1. Introduction
Amyotrophic lateral sclerosis (ALS), the most common motor neuron disease1, and frontotemporal dementia (FTD)2, the most frequent presenile onset neurodegenerative disorder, now belong to a class of neurological conditions caused by repeat expansions, owing to the 2011 discovery that an expanded hexanucleotide (GGGGCC) repeat within a non-coding region of the chromosome 9 open reading frame 72 (C9orf72) gene is the major genetic cause of both disorders, collectively referred to as c9FTD/ALS (OMIM *614260). Because repeat expansions causing other neurodegenerative diseases are known to alter epigenetic mechanisms while simultaneously leading to toxic RNA gain-of-function (GOF) and protein loss-of-function (LOF)3, researchers now seek to elucidate whether GOF and/or LOF mechanisms contribute to c9FTD/ALS. Studies from our group as well as from others led to the finding that sense and antisense RNA transcripts containing the expanded repeat are produced and form nuclear RNA foci, which have the capacity to sequester RNA-binding proteins and impair their function4. It was also shown that the bidirectionally transcribed expanded repeat can become the template for the synthesis of aggregation-prone “c9RAN proteins” by repeat-associated non-ATG (RAN) translation5–7. While these two pathological mechanisms support the RNA GOF hypothesis, new research also demonstrates that decreased expression of one or multiple C9orf72 transcript variants is detected in the frontal cortex, motor cortex, cerebellum and cervical spinal cord in c9FTD/ALS6,8–12, as well as in c9FTD/ALS lymphoblastoid cell lines10 and neuronal lines differentiated from c9FTD/ALS induced pluripotent stem cells11,13. The consequence of C9orf72 downregulation has been evaluated by knocking-down the C9orf72 orthologue in zebrafish, which led to both altered morphology of motor neuron axons and locomotor deficits, a phenotype rescued by overexpression of human C9orf7214. Of note, the fact that C9orf72 mouse orthologue is enriched in brain regions susceptible to degenerate in ALS and FTD15, suggests that sufficient C9orf72 expression is critical for neuronal survival. Deciphering the cause of decreased C9orf72 expression, and whether C9orf72 LOF contributes to c9FTD/ALS pathogenesis are questions important to the field, but limitations with currently available C9orf72 antibodies prevent adequate protein quantification analyses. Recently, we reported that C9orf72 mRNA levels are reduced in frontal cortices and cerebella of C9orf72 expanded repeat carriers as a result of histone trimethylation at residues H3K9, H3K27, H3K79, and H4K2016. We also provided direct evidence that aberrant methylation of these lysine residues plays a significant role in the pathophysiology of c9FTD/ALS by demonstrating that methylation inhibition by 5-aza-2-deoxycytidine treatment not only increases C9orf72 mRNA expression in c9FTD/ALS-derived fibroblasts, but also reduces the aberrant trimethylation of histone H3K9 bound to C9orf7216. In addition, a recent study reported DNA hypermethylation of the C9orf72 promoter region, at the 5' end of the repeat expansion, in blood, frontal cortex, and spinal cord of about 40% of c9ALS cases. While hypermethylation was reported to be consistently present across these tissues in seven patients, some variation in terms of the specific number of CpG sites methylated was observed17. Moreover, it is still unknown whether such hypermethylation is present to a similar extent in c9FTD patients, whether the number of methylated sites varies across brain regions, which might correlate with disease phenotype, and whether DNA hypermethylation occurs concomitantly with aberrant histone methylation. To shed new light on these questions, we sought to examine whether the cerebella of c9FTD and c9ALS patients, a brain region affected in both diseases and previously reported by our group to be aberrantly methylated at histone residues H3K9, H3K27, H3K79, and H4K2016,18, also show hypermethylation of the CpG island located at the 5' end of the repeat expansion.
2. Results
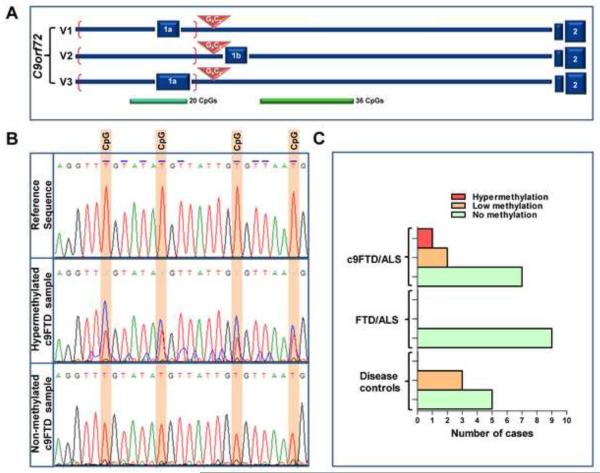
We previously reported that aberrant histone methylation in the cerebellum leads to reduced C9orf72 mRNA expression in this brain region. However, whether or not DNA hypermethylation of the CpG island at the 5' end of the repeat expansion (Figure 1A) contributes to the reduction or has the ability to modulate disease phenotype is still unknown. To explore this question, we isolated DNA from the cerebellum of four ALS and six FTD patients carrying the C9orf72 expansion, termed c9ALS and c9FTD respectively, four ALS and five FTD cases lacking the C9orf72 expansion, and eight disease control participants (clinical information in Table 1). DNA was treated with bisulfite, which effectively converts cytosine residues to uracils without converting the 5-methylcytosine residues. Subsequently after PCR amplification, sequence analysis was performed; all uracil residues were read as thymines and all 5-methylcytosine residues were read as cytosines. Methylation analysis revealed the majority of the disease control cases lacked methylated sites; however, one or two of the 26 CpG sites evaluated were methylated in three cases. As a result, we categorized samples having more than three methylated sites as being hypermethylated, as previously suggested17. Of note, we found only one sample in our cohort of pathogenic repeat carriers to be hypermethylated with 16 different CpG sites methylated (Table 1 and Figure 1B). Of interest, this sample was obtained from an FTD patient, the first c9FTD case exhibiting hypermethylation of the CpG promoter region reported so far. Two other expansion repeat carriers were found to have one CpG site methylated. No methylation was found in the cerebella of ALS and FTD patients carrying normal alleles (Table 1 and Figure 1C). The cytosine located 167 base pairs before the annotated C9orf72 gene (NG_031977.1) has been found to be the most commonly methylated, and was present in five different samples out of the six found methylated.
Figure 1.
The C9orf72 promoter is hypermethylated in one c9FTD patient. (A) Schematic representation showing the three known transcript variants of C9orf72; V1: NM_145005.5, V2: NM_018325.3, V3: NM_001256054.1. Red triangles mark the repeat location. Red brackets mark the amplified region after the first PCR run. The two CpG islands located at the 5' and 3' end of the repeat region are shown in green. (B) Chromatograms showing sequence amplification of the CpG island located at the 5' end of the repeat region after bisulfite treatment of the DNA isolated from cerebellum. The top panel shows the reference sequence. The middle panel shows a section of the sequence obtained from the c9FTD patient hypermethylated at the C9orf72 promoter region (c9FTD/ALS sample # 8, see Table 1). The last panel shows a section of the sequence obtained from a c9FTD patient in whom no methylated sites were identified (c9FTD/ALS sample #4, see Table 1). Blue line over thymines in the reference sequence marks sites of bisulfite conversion from cytosines. CpG sites are highlighted in orange. (C) Bar graph representing the number of samples showing hypermethylation, low methylation, or no methylation of the CpG island located at the 5' end of the repeat region in cerebellum of each group.
Table 1.
Clinical, genetic, and epigenetic information on study participants.
| Group | Sample | Sample type | Family history | Mutation | Pathological diagnosis | Sex | Age of onset (years) | Duration (months) | Trimethylated H3K9, H3K27, H3K79, H4K20 | Number of cerebellum methylated CpG sites (/26) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Frontal cortex | Cerebellum | ||||||||||
| c9FTD/c9ALS | 1 | ALS | no | C9orf72 repeat expansion | ALS/PA | F | 61 | 33 | yes | yes | 0 |
| 2 | ALS | no | C9orf72 repeat expansion | ALS | M | 56 | 26 | yes | yes | 1 | |
| 3 | ALS | no | C9orf72 repeat expansion | ALS | M | 52 | 76 | yes | yes | 0 | |
| 4 | ALS | Cognitive impairments/dementia | C9orf72 repeat expansion | ALS | M | 49 | 49 | yes | yes | 0 | |
| 5 | FTD | Dementia | C9orf72 repeat expansion | FTLD-U/HpScl | M | 67 | 98 | yes | yes | 16 | |
| 6 | FTD | AD/FTD | C9orf72 repeat expansion | FTLD-U | M | 70 | 28 | yes | yes | 0 | |
| 7 | FTD | FTD | C9orf72 repeat expansion | FTLD-U | M | 74 | 93 | yes | yes | 1 | |
| 8 | FTD | no | C9orf72 repeat expansion | FTLD-U/HpScl/PA | M | 62 | 131 | yes | yes | 0 | |
| 9 | FTD | ALS/FTD/Neurodegeneration | C9orf72 repeat expansion | FTLD/MND/HpScl | M | <60 | 60 | yes | yes | 0 | |
| 10 | FTD | Dementia Neurodegeneration | C9orf72 repeat expansion | FTLD-U/HpScl | M | 68 | 71 | yes | yes | 0 | |
| FTD/ALS | 1 | ALS | no | unknown | ALS/PA | F | 60 | 15 | no | no | 0 |
| 2 | ALS | no | unknown | ALS/heterotopia | M | 45 | 58 | no | no | 0 | |
| 3 | ALS | no | unknown | ALS | M | 50 | 47 | no | no | 0 | |
| 4 | ALS | no | unknown | ALS/SC | M | 53 | 65 | no | no | 0 | |
| 5 | FTD | Neurodegeneration | unknown | FTLD-U/HpScl/AGD | M | 63 | 51 | no | no | 0 | |
| 6 | FTD | no | unknown | FTLD-U/HpScl/AGD/VaD | M | 73 | 105 | no | no | 0 | |
| 7 | FTD | Neurodegeneration | unknown | FTLD-U/HpScl/PA/BLBD | M | 74 | 45 | no | no | 0 | |
| 8 | FTD | no | unknown | FTLD-U/VaD/AGD | M | 65 | 18 | no | no | 0 | |
| 9 | FTD | no | unknown | FTLD-U/Limbic gliosis | M | 66 | 122 | no | no | 0 | |
| Disease controls | 1 | n/a | ET | unknown | PA/CVA | M | 72 | 112 | no | no | 0 |
| 2 | n/a | no | unknown | PA/VaD | F | n/a | n/a | no | no | 0 | |
| 3 | n/a | ET | unknown | AGD/PA | F | 82 | 22 | no | no | 0 | |
| 4 | n/a | no | unknown | AGD/Fahr | M | 72 | 50 | no | no | 0 | |
| 5 | n/a | Dementia | n/a | Normal | F | 64 | 9 | no | no | 1 | |
| 6 | n/a | unavailable | unknown | VaD | M | 63 | 79 | no | no | 2 | |
| 7 | n/a | Neurodegeneration | unknown | VaD/HpScl | M | 63 | 100 | no | no | 2 | |
| 8 | n/a | ALS/dementia | unknown | VaD/SC | M | unknown | unknown | no | no | 0 | |
AD: Alzheimer's Disease, AGD: Argyrophilic Grain Disease, ALS: Amyotrophic Lateral Sclerosis, BLBD: Brainstem Lewy Body Disease, CVA: Cerebral Vascular Accident, ET: Essential Tremor, Fahr: Fahr's Syndrome, FTLD-U: Frontotemporal Lobar Degeneration with Ubiquitin inclusions, HpScl: Hippocampal Sclerosis, ILBD: Incidental Lewy Body Disease, MND: Motor Neuron Disease, PA: Pathological Aging, SC: Sydenham's chorea, VaD: Vascular Dementia. M: male, F: female, n/a: non-applicable.
3. Discussion
In this study, we evaluated aberrant DNA methylation of the C9orf72 promoter region in cerebella of both pathogenic repeat carriers and carriers of normal alleles. Out of the ten c9FTD/ALS samples tested, only one was found to be hypermethylated, representing 10% of our small cohort and approximately 17% of the c9FTD patients included in this study. The hypermethylated sample was from a patient diagnosed with FTD at 67 years of age, with disease progression lasting 98 months (Table 1). The clinical profile of this patient was similar to the one of other non-methylated FTD samples included in this group. Of the c9ALS samples, none were hypermethylated, with the caveat being that only four patients were tested in this cohort. Future studies will be needed to validate whether the proportion of c9ALS patients exhibiting hypermethylation in cerebellum differs from that of c9FTD.
We previously demonstrated that all c9FTD/ALS cases included in this study exhibit reduced C9orf72 mRNA expression as a result of aberrant histone methylation at several lysine residues16. Here, we demonstrate that 90% of these cases are not hypermethylated at the 5' end of the repeat expansion, further suggesting that aberrant histone methylation alone might be sufficient to induce reduced C9orf72 mRNA expression. While histone methylation is generally perceived as a dynamic process, which induces short-term gene silencing through regulation of several histone-modifying complexes, DNA methylation is considered to be very stable. As interactions exist between all epigenetic markers, changes in histone methylation can induce de novo somatic DNA methylation, this way leaving stable fingerprints in the epigenome. Sporadic DNA hypermethylation of the C9orf72 promoter region may represent such de novo fingerprints, thus explaining why DNA hypermethylation is present in only a subgroup of c9FTD/ALS patients and why the number of methylated sites slightly varies across tissues. On the other hand, DNA hypermethylation at other sites near the C9orf72 locus could induce aberrant histone methylation, leading to decreased expression. For example, it is possible that CpG islands located in the promoter region of nearby genes, or other CpG sites either surrounding or within C9orf72, are also methylated. In addition, as the repeat expansion creates a new CpG island, it is highly likely that the repeat itself is methylated. The significant heterogeneity in phenotypes found among family members carrying a pathogenic C9orf72 repeat expansion may alternatively be attributable to the number of methylated CpG sites, as it was previously reported that level of DNA methylation is inversely associated with disease duration17, but several large cohorts of patients need to be evaluated to further explore this hypothesis. Such phenotypic variation is believed to be the result of modifier genes, but it can also arise from different epitypes, which defines unique epigenetic status at different location for each individual, and influence disease progression and penetrance. While expanded repeats have been shown to modify the epitype in other diseases3, other factors, including environmental exposure to toxins, diet, and aging, are all known to influence epigenetic changes19,20 and might in turn act as disease modifiers. Interestingly, levels of proteins involved in methylation processes, such as DNMT1, DNMT3a, and 5-methylcytosine, have all been reported to be up-regulated in motor neurons of ALS patients21. This suggests that epigenetic changes might not be restricted to pathogenic C9orf72 repeat carriers and might possibly have a greater role in ALS and FTD than previously believed.
While the underlying mechanisms responsible for neurodegeneration in FTD and ALS still need to be clearly defined, recent advances have permitted the identification of several contributors to disease. As in other neurodegenerative diseases, c9FTD/ALS is characterized by the accumulation of sense and antisense RNA foci, sequestration of select RNA-binding proteins leading to transcriptome defects, generation of sense and antisense c9RAN proteins, and epigenetic changes4. Whether these events are a primary mechanism in c9FTD/ALS or a consequence of other disturbed cell functions is still unknown. However, understanding the role of epigenetic events in the disease cascade may greatly aid in the identification of biomarkers and the development of future therapeutics.
4. Experimental Procedure
Standard Protocol Approvals, Registrations, and Patient Consents
Protocols were approved by the institutional review board and ethics committee on human and animal experimentation of Mayo Clinic. All participants or authorized family members gave written informed consent after which participant information was collected.
Subjects
Participants in this study were recruited at Mayo Clinic Florida and were independently ascertained by trained neurologists. Cerebella tissues were obtained after post-mortem analyses from four ALS and six FTD patients carrying the C9orf72 expansion, four ALS and five FTD cases without C9orf72 expansion, and eight disease control participants (clinical information in Table 1).
Bisulfite treatment and methylation analysis
DNA was extracted from cerebellum using the Wizard genomic DNA purification kit (Promega, Madison, WI, USA) as per manufacturers' instructions. As bisulfite converts cytosine residues to uracils, which are subsequently read as thymines while leaving the 5-methylcytosine residues unconverted, bisulfite treatment of 1 μg of cerebellum DNA was conducted using Epi Tech Bisulfite kit (QIAGEN) as per manufacturers' instructions. Semi-nested polymerase chain reaction (PCR) amplification of the CpG island 5' of the repeat expansion was conducted as previously reported17. Briefly, forward bisulfite primer 5'TTATTAGGGTTTGTAGTGGAGTTTT3' and reverse bisulfite primer 5'AAATCTTTTCTTATTCACCCTCAAC3' were used for the first PCR amplification, followed by a second amplification using internal forward primer 5'TATTAGGGTTTGTAGTGGAGTTTT3' and internal reverse primer 5'CCACACCTACTCTTACTAAACCC3'. 2 μl of the bisulfite converted DNA was amplified with the FastStart PCR Master Mix (Roche, Basel, Switzerland). For each sample, a denaturation step of 4 minutes was first performed at 95°C. Then a touchdown protocol was used which consisted of an initial cycle of 30 seconds denaturation at 95°C, 30 seconds annealing at 68°C for the first reaction and at 67°C for the nested reaction, and 3 minutes elongation at 72°C. This was followed by ten cycles in which the annealing temperature was decreased by 1.0°C per cycle. Then, a 30 seconds denaturation at 95°C, followed by a 30 seconds annealing at 58°C for the first reaction and at 57°C for the nested reaction, and 3 minute elongation at 72°C were run for 30 or 20 cycles respectively. A final extension at 72°C was performed for 7 minutes. Samples were electrophoresed in a 2% agarose gel. PCR products were purified using AMPure (Agencourt Bioscience, Beverly, MA, USA) then sequenced in both directions with the internal primer pair using Big Dye Terminator v.3.1 Cycle Sequencing kit (Applied Biosystems). Sequencing reactions were purified using CleanSEQ (Agencourt Bioscience) and analyzed by ABI3730 Genetic Analyzer. Sequences were read and analyzed using the integrated DNA methylation tool provided with the Mutation Surveyor software v4.0.7 (Softgenetics, State College, PA, USA). Samples included in the analysis showed a bisulfite conversion rate >95%.
Highlights
First assessment of DNA hypermethylation in cerebellum of c9FTD and c9ALS patients
Only 10% of histone trimethylated cases show DNA hypermethylation in cerebellum
Novel report of a c9FTD case hypermethylated at the C9orf72 promoter region
Acknowledgements
We would like to thank the patients involved in this study as well as acknowledge the technical support of Caroline T. Stetler, Mercedes Prudencio, Amanda M. Liesinger, and Linda G. Rousseau from Mayo Clinic, as well as Brent DiGiorgio from Softgenetics. This work was supported by Mayo Clinic Foundation (LP), National Institutes of Health/National Institute on Aging [R01AG026251 (LP)], National Institutes of Health/National Institute of Neurological Disorders and Stroke [R01 NS 063964-01 (LP), R01 NS077402 (LP), ES20395-01 (LP)], Amyotrophic Lateral Sclerosis Association (LP, POB), the Canadian Institutes of Health Research (VVB), and the Siragusa Foundation (VVB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure VVB contributed to the conception and design, acquisition of data, analysis and interpretation of data, and drafting the article. POB contributed to the acquisition of data, analysis and interpretation of data, and critical revision of the manuscript for important intellectual content. TFG contributed to analysis and interpretation of data, and critical revision of the manuscript for important intellectual content. MEM contributed to the acquisition of clinical data, and critical revision of the manuscript for important intellectual content. DD contributed by providing brain tissues and critical revision of the manuscript for important intellectual content. LP contributed to the conception and design, interpretation of data, critical revision of the manuscript for important intellectual content, acquisition of funding, and supervision of the study. All authors gave final approval of the version to be published. In addition, all authors have participated sufficiently in the work to take public responsibility for appropriate portions of the content.
Conflict of Interests All authors declare no conflict of interest.
References
- 1.Johnston CA, et al. Amyotrophic lateral sclerosis in an urban setting: a population based study of inner city London. Journal of neurology. 2006;253:1642–3. doi: 10.1007/s00415-006-0195-y. [DOI] [PubMed] [Google Scholar]
- 2.Galimberti D, Scarpini E. Genetics and biology of Alzheimer's disease and frontotemporal lobar degeneration. Int J Clin Exp Med. 2010;3:129–43. [PMC free article] [PubMed] [Google Scholar]
- 3.Belzil VV, Gendron TF, Petrucelli L. RNA-mediated toxicity in neurodegenerative disease. Mol Cell Neurosci. 2012;56C:406–419. doi: 10.1016/j.mcn.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gendron TF, Belzil V, Zhang YJ, Petrucelli L. Mechanisms of Toxicity in C9FTLD/ALS. Acta neuropathologica. 2014 doi: 10.1007/s00401-013-1237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ash PE, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–46. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mori K, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–8. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 7.Gendron TF, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta neuropathologica. 2013;126:829–44. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gijselinck I, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11:54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 9.Belzil VV, et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126:895–905. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciura S, et al. Loss of function of C9orf72 causes motor deficits in a zebrafish model of Amyotrophic Lateral Sclerosis. Ann Neurol. 2013 doi: 10.1002/ana.23946. [DOI] [PubMed] [Google Scholar]
- 11.Donnelly CJ, et al. RNA Toxicity from the ALS/FTD C9ORF72 Expansion Is Mitigated by Antisense Intervention. Neuron. 2013;80:415–28. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fratta P, et al. Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia. Acta Neuropathol. 2013;126:401–9. doi: 10.1007/s00401-013-1147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almeida S, et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126:385–99. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciura S, et al. Loss of function of C9orf72 causes motor deficits in a zebrafish model of Amyotrophic Lateral Sclerosis. Annals of neurology. 2013 doi: 10.1002/ana.23946. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki N, et al. The mouse C9ORF72 ortholog is enriched in neurons known to degenerate in ALS and FTD. Nature neuroscience. 2013;16:1725–7. doi: 10.1038/nn.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belzil VV, et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta neuropathologica. 2013;126:895–905. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xi Z, et al. Hypermethylation of the CpG Island Near the GC Repeat in ALS with a C9orf72 Expansion. American Journal of Human Genetics. 2013 doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prell T, Grosskreutz J. The involvement of the cerebellum in amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener. 2013 doi: 10.3109/21678421.2013.812661. [DOI] [PubMed] [Google Scholar]
- 19.Cencioni C, et al. Oxidative stress and epigenetic regulation in ageing and age-related diseases. Int J Mol Sci. 2013;14:17643–63. doi: 10.3390/ijms140917643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu H, Liu X, Deng Y, Qing H. DNA methylation, a hand behind neurodegenerative diseases. Front Aging Neurosci. 2013;5:85. doi: 10.3389/fnagi.2013.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chestnut BA, et al. Epigenetic regulation of motor neuron cell death through DNA methylation. J Neurosci. 2011;31:16619–36. doi: 10.1523/JNEUROSCI.1639-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]