/c70 protein and the CHIP E3 ligase. J Biol Chem. 2013;288:31069–31079. doi: 10.1074/jbc.M113.479345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440:551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cyr DM, Höhfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci. 2002;27:368–375. doi: 10.1016/s0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 26**.Shi Y, Wang J, Li JD, Ren H, Guan W, He M, Yan W, Zhou Y, Hu Z, Zhang J, et al. Identification of CHIP as a Novel Causative Gene for Autosomal Recessive Cerebellar Ataxia. PLoS One. 2013;8:e81884. doi: 10.1371/journal.pone.0081884. This study establishes a link between mutations in CHIP and cerebral ataxia and provides evidence that disease mutant CHIP proteins are ineffective at mediating the degradation of N-methyl-D-aspartate receptors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin CH, MacGurn JA, Chu T, Stefan CJ, Emr SD. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell. 2008;135:714–725. doi: 10.1016/j.cell.2008.09.025. [DOI] [PubMed] [Google Scholar]
- 28.Nikko E, Pelham HRB. Arrestin-Mediated Endocytosis of Yeast Plasma Membrane Transporters. Traffic. 2009;10:1856–1867. doi: 10.1111/j.1600-0854.2009.00990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nikko E, Sullivan JA, Pelham HRB. Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep. 2008;9:1216–1221. doi: 10.1038/embor.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30**.Zhao Y, MacGurn JA, Liu M, Emr S. The ART-Rspubiquitin ligase network comprises a plasma membrane quality control system that protects yeast cells from proteotoxic stress. Elife. 2013;2:e00459. doi: 10.7554/eLife.00459. This study demonstrates a role for the ART-Rspnetwork in the recognition and downregulation of multiple PM proteins in response to various proteotoxic stresses. The authors also demonstrate the accumulation of misfolded PM proteins at the cell surface is toxic and that sequential membrane protein quality control mechanisms work together to protect the cell in conditions that challenge PM protein folding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31**.Keener JM, Babst M. Quality control and substrate-dependent downregulation of the nutrient transporter Fur4. Traffic. 2013;14:412–427. doi: 10.1111/tra.12039. This study provides evidence that recognition of misfolded Fur4, the yeast uracil transporter, is mediated by Rspand is based on sensing conformational flux of a specific peptide away from the PM. The authors provide evidence that this mechanism of recognition is similar to substrate- induced conformational changes that also drive downregulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimamura T, Weyand S, Beckstein O, Rutherford NG, Hadden JM, Sharples D, Sansom MS, Iwata S, Henderson PJ, Cameron AD. Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science. 2010;328:470–473. doi: 10.1126/science.1186303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weyand S, Shimamura T, Yajima S, Suzuki S, Mirza O, Krusong K, Carpenter EP, Rutherford NG, Hadden JM, O'Reilly J, et al. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science. 2008;322:709–713. doi: 10.1126/science.1164440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reyes N, Ginter C, Boudker O. Transport mechanism of a bacterial homologue of glutamate transporters. Nature. 2009;462:880–885. doi: 10.1038/nature08616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35*.Rosenbaum JC, Fredrickson EK, Oeser ML, Garrett-Engele CM, Locke MN, Richardson LA, Nelson ZW, Hetrick ED, Milac TI, Gottschling DE, et al. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell. 2011;41:93–106. doi: 10.1016/j.molcel.2010.12.004. This study presented evidence for the recognition of misfolded proteins by intrinsically disordered regions of the San1 E3 ubiquitin ligase, which provides a critical mechanism for nuclear protein quality control. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fredrickson EK, Rosenbaum JC, Locke MN, Milac TI, Gardner RG. Exposed hydrophobicity is a key determinant of nuclear quality control degradation. Mol Biol Cell. 2011;22:2384–2395. doi: 10.1091/mbc.E11-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bardwell JC, Jakob U. Conditional disorder in chaperone action. Trends Biochem Sci. 2012;37:517–525. doi: 10.1016/j.tibs.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.Reichmann D, Xu Y, Cremers CM, Ilbert M, Mittelman R, Fitzgerald MC, Jakob U. Order out of disorder: working cycle of an intrinsically unfolded chaperone. Cell. 2012;148:947–957. doi: 10.1016/j.cell.2012.01.045. This study demonstrates that Hsp33 can undergo a redox-dependent order-to-disorder transition in order to bind misfolded client proteins via an intrinsic disordered domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39**.Kolb AR, Needham PG, Rothenberg C, Guerriero CJ, Welling PA, Brodsky JL. ESCRT regulates surface expression of the Kir2.1 potassium channel. Mol Biol Cell. 2014;25:276–289. doi: 10.1091/mbc.E13-07-0394. In this study, the authors use yeast genetic screening to identify a role for the ESCRT pathway in the quality control of Kir2.1, a human potassium channel. The data presented illustrate how multiple quality control pathways contribute to Kir2.1 abundace at the cell surface. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40**.Zhang ZR, Bonifacino JS, Hegde RS. Deubiquitinases sharpen substrate discrimination during membrane protein degradation from the ER. Cell. 2013;154:609–622. doi: 10.1016/j.cell.2013.06.038. This investigation of the recognition and ubiquitination of misfolded integral membrane proteins at the ER provides evidence of a role for DUBs in quality control by promoting the distinction between native and non-native states of integral membrane proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lam MH, Urban-Grimal D, Bugnicourt A, Greenblatt JF, Haguenauer-Tsapis R, Emili A. Interaction of the deubiquitinating enzyme Ubp1 and the e3 ligase Rsp5 is required for transporter/receptor sorting in the multivesicular body pathway. PLoS One. 2009;4:e4259. doi: 10.1371/journal.pone.0004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kee Y, Muñoz W, Lyon N, Huibregtse JM. The deubiquitinating enzyme Ubpmodulates Rsp5-dependent Lys63-linked polyubiquitin conjugates in Saccharomyces cerevisiae. J Biol Chem. 2006;281:36724–36731. doi: 10.1074/jbc.M608756200. [DOI] [PubMed] [Google Scholar]
- 43.Kee Y, Lyon N, Huibregtse JM. The Rspubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J. 2005;24:2414–2424. doi: 10.1038/sj.emboj.7600710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quan S, Bardwell JC. Chaperone discovery. Bioessays. 2012;34:973–981. doi: 10.1002/bies.201200059. [DOI] [PubMed] [Google Scholar]
- 45.Lauwers E, Grossmann G, André B. Evidence for coupled biogenesis of yeast Gap1 permease and sphingolipids: essential role in transport activity and normal control by ubiquitination. Mol Biol Cell. 2007;18:3068–3080. doi: 10.1091/mbc.E07-03-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kandror O, Bretschneider N, Kreydin E, Cavalieri D, Goldberg AL. Yeast adapt to near-freezing temperatures by STRE/Msn2,4-dependent induction of trehalose synthesis and certain molecular chaperones. Mol Cell. 2004;13:771–781. doi: 10.1016/s1097-2765(04)00148-0. [DOI] [PubMed] [Google Scholar]
- 47.Benaroudj N, Lee DH, Goldberg AL. Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J Biol Chem. 2001;276:24261–24267. doi: 10.1074/jbc.M101487200. [DOI] [PubMed] [Google Scholar]
- 48.Lee DH, Goldberg AL. Proteasome inhibitors cause induction of heat shock proteins and trehalose, which together confer thermotolerance in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:30–38. doi: 10.1128/mcb.18.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamoto A, Mizukami Y, Sakurai H. Identification of a novel class of target genes and a novel type of binding sequence of heat shock transcription factor in Saccharomyces cerevisiae. J Biol Chem. 2005;280:11911–11919. doi: 10.1074/jbc.M411256200. [DOI] [PubMed] [Google Scholar]
- 50.Matsumoto R, Akama K, Rakwal R, Iwahashi H. The stress response against denatured proteins in the deletion of cytosolic chaperones SSA1/is different from heat-shock response in Saccharomyces cerevisiae. BMC Genomics. 2005;6:141. doi: 10.1186/1471-2164-6-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berry DB, Gasch AP. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol Biol Cell. 2008;19:4580–4587. doi: 10.1091/mbc.E07-07-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones DL, Petty J, Hoyle DC, Hayes A, Oliver SG, Riba-Garcia I, Gaskell SJ, Stateva L. Genome-wide analysis of the effects of heat shock on a Saccharomyces cerevisiae mutant with a constitutively activated cAMP-dependent pathway. Comp Funct Genomics. 2004;5:419–431. doi: 10.1002/cfg.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tai SL, Daran-Lapujade P, Walsh MC, Pronk JT, Daran JM. Acclimation of Saccharomyces cerevisiae to low temperature: a chemostat-based transcriptome analysis. Mol Biol Cell. 2007;18:5100–5112. doi: 10.1091/mbc.E07-02-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welker S, Rudolph B, Frenzel E, Hagn F, Liebisch G, Schmitz G, Scheuring J, Kerth A, Blume A, Weinkauf S, et al. Hsp12 is an intrinsically unstructured stress protein that folds upon membrane association and modulates membrane function. Mol Cell. 2010;39:507–520. doi: 10.1016/j.molcel.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 56.Kastenmayer JP, Ni L, Chu A, Kitchen LE, Au WC, Yang H, Carter CD, Wheeler D, Davis RW, Boeke JD, et al. Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res. 2006;16:365–373. doi: 10.1101/gr.4355406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y, Chang A. Quality control of a mutant plasma membrane ATPase: ubiquitylation prevents cell-surface stability. J Cell Sci. 2006;119:360–369. doi: 10.1242/jcs.02749. [DOI] [PubMed] [Google Scholar]


