Abstract
Purpose
The purpose of this study was to show that healthy adult human ovaries can be a source of cells showing typical MSCs characteristics under in vitro conditions.
Methods and results
The cells, which were isolated from ovarian cortex tissue and named putative ovarian mesenchymal stem cells (PO-MSCs), were compared to bone marrow-derived MSCs (BM-MSCs) and to adult human dermal fibroblasts (HDFs). The results of a gene expression analysis using the Human Mesenchymal Stem Cell RT² Profiler™ PCR Array revealed that PO-MSCs were different than fibroblasts. They expressed most of the analyzed genes as BM-MSCs, although some genes were differentially expressed. However, the heterogeneity of PO-MSCs samples was revealed. The PO-MSCs expressed the characteristic genes related to MSCs, such as CD105, CD44, CD90, M-CAM, CD73 and VCAM1. In addition, the expression of markers CD44, CD90, M-CAM and STRO-1 was confirmed in PO-MSCs using immunocytochemistry. The PO-MSCs showed multipotent character, since they were able to differentiate into the cells of adipogenic, osteogenic, neural and pancreatic lineage.
Conclusions
Healthy adult human ovaries can harbour an interesting population of cells showing typical MSCs characteristics under in vitro conditions and for this reason we named these cells putative MSCs. These cells express genes encoding main MSCs markers and have an interesting differential potential. Based on these results, we propose PO-MSCs as a novel type of MSCs which share some similarities with BM-MSCs. Nevertheless they show distinct and specific characteristics and are not fibroblasts.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-014-0254-8) contains supplementary material, which is available to authorized users.
Keywords: Ovary, Mesenchymal stem cells, Bone marrow, Gene expression, Multipotency
Introduction
Mesenchymal stem cells (MSCs) are one of the most common types of cells that arise in the field of stem cell research and also in the field of regenerative medicine and cellular therapy. The term MSCs is widely used and can describe cells isolated from different tissues. From developmental perspective it is believed, that MSCs originate from mesodermal germ layer. They have been isolated from many tissues, such as adipose tissue [1], bone marrow [2], skeletal muscle [3], deciduous dental pulp [4], synovium [5], Wharton’s jelly [6], umbilical cord [7] and umbilical cord blood [8]. Regardless of the source, these cells should meet the requirements recommended by the International Society for Cellular Therapy [9]. They proposed that the cells must be able to adhere to a plastic well surface, to differentiate into osteoblasts, chondrocytes and adipocytes. Additionally, the cells should express CD105, CD73, CD90/THY-1 and should not express CD45, CD34, CD14, CD19 and HLA-DR markers. Recent findings have shown that these recommendations are not sufficient anymore, because some differences exist among MSCs derived from different sources with regard to the type of tissue and species [10]. Some of the proposed markers can also be expressed in non-MSCs; for instance, the CD90 marker can be expressed by tumor initiating-cells [11] or male germ-line stem cells [12]. In addition, MSCs can also express some markers of pluripotency [13–16] and differentiate not only into cells of mesodermal lineage, but also into cells of ectodermal [17] or endodermal lineage [18]. For these reasons Keating [19] put forth ten proposals to reduce confusion in the field. These proposals include a revision of MSCs terminology, surface marker profile and in vitro differentiation potential, specification of the source (tissue and species) and re-evaluation of transcriptome, proteome and secretome. Regardless of the lack of clarity, the research of MSCs is rapidly continuing and the cells are being translated step-by-step into the clinics.
Despite the fact that MSCs have been isolated from a large number of tissues, they have not been isolated from healthy human adult ovaries until now. So far they have only been studied in relation to human ovarian cancer; McLean et al. [20] discovered the presence of ovarian cancer-associated MSCs, which promoted tumor growth and increased the number of cancer stem cells. For this reason, and based on our previous study [21] confirming that a small proportion of cells were positive for a set of MSCs markers in ovarian cell cultures, we decided to explore MSCs from healthy human adult ovaries (indicated as PO-MSCs) and characterize them according to today’s interpretation of MSCs, as well as compare them with other types of cells of mesodermal origin: bone marrow-derived MSCs and human adult dermal fibroblasts.
Methods
Adult ovarian cortex biopsies were retrieved from seven patients (mean age of 51.6 years, range: 41–75 years) surgically treated at the University Medical Centre Ljubljana, due to different gynecological reasons (non-ovarian cancer, ovariectomy in breast cancer prevention) and with the written consent of patients. This study was approved by the Slovenian Medical Ethical Committee (no. 135/09/09). From each patient one ovarian cell culture was established. Of these cultures, four different cultures were analyzed for the expression of MSC-sprecific and MSC-related genes, three different cultures were analyzed using immunocytochemistry and one using flow-cytometry, and additional four different cultures were exposed to differentiation media to evaluate the differentiation potential of isolated cells.
Isolation of cells from ovarian cortex biopsies
The ovarian cells were isolated as described previously [21], except for the two changes: gelatin was left out as a supportive layer for cells to attach the plastic dish bottom when setting up the culture, and the concentration of fetal bovine serum (FBS) in the culture medium was lowered from 20 to 10 %. Briefly, ovarian cortex biopsies were cut into small pieces using a sterile scalpel and then incubated in 0.6 mg/ml collagenase type XI (Sigma-Aldrich) for 10 min. After centrifugation at 1,700 rpm for 8 min, they were again incubated in a mixture of prepared hyaluronidase (SynVitro Hydase, Origio) and 0.6 mg/ml collagenase type XI. After 10 min, the enzymes were inactivated using a 10 % fetal bovine serum (FBS, Gibco) and cells were washed via centrifugation in a basic medium consisting of DMEM/F12, supplemented with 3.7 g/l NaHCO3, 1 % penicillin/streptomycin (all reagents from Sigma-Aldrich) and with the pH adjusted to 7.4 by 1 M NaOH. The washed cells were resuspended in a basic medium with 10 % FBS, passed through a 70-μm cell strainer (BD Falcon) and poured into 12-well culture plates (TPP). The cells were cultured in a CO2-incubator (37 °C, 6 % CO2 in air) and passage every 14–18 days using 0.15 % trypsin (Sigma-Aldrich).
Culture of BM-MSCs and HDFs
PO-MSCs were compared to BM-MSCs to explore their stemness and the potential character of mesenchymal stem cells and to HDFs to exclude that they are fibroblasts because of a comparable morphology. For comparison a commercially available cell lines of BM-MSCs and HDFs were used. BM-MSCs were provided by Chemicon (Millipore, cat. no. SCC034). They were cultured in a Mesenchymal stem cell expansion medium provided by the same producer (cat. no. SCM015). HDFs were provided by Cascade Biologics (Invitrogen, cat. no. C-013-5C). In the first five passages, HDFs were cultured in Medium 106 supplemented with low serum growth supplement (both Invitrogen) and in subsequent passages in a basic medium with 10 % FBS.
Immunocytochemistry
Using immunocytochemistry we analyzed the expression of some MSCs cell surface markers recommended by the guidelines of the International Society for Cell Therapy [9]. A set of primary antibodies included in the Human Mesenchymal Stem Cell Characterization Kit (Millipore, cat. no. SCR067) was used for this purpose; a mouse anti-M-CAM monoclonal antibody (clone P1H12, diluted 1:500), a mouse anti-THY-1 (CD90) monoclonal antibody (clone F15-42-1, diluted 1:500), a mouse anti-CD14 monoclonal antibody (clone 2D-15C, diluted 1:500), a mouse anti-CD19 monoclonal antibody (clone FMC63, diluted 1:500), a mouse anti-STRO-1 monoclonal antibody (clone STRO-1, diluted 1:500) and a mouse anti-H-CAM/CD44 monoclonal antibody (clone F10-44-2). Firstly, the cells were fixed using 4 % paraformaldehyde (PFA) for 10 min, then incubated for 20 min in 10 % FBS and for 1 hour in primary antibodies (except STRO-1). After a thorough washing, the cells were incubated for 30 min in goat anti-mouse secondary antibodies conjugated to Alexa Fluor 488 (1:200, Molecular Probes). After another thorough washing, the cells were counterstained with DAPI. As a negative control, the primary antibodies were omitted from the procedure and replaced with 1 % FBS. To estimate the proportion of stained cells two different independant persons counted 200 cells per slide with immunostained cells and calculated the percentage of positive cells.
For the confirmation of STRO-1 (these primary antibodies are IgM), ß-tubulin III, S100, NeuN, glucagon and insulin expression, an ABC method was performed using diaminobenzidine as a substrate. Cells were fixed in a 4 % PFA, permeabilized with 0.2 % Triton X-100 when intracellular protein was analyzed, and to block endogenous peroxidases the cells were incubated in 3 % H2O2. After an incubation with 10 % FBS, the cells were incubated for 1 hour in primary antibodies (a mouse anti-STRO-1 monoclonal antibody, clone STRO-1, diluted 1:500), a mouse anti-ß-tubulin III monoclonal antibody (clone TUJ1, diluted 1:1,000, Covance), a rabbit anti-S100 polyclonal antibody (diluted 1:500, DakoCytomation), a mouse anti-NeuN monoclonal antibody (clone A60, diluted 1:100, Millipore), a rabbit anti-insulin (H-86) polyclonal antibody (diluted 1:200, SantaCruz) and a rabbit anti-glucagon polyclonal antibody (diluted 1:1,000, DakoCytomation) and then for 30 min in biotin-conjugated secondary antibodies (polyclonal rabbit anti-mouse immunoglobulins (1:400, DakoCytomation) or polyclonal goat anti-rabbit immunoglobulins (1:600, DakoCytomation)). After thoroughly washing the cells, they were incubated in an ABC reagent (Vectastain Elite ABC Kit, Vector Laboratories) for 20 min and then in diaminobenzidine substrate until the cells were stained brown (but for no more than 5 min). As a negative control the primary antibodies were omitted from the procedure and replaced with 1 % FBS.
Flow-cytometry on the expression of marker CD105
The PO-MSCs culture was analyzed by using FITC (fluorescein isothiocyanate)-conjugated antibodies against CD105 (EuroClone). Mouse IgG1 conjugated with FITC antibodies (BD Pharmingen™) was used as an isotype control. To obtain the single cell suspension, the trypsin was used to harvest cells from the cell culture. The sample was analyzed by using FACSCalibur (BD) and the data by using BD CellQuest Pro Software.
Alkaline phosphatase staining
To detect alkaline phosphatase (AP) activity the commercially available Alkaline Phosphatase Detection Kit (Millipore) was used according to the manufacturer’s instructions. Briefly, the cells from seven different ovarian cell cultures were fixed for 1 min using 4 % paraformaldehyde, washed with PBS and then incubated for 15 min in a mixture of Fast Red Violet, Naphthol AS-BI phosphate solution and water (2:1:1 ratio). After washing with PBS, the cells were observed under an inverted microscope (Hoffman illumination). The cells expressing AP activity were stained pink to red.
Differentiation of cells into cells of adipogenic, osteogenic, neural and pancreatic lineage
Four differentiation protocols were used to differentiate PO-MSCs. To induce adipogenic differentiation an induction medium was used as previously described [22]. The cells were cultured in a medium consisting of hESC medium (DMEM/F12, 20 % KnockOut Serum Replacement (Gibco), 1 mM L-glutamine (PAA), 1 % non-essential amino acids (PAA), 0.1 mM 2-mercaptoethanol (Invitrogen), 13 mM HEPES, 8 ng/ml human basic fibroblast growth factor (bFGF, Sigma-Aldrich) and 1 % penicillin/streptomycin) and 20 % follicular fluid serum retrieved from the in vitro fertilization program. The differentiation medium was changed in 3–4 days. After 2 weeks (for PO-MSCs) and after 3 weeks (for BM-MSCs and HDFs) the cells were fixed in a 4 % PFA for 20 min and incubated for 10 min in an Oil Red O working solution. After thorough washes, the cells were observed under an inverted microscope (Hoffman illumination) on lipid droplets, which were stained red.
Osteogenic differentiation was induced using the well known osteogenic differentiation medium [1]. It consisted of DMEM low glucose, L-glutamin, FBS, dexamethasone (Sigma), L-ascorbic acid 2-phosphate (Sigma), β-Glycerophosphate (Sigma) and penicillin/streptomycin. To confirm successful differentiation the cell culture was stained using the von Kossa protocol after 16–19 days of differentiation. The cells were fixed in a 4 % PFA, incubated in 2 % silver nitrate in the dark for 10 min, washed with distilled water and exposed to UV-light for 25 min. After washing, the cells were observed under an inverted microscope (Hoffman illumination) to detect the calcium deposits, which were stained black.
To induce neural differentiation, the cells were cultured in DMEM/F12, supplemented with 80 ng/ml human basic FGF, 30 μM forskolin, 2 % FBS, 1 % non-essential amino acids and 1 % ITS (Insulin-Transferrin-Selenium) [21]. After 3–4 weeks the cells were analyzed using immunocytochemistry for the expression of markers ß-tubulin III, S100 and NeuN.
To induce pancreatic differentiation the cells were cultured according to the Chandra et al. protocol [23], which was slightly modified. Briefly, the cells were cultured for 2 days in an SFM medium (serum free medium; DMEM/F12, 1 % ITS, 1 % BSA) supplemented with 4 nM activin A, 50 μM 2-mercaptoethanol and 2 ng/ml bFGF. On the third day the medium was changed to SFM supplemented with 0.3 mM taurine and on the fifth day to SFM supplemented with 3 mM taurine, 1 mM nicotinamide and 1 % non-essential amino acids. After 10–14 days the cells were analyzed by using dithizon and immunocytochemistry to check the expression of insulin and glucagon.
Gene expression analysis
Human Mesenchymal Stem Cell RT² Profiler™ PCR Array (PAHS-082, SABiosciences, Qiagen) was used to evaluate the expression of 84 specific genes. Four different samples of PO-MSCs cultures (samples 1–4), a sample of BM-MSCs and a sample of HDFs were analyzed. The total RNA was isolated from 105 to 106 cells using the miRNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. cDNA was synthesized from 500 ng of the total RNA using the RT2 First Strand Kit (Qiagen), which includes the additional removal of genomic DNA from the RNA sample and a specific control of reverse transcription. The quality of isolated RNA was also evaluated using RT2 RNA QC PCR Arrays (Qiagen) according to the manufacturer’s instructions. This test includes various measures allowing to control the presence of reverse transcription and PCR inhibitors, contamination with genomic DNA and contamination with DNA during the procedure.
After all the control tests, the samples were analyzed using the RT² Profiler™ PCR Array. Altogether 84 different genes were simultaneously amplified in each sample. Five housekeeping (reference) genes (B2M, HPRT1, RPL13A, GAPDH, ACTB), genomic DNA contamination control, reverse transcription control and positive PCR control were included in each PCR array. A melting curve analysis was performed to verify that the product consisted of a single amplicon. PCR arrays were performed in 384-well plates on a LightCycler 480 instrument (Roche Applied Science). Briefly, the reaction mix was prepared from 2× SABiosciences RT2 qPCR Master Mix and 102 μl of sample cDNA. 10 μl of this mixture was added into each well of the PCR Array. The data were analyzed via Roche LightCycler 480 software and the Ct values were extracted for each gene. The thresholds and baselines were set according to the manufacturer’s instructions (SABioscinces, Qiagen). The data were analyzed using software supplied by Qiagen (http://www.sabiosciences.com/pcr/arrayanalysis.php) and statistical comparisons of gene expression differences were performed using a linear model approach implemented in a limma package [24] for the Bioconductor 2.13 environment [25]. Initially, estimates of relative gene expression (compared to BM-MSCs and HDFs) were calculated using the ΔCt method with reference genes as endogenous controls. Afterwards, the values were log-transformed and the values fit to the linear model, comparing pairwise differences between classes of cells analyzed in the study. The fold changes were estimated by subtracting the log2 ΔCt expression values between the two compared classes, in accordance with the ΔΔCt approach [26]. Significance values were estimated and followed. In addition, they were adjusted for multiple testing according to the Benjamini-Hoechberg false-discovery rate (FDR) [27]. Ultimately, p-values under 0.05 and FDR values falling below 0.2 were used as criteria to determine differentially expressed genes between each pair of classes tested.
Results
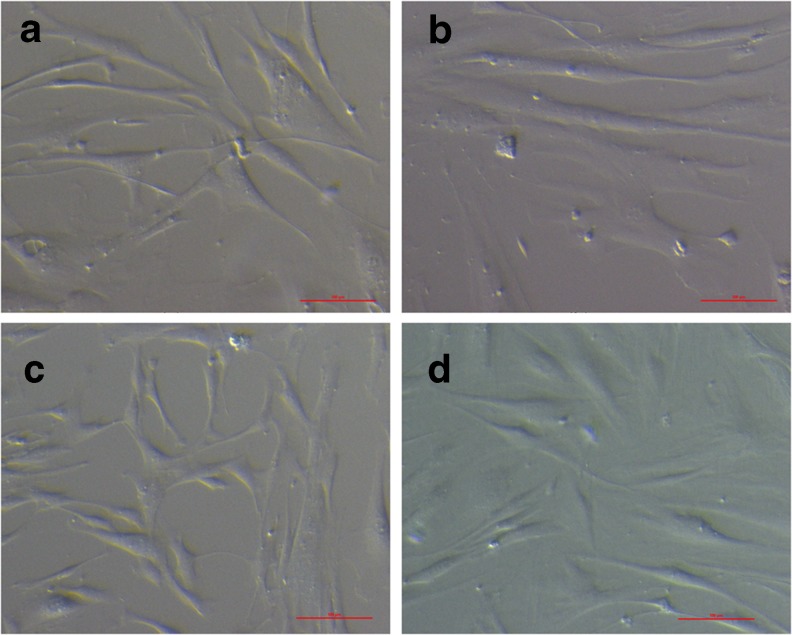
In this report we describe the similarities and differences between in vitro cultured cells from ovarian cortex showing characteristics of MSCs (indicated as PO-MSCs), BM-MSCs and HDFs. Ovarian cells were derived from 7 different patients. Most of the derived cells present in the ovarian cell cultures had morphology similar to fibroblasts and BM-MSCs (Fig. 1). Although there were also some other cell types present in the ovarian cell cultures of the initial passages (probably epithelial cells from ovarian surface epithelium, blood cells and others), they usually disappeared after the first or the second passage. Like in our previous study [21], typical cell colonies were observed but their number was significantly lower in this study (never more than five on the whole cell culture plate and sometimes even none) probably due to modifications of the culture system described in the materials and methods.
Fig. 1.
The morphology of cells derived from different sources. (a, b) PO-MSCs, (c) BM-MSCs, (d) HDFs. Scale bar: 100 μm
Expression of cell surface markers of mesenchymal stem cells analyzed by using immunocytochemistry
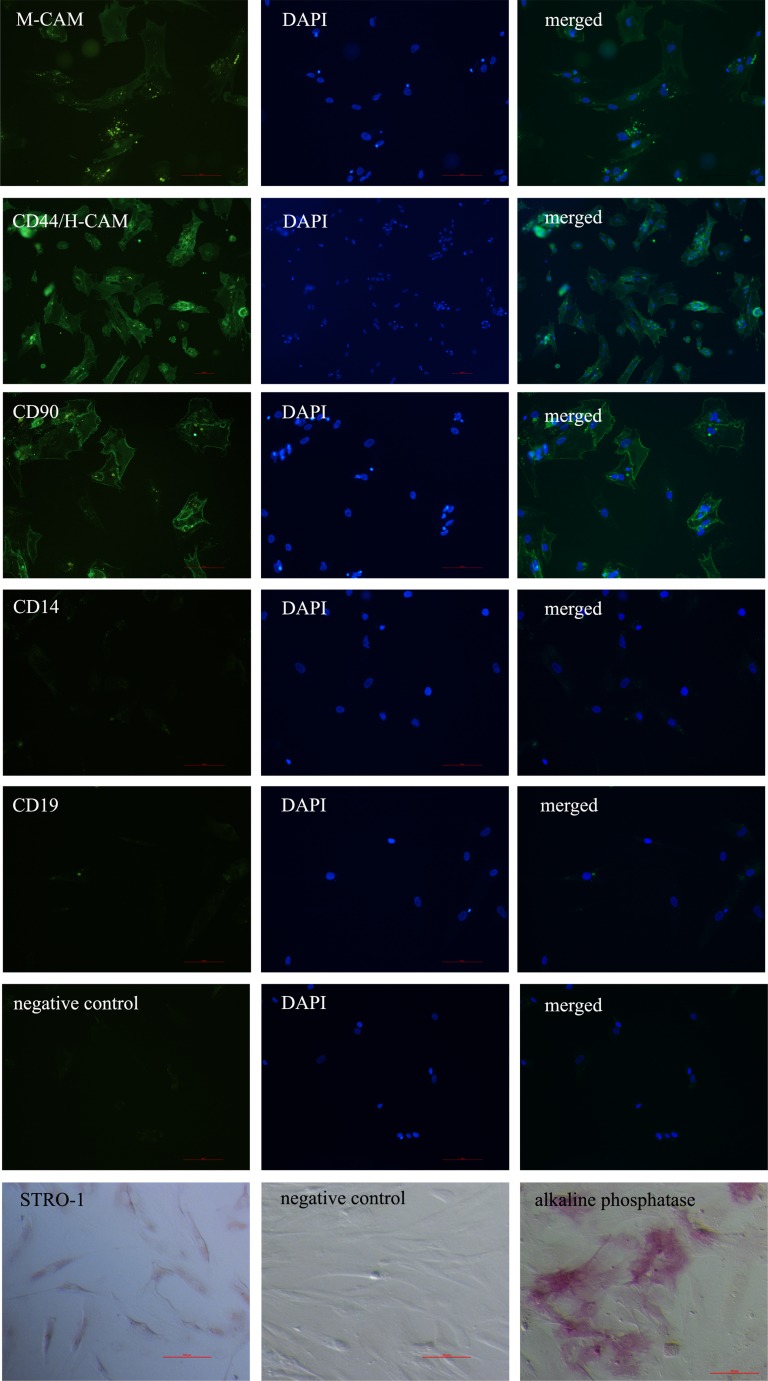
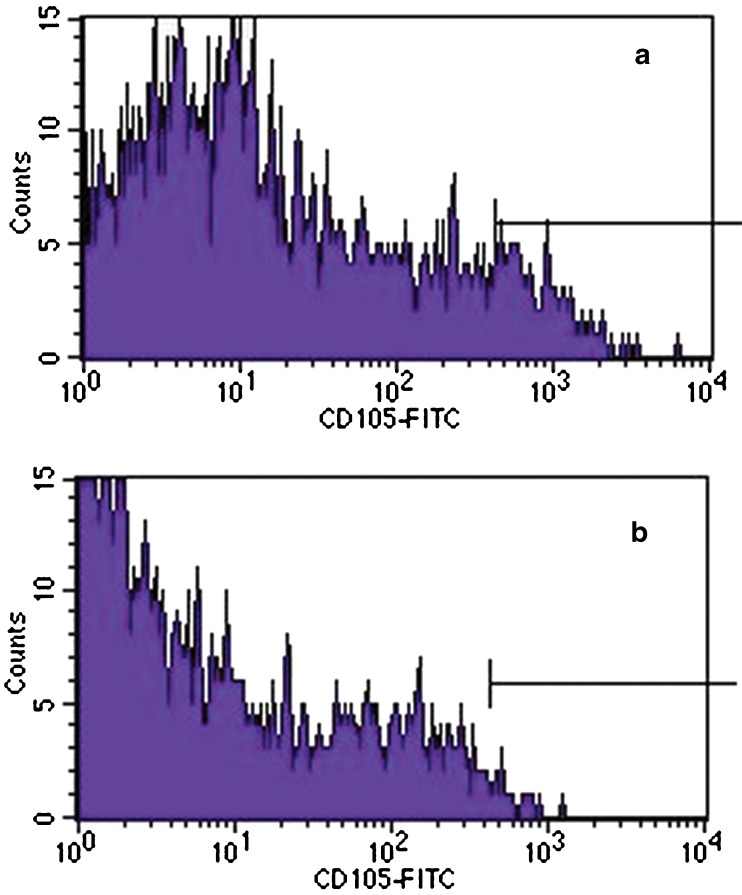
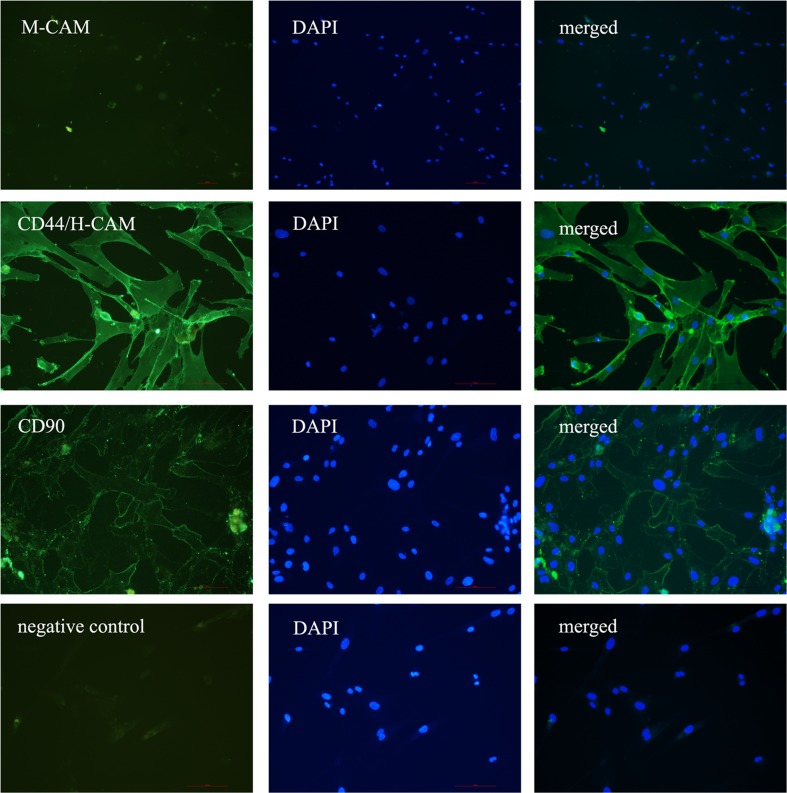
In all three different PO-MSCs cultures that we tested, the markers of mesenchymal stem cells M-CAM, CD44, THY-1/CD90 and STRO-1 were expressed, as revealed by immunocytochemistry. The proportion of positive cells varied between different PO-MSCs cultures, except for the marker CD44, where in all the tested cell cultures the majority of cells representing 96.7 ± 2.9 % of all cells on average were CD44 positive. By our estimation, compared to marker CD44, lower proportions of other analyzed markers were determined: 56.7 ± 15.3 % of cells were THY-1/CD90-positive, 31.7 ± 23.6 % of cells were M-CAM-positive, and 10.0 ± 8.7 % of cells were STRO-1-positive (Fig. 2, summarized in Supplemental Table 1). Additionally, the PO-MSCs cultures were negative for CD14 and CD19 markers (Fig. 2). Female age did not seem to affect the expression of MSCs markers in ovarian cultures, as can be seen in Supplemental Table 1. Conversely, the ovarian cell culture in a woman diagnosed with endometrioma on both ovaries was characterized by a low proportion of M-CAM-positive cells in comparison with ovarian cell cultures in other women. According to the results obtained by flow-cytometry, PO-MSCs cultures also contained approximately 6 % of CD105-positive cells (Fig. 3). Additionally, all seven analyzed PO-MSCs cultures revealed the presence of cells positive for alkaline phosphatase staining (Fig. 2).
Fig. 2.
Positive staining of PO-MSCs for the expression of markers M-CAM, CD44/H-CAM and CD90 and negative staining for markers CD14 and CD19 as revealed by immuno-fluorescence, positive staining of STRO-1 as revealed by immunocytochemistry, and positive staining on alkaline phosphatase activity. Scale bar: 100 μm
Fig. 3.
Flow cytometry analysis of ovarian cortex cell culture. (a) A subpopulation of cells expressing a mesenchymal stem cell marker -CD105. (b) Isotype control
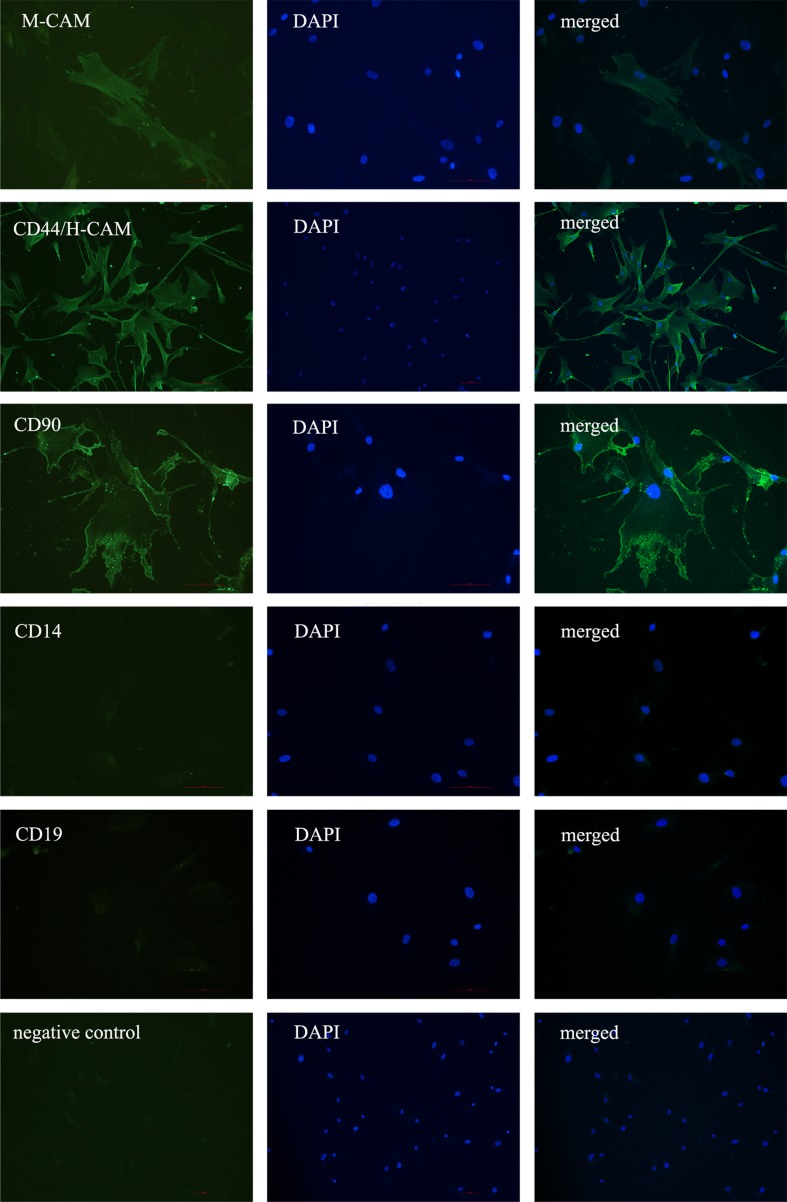
Furthermore, we have analyzed BM-MSCs and HDFs for the expression of MSCs cell surface markers (M-CAM, CD44, THY-1/CD90 and STRO-1) using immunocytochemistry. In comparison with PO-MSCs cultures, there was a lower proportion (5–10 %) of M-CAM and THY-1/CD90 positive cells determined in the BM-MSCs culture (Fig. 4). Similarly to PO-MSCs cultures most of BM-MSCs were positive for marker CD44/H-CAM and negative for CD14 and CD19. The HDFs showed a different pattern of immunostaing, where most of the cells were positive for CD44 and around half for the THY-1/CD90 marker (Fig. 5). The HDFs were negative for CD14, CD19 and M-CAM markers.
Fig. 4.
Positive staining of BM-MSCs for the expression of markers M-CAM, CD44/H-CAM and CD90 and negative staining for markers CD14 and CD19 as revealed by immuno-fluorescence. Scale bar: 100 μm
Fig. 5.
Positive staining of HDFs for the expression of markers CD44/H-CAM and CD90 and negative staining for M-CAM, as revealed by immuno-fluorescence. Scale bar: 100 μm
Differentiation potential of putative ovarian mesenchymal stem cells (PO-MSCs) in comparison with BM-MSCs and HDFs
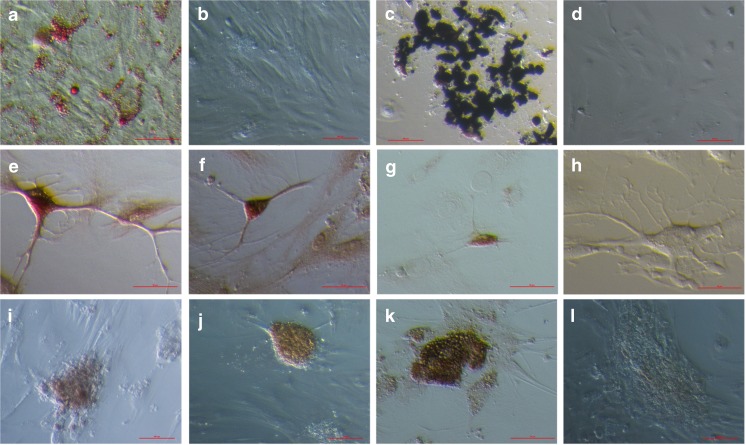
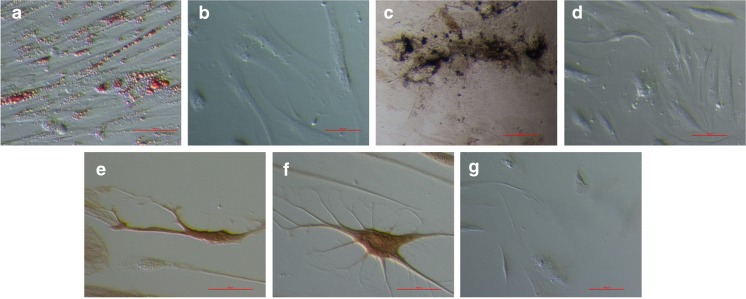
PO-MSCs were differentiated into cells of four different cell lineages – adipogenic, osteogenic, neural and pancreatic - representing all three germ layers (mesoderm, ectoderm and endoderm) (results are summarized in Supplemental Table 1). Adipogenic differentiation was confirmed by positive Oil Red O staining. In all three differentiated PO-MSCs cultures and in BM-MSCs, red-stained lipid droplets were observed (Figs. 6a, b and 7a, b). The proportion of differentiated cells varied between cultures, but in some PO-MSCs cultures we observed up to half of cells containing lipid droplets. In BM-MSCs no more than a third of cells in a culture contained lipid droplets. Osteogenic differentiation was confirmed in all tested PO-MSCs and BM-MSCs cell cultures via positive von Kossa staining (Figs. 6c, d and 7c, d). The proportion of differentiated cells varied between different PO-MSCs cultures, in some of the PO-MSCs cultures up to half of cells showed osteogenic differentiation in the culture, while in BM-MSCs cultures; it was around 20 % of differentiated cells. Interestingly, in the ovarian cell culture of the oldest, 75 years old patient, a high proportion of cells was differentiated into adipogenic and osteogenic cells (50 % and 50 %); the differentiation potential of these ovarian cells seems to be higher than in younger women. Neural differentiation (ectoderm) was confirmed using immunocytochemistry, where differentiated PO-MSCs were positively stained for the markers ß-tubulin III, S100 and NeuN (Fig. 6e–h), and differentiated BM-MSCs were positively stained for ß-tubulin III and S100 (Fig. 7e–g). The proportion of cells that were differentiated in neural lineage was low in both types of cells: only around 1–2 % of cells were determined as differentiated in the tested cultures. Pancreatic differentiation (endoderm) was confirmed using dithizone staining and immunocytochemistry. Differentiated PO-MSCs, which formed islet-like structures (5–10 islet-like structures per well (the diameter of the well is 1 cm) positively stained on dithizon (Fig. 6i). With the immunocytochemistry, the very same cell cultures were also shown as positive for insulin and glucagon, as can be seen in Fig. 6j–l. On the other hand, the HDFs did not differentiate by the used protocols and were all negative for applied tests.
Fig. 6.
In vitro differentiated PO-MSCs into (a) adipogenic cells (Oil Red O staining), (b) negative control (Oil Red O staining of PO-MSCs cultured in a basic medium with FBS, into (c) osteogenic cells (von Kossa staining), (d) negative control (von Kossa staining of PO-MSCs cultured in DMEM low glucose, L-glutamin and FBS). Differentiation into neuronal-like cells confirmed by immunocytochemistry on the expression of (e) β-tubulin III, (f) S-100, (g) NeuN and (h) negative control. Differentiation into pancreatic-like cells confirmed by (i) dithizon staining and immunocytochemistry on the expression of (j) insulin, (k) glucagon and (l) negative control. Scale bar: (a, c, e, f, g, h, k) 50 μm and for (b, d, e, i, j) 100 μm
Fig. 7.
In vitro differentiated BM-MSCs into (a) adipogenic cells (Oil Red O staining), (b) negative control (Oil Red O staining of BM-MSCs cultured in Mesenchymal stem cell medium), into (c) osteogenic cells (von Kossa staining), (d) negative control (von Kossa staining of BM-MSCs cultured in DMEM low glucose, L-glutamin and FBS). Differentiation into neuronal-like cells confirmed by immunocytochemistry on the expression of (e) β-tubulin III and (f) S-100. (g) negative control for immunocytochemistry. Scale bar: (a, e, f) 50 μm and for (b–d, g) 100 μm
Gene expression profiling by the human mesenchymal stem cell RT² profiler™ PCR array
PO-MSCs, BM-MSCs and HDFs were analyzed using the Human Mesenchymal Stem Cell RT² Profiler™ PCR Array for the expression of 84 specific genes. The results are presented as the fold-change with calculated t-test values, p-values and FDR-values for gene expressions of PO-MSCs in comparison with the BM-MSCs and HDFs, as described in the section on materials and methods. To gain a better overview the results were grouped together based on gene function according to the manufacturer’s recommendations. Because the expression of a few genes may be important in various processes, they were classified into several groups at the same time. The analyzed genes were grouped as MSC-specific genes and genes associated with stemness (pluripotency) (Table 1), other genes associated with MSCs (Table 2) and MSCs differentiation markers specific for osteogenesis, adipogenesis, chondrogenesis, myogenesis and tenogenesis (Table 3).
Table 1.
Expression of MSCs specific genes and genes associated with stemness in PO-MSCs compared to BM-MSCs and HDFs
| PO-MSCs vs. BM-MSCs | PO-MSCs vs. HDFs | |||||||
|---|---|---|---|---|---|---|---|---|
| Fold change | t-test value | p value | FDR value | Fold change | t-test value | p value | FDR value | |
| MSCs-specific genes | ||||||||
| ALCAM | 1.25 | 0.37 | 0.732 | 0.854 | 10.25 | 3.75 | 0.018* | 0.102 |
| ANPEP | 0.09 | −3.45 | 0.024* | 0.200 | 0.06 | −3.91 | 0.016* | 0.097 |
| CASP3 | 0.88 | −0.23 | 0.828 | 0.926 | 1.47 | 0.68 | 0.531 | 0.673 |
| CD44 | 0.90 | −0.13 | 0.900 | 0.941 | 0.69 | −0.48 | 0.658 | 0.759 |
| ENG (CD105) | 0.81 | −0.49 | 0.651 | 0.791 | 2.47 | 2.08 | 0.102 | 0.275 |
| ERBB2 | 1.28 | 0.64 | 0.553 | 0.745 | 2.44 | 2.33 | 0.077 | 0.217 |
| ITGA6 | 3.61 | 1.72 | 0.157 | 0.366 | 4.12 | 1.90 | 0.127 | 0.315 |
| ITGAV | 1.29 | 0.99 | 0.376 | 0.598 | 2.31 | 3.25 | 0.029* | 0.120 |
| M-CAM | 3.26 | 1.08 | 0.336 | 0.574 | N/A | N/A | N/A | N/A |
| NT5E (CD73) | 1.01 | 0.01 | 0.991 | 1.000 | 1.20 | 0.24 | 0.818 | 0.860 |
| PDGFRB | 1.31 | 0.49 | 0.650 | 0.791 | 0.66 | −0.73 | 0.501 | 0.673 |
| THY1 (CD90) | 6.73 | 2.11 | 0.099 | 0.288 | 0.40 | −1.03 | 0.359 | 0.571 |
| VCAM1 | 3.11 | 0.66 | 0.542 | 0.744 | N/A | N/A | N/A | N/A |
| Genes associated with stemness | ||||||||
| FGF2 | 0.71 | −0.99 | 0.375 | 0.598 | 1.20 | 0.52 | 0.630 | 0.751 |
| LIF | 1.80 | 0.81 | 0.459 | 0.669 | N/A | N/A | N/A | N/A |
| POU5F1 | 4.40 | 4.85 | 0.007* | 0.169 | 2.25 | 2.65 | 0.054 | 0.167 |
Differentially expressed genes marked with asterisk (*) have p-values under 0.05. (N/A-not expressed)
Table 2.
Expression of MSCs-associated genes in PO-MSCs compared to BM-MSCs and HDFs
| PO-MSCs vs. BM-MSCs | PO-MSCs vs. HDFs | |||||||
|---|---|---|---|---|---|---|---|---|
| Fold change | t-test value | p value | FDR value | Fold change | t-test value | p value | FDR value | |
| MSCs-associated genes | ||||||||
| ANXA5 | 1.46 | 0.66 | 0.542 | 0.744 | 0.78 | −0.43 | 0.686 | 0.759 |
| BDNF | 2.49 | 2.66 | 0.053 | 0.219 | 47.34 | 11.26 | 0.0003* | 0.008 |
| BGLAP | 0.61 | −2.34 | 0.076 | 0.259 | 1.26 | 1.07 | 0.340 | 0.568 |
| COL1A1 | 1.51 | 0.89 | 0.419 | 0.638 | 15.83 | 5.99 | 0.003* | 0.051 |
| CTNNB1 | 1.82 | 2.38 | 0.073 | 0.259 | 0.90 | −0.44 | 0.680 | 0.759 |
| EGF | 0.93 | −0.15 | 0.890 | 0.941 | N/A | N/A | N/A | N/A |
| GTF3A | 1.79 | 1.67 | 0.166 | 0.374 | 4.15 | 4.08 | 0.014* | 0.093 |
| HGF | 2.09 | 1.00 | 0.371 | 0.598 | 39.26 | 4.97 | 0.007* | 0.072 |
| ICAM1 | 2.27 | 0.73 | 0.502 | 0.717 | 2.29 | 0.74 | 0.499 | 0.673 |
| IGF1 | 116.16 | 12.09 | 0.032* | 0.200 | N/A | N/A | N/A | N/A |
| IL1B | 0.55 | −2.10 | 0.121 | 0.327 | N/A | N/A | N/A | N/A |
| IL6 | 0.33 | −2.88 | 0.042* | 0.219 | 10.48 | 6.07 | 0.003* | 0.051 |
| ITGB1 | 4.72 | 6.74 | 0.002* | 0.072 | 3.10 | 4.91 | 0.007* | 0.072 |
| KITLG | 1.17 | 0.22 | 0.836 | 0.926 | 0.34 | −1.48 | 0.209 | 0.441 |
| MITF | 1.91 | 0.93 | 0.401 | 0.623 | 1.36 | 0.44 | 0.680 | 0.759 |
| MMP2 | 0.39 | −1.93 | 0.123 | 0.327 | 0.23 | −3.02 | 0.036* | 0.141 |
| NES | 2.25 | 0.44 | 0.680 | 0.807 | N/A | N/A | N/A | N/A |
| NUDT6 | 0.71 | −0.49 | 0.647 | 0.791 | 1.22 | 0.28 | 0.793 | 0.860 |
| PIGS | 1.09 | 0.21 | 0.846 | 0.926 | 1.88 | 1.44 | 0.219 | 0.441 |
| SLC17A5 | 0.85 | −0.29 | 0.789 | 0.905 | 0.44 | −1.44 | 0.220 | 0.441 |
| TGFB3 | 4.24 | 3.08 | 0.034* | 0.200 | 4.84 | 3.36 | 0.026* | 0.120 |
| VEGFA | 1.91 | 3.27 | 0.029* | 0.200 | 9.78 | 11.55 | 0.0002* | 0.008 |
| VIM | 1.30 | 0.48 | 0.655 | 0.791 | 2.29 | 1.53 | 0.197 | 0.441 |
Differentially expressed genes marked with asterisk (*) have p-values under 0.05. (N/A-not expressed)
Table 3.
Expression of genes involved in various differentiation processes (chondrogenesis, osteogenesis, adipogenesis, tenogenesis, myogenesis) in PO-MSCs compared to BM-MSCs and HDFs
| PO-MSCs vs. BM-MSCs | PO-MSCs vs. HDFs | |||||||
|---|---|---|---|---|---|---|---|---|
| Fold change | t-test value | p value | FDR value | Fold change | t-test value | p value | FDR value | |
| Genes associated with chondrogenesis | ||||||||
| ABCB1 | 15.45 | 6.96 | 0.062 | 0.242 | N/A | N/A | N/A | N/A |
| BMP4 | 2.46 | 1.45 | 0.217 | 0.427 | 1.41 | 0.56 | 0.606 | 0.737 |
| BMP6 | 0.38 | −1.30 | 0.260 | 0.467 | 2.56 | 1.26 | 0.272 | 0.469 |
| GDF5 | 6.63 | 3.13 | 0.033* | 0.200 | 0.65 | −0.72 | 0.508 | 0.673 |
| GDF6 | 8.89 | 1.76 | 0.149 | 0.359 | 8.30 | 1.71 | 0.159 | 0.379 |
| HAT1 | 1.60 | 1.54 | 0.195 | 0.401 | 1.51 | 1.36 | 0.242 | 0.451 |
| KAT2B | 3.04 | 4.29 | 0.011* | 0.198 | 0.83 | −0.71 | 0.516 | 0.673 |
| SOX9 | 0.56 | −0.48 | 0.655 | 0.791 | 11.61 | 2.04 | 0.108 | 0.278 |
| ITGB1 | 4.72 | 6.74 | 0.002* | 0.072 | 3.10 | 4.91 | 0.007* | 0.072 |
| TGFB1 | 1.77 | 1.44 | 0.220 | 0.427 | 3.15 | 2.88 | 0.042* | 0.145 |
| Genes associated with osteogenesis | ||||||||
| BMP6 | 0.38 | −1.30 | 0.260 | 0.467 | 2.56 | 1.26 | 0.272 | 0.469 |
| HDAC1 | 2.07 | 1.90 | 0.126 | 0.327 | 0.69 | −0.98 | 0.381 | 0.590 |
| PTK2 | 1.87 | 1.19 | 0.298 | 0.522 | 2.09 | 1.39 | 0.232 | 0.449 |
| RUNX2 | 3.04 | 1.59 | 0.184 | 0.389 | 2.09 | 1.05 | 0.348 | 0.568 |
| SMURF1 | 1.87 | 1.31 | 0.256 | 0.467 | 1.90 | 1.34 | 0.247 | 0.451 |
| SMURF2 | 0.99 | −0.01 | 0.989 | 1.000 | 2.88 | 1.51 | 0.202 | 0.441 |
| Genes associated with adipogenesis | ||||||||
| PPARG | 1.73 | 1.31 | 0.256 | 0.467 | 1.11 | 0.25 | 0.812 | 0.860 |
| RHOA | 0.67 | −0.50 | 0.640 | 0.791 | 0.90 | −0.14 | 0.894 | 0.924 |
| RUNX2 | 3.04 | 1.59 | 0.184 | 0.389 | 2.09 | 1.05 | 0.348 | 0.568 |
| Genes associated with tenogenesis | ||||||||
| GDF15 | 0.36 | −1.60 | 0.180 | 0.389 | 8.13 | 3.26 | 0.029* | 0.120 |
| SMAD4 | 3.45 | 3.42 | 0.025* | 0.200 | 0.97 | −0.08 | 0.939 | 0.954 |
| TGFB1 | 1.77 | 1.44 | 0.220 | 0.427 | 3.15 | 2.88 | 0.042* | 0.145 |
| Gene associated with myogenesis | ||||||||
| JAG1 | 2.54 | 0.82 | 0.454 | 0.669 | 4.24 | 1.28 | 0.268 | 0.469 |
Differentially expressed genes marked with asterisk (*) have p-values under 0.05. (N/A-not expressed)
MSC-specific genes and genes associated with stemness
Out of 19 genes classified as MSC-specific, 13 of them were expressed in PO-MSCs (Table 1), including the most important markers CD44, CD105, CD73, CD90 and M-CAM. Three of these genes were differentially expressed when compared to BM-MSCs and HDFs: ANPEP (p = 0.024, FDR = 0.200 and 0.016, 0.097) was down-regulated in both comparisons, and ALCAM (p = 0.018, FDR = 0.102) and ITGAV (p = 0.029, FDR = 0.120) were up-regulated when PO-MSCs were compared to HDFs. A set of six genes (BMP2, FUT4, FZD9, KDR, NGFR, PROM1) was not expressed in any of the analyzed sample. Additionally, M-CAM and VCAM1 were not expressed in HDFs. Out of eight genes that were classified as stemness genes, only the expression of three genes was detected: FGF2, LIF and POU5F1 (Table 1). Of these, POU5F1 (p = 0.007, FDR = 0.169) was up-regulated in PO-MSCs when compared to BM-MSCs. The expression of other genes such as SOX-2, TERT, ZFP42 and WNT3A that are usually associated with pluripotency, was not detected in any of the analyzed samples (PO-MSCs, BM-MSCs and HDFs).
MSCs-associated genes
This group of genes is the largest, containing 32 genes, and at the same times the most diverse. Twenty-three of these genes were expressed in PO-MSCs samples (Table 2), of which ten were differentially expressed when compared to BM-MSCs and HDFs: IGF-1 (p = 0.032, FDR = 0.200), ITGB1 (p = 0.002, FDR = 0.072), TGFB3 (p = 0.034, FDR = 0.200) and VEGFA (p = 0.029, FDR = 0.200) were up-regulated in PO-MSCs when compared to BM-MSCs, and BDNF(p = 0.0003, FDR = 0.008), COL1A1 (p = 0.003, FDR = 0.051), GTF3A (p = 0.014, FDR = 0.093), HGF (p = 0.007, 0.072), IL6 (p = 0.003, FDR = 0.051), ITGB1 (p = 0.007, FDR = 0.072), TGFB3 (p = 0.026, FDR = 0.120) and VEGFA (p = 0.0002, FDR = 0.008) when compared to HDFs. In PO-MSCs we see that IL6 (p = 0.042, 0.219) was down-regulated when compared to BM-MSCs, and MMP2 (p = 0.036, FDR = 0.141) when compared to HDFs. There were also nine genes that were not expressed in PO-MSCs, BM-MSCs or in HDFs: BMP7, CSF2, CSF3, FUT1, IFNG, IL10, PTPRC, TNF and VWF. Additionally, there were two genes which were not expressed in HDFs only: EGF, NES.
MSCs differentiation markers
The expression of genes for several lineage-specific markers for osteogenesis, adipogenesis, chondrogenesis, myogenesis and tenogenesis was tracked (Table 3). Ten of 13 genes associated with chondrogenesis were expressed in PO-MSCs, of which four were differentially expressed: GDF5 (p = 0.033, FDR = 0.200) and KAT2B (p = 0.011, FDR = 0.198) were up-regulated and ITGB1 (p = 0.002, FDR = 0.072) down-regulated in PO-MSCs when compared to BM-MSCs and ITGB1 (p = 0.007, FDR = 0.072) and TGFB1 (p = 0.042, FDR = 0.145) were up-regulated when compared to HDFs. The genes BMP2, GDF7 and ITGAX were not expressed in any of the analyzed samples. All three genes associated with tenogenesis were expressed in PO-MSCs, BM-MSCs and in HDFs, of which SMAD4 (p = 0.025, FDR = 0.200) was up-regulated in PO-MSCs when compared to BM-MSCs, and GDF15 (p = 0.029, FDR = 0.120) and TGFB1 (p = 0.042, FDR = 0.145) were up-regulated when compared to HDFs. The expression of genes associated with osteogenesis showed that 6 of 11 genes were expressed in PO-MSCs, BM-MSCs and HDFs, but none of them were differentially expressed. The genes BMP2, FGF10, HNF1A, KDR and TBX5 were not expressed in any sample. Similarly, none of the genes associated with adipogenesis were differentially expressed, although all three tested genes were expressed (PPARG, RHOA, RUNX2). In the end, one myogenesis-associated gene (JAG1) was also expressed in all samples but with no statistical significance, and the other gene (NOTCH1) was not expressed in any sample.
Discussion
The results of this study for the first time indicate the presence of putative mesenchymal stem cells in healthy adult ovaries in humans. Our previous study [21] revealed that some of the in vitro cultured cells from ovarian cortex express a specific pattern of pluripotency and pluripotent stem cell markers; a proportion of ovarian cell colonies expressed both the markers of pluripotent and multipotent/mesenchymal stem cells. These observations encouraged us to focus on other cells present in the ovarian cortex cell culture, which by morphology resemble fibroblasts. The results of this study show that cells present in ovarian cortex cell cultures express a wide range of MSCs and MSCs-associated markers and have an interesting differentiation potential. We named these cells putative ovarian mesenchymal stem cells (PO-MSCs) and compared them with a commercially available BM-MSCs line and HDFs cell line. BM-MSCs were used for comparison since they are currently the most researched and widely used MSCs and HDFs were used to show that our populations of isolated cells from ovaries are not usual fibroblasts, even though we are aware of HDFs controversy in MSCs studies [28, 29].
Our results indicate that healthy adult human ovaries contain specific cells showing MSCs characteristics. As previously mentioned, MSCs have been isolated from a wide range of tissues [1–8], but caution is necessary because some differences exist between them. Diverse studies have shown distinct gene expression profiles for MSCs depending on their tissue source [30] and also different differentiation potential for these MSCs [31, 32]. Similarly, in our study the results showed differences in the gene expression characteristics of PO-MSCs and BM-MSCs. However, when the most important MSCs markers were examined, such as CD105, CD73, M-CAM, CD90, CD44 or VCAM1, all were expressed in PO-MSCs. Additionally, using immunocytochemistry, we confirmed the expression of selected MSCs markers in PO-MSCs. Due to the small number of patients included in this study and because each experiment was not performed on all seven patients, which represents a limitation, it is also reasonable to mention the observed heterogeneity of PO-MSCs samples obtained from different patients, thus the heterogeneity of MSCs could also be expected. This is in accordance with the BM-MSCs study which included 61 patients and showed the high inter-donor variability, which was high even in the BM-MSCs obtained from same-age donors [33]. It could be expected that aging negatively affects the characteristics of MSCs and has an impact on gene expression, although we did not observe this. Some studies have shown that aging increased senescence and reduced cell viability, proliferation and differentiation potential [34–36]. Furthermore, when performing in vitro studies, the long-term expansion of MSCs had a significantly negative impact on the characteristics of MSCs, regardless of donor age [35, 37]. Additionally, it is postulated that MSCs consist of different cell sub-populations and that the results (gene expression, immunocytochemistry) may vary depending on the ratio between these sub-populations [38]. This is probably also the reason for significant differences between MSCs clones, as has been reported several times before [39–43]. Interestingly, our study shows that ovarian pathologies (e.g. ovarian endometrioma) affecting the characteristics of PO-MSCs are not excluded.
Besides the genes related to MSCs, PO-MSCs also expressed three genes related to pluripotent stem cells: FGF2, LIF and POU5F1. It is important that these data are interpreted with caution. From existing literature it is known that primers for POU5F1 can be unreliable [44]. Moreover, the expression of FGF2 could also be associated with MSCs and not only with pluripotency [45]. On the other hand, PO-MSCs did not express some other important pluripotency-related genes, e.g. SOX-2 and TERT; therefore, we may conclude that PO-MSCs cannot be associated with pluripotency at this point.
Furthermore, in PO-MSCs, several genes related to differentiation processes were expressed, although only four genes (GDF5, KAT2B, ITGB1, SMAD4) were differential in a comparison of PO-MSCs with BM-MSCs. This could be partly explained by the fact that whole ovarian cell cultures were harvested for gene expression profile analysis, and the presence of other cell types therefore cannot be excluded. The analyzed cells might consist of a heterogeneous population of cells: from non-differentiated to partially or fully differentiated cells, which could be a natural condition of the ovary or a consequence of a suboptimal culture system. In order to avoid such dilemmas, it is proposed that PO-MSCs would be sorted from the tissue or cell cultures using MACS or FACS based on the expression of relevant markers; our results show that CD44 and CD73 are definitely ideal markers to do that.
The results of this study indicate that PO-MSCs are multipotent cells, since they are capable of differentiation into cell lineages of mesodermal (adipogenic and osteogenic lineage), ectodermal (neural lineage) and endodermal (pancreatic lineage) origin. It is well known that one of the main characteristics of MSCs is their differentiation into the cell lineages of mesodermal origin [9], while the differentiation into the cell lineages of endo- or ectodermal origin is still a matter of debate and criticism [46–48]. Regardless of these concerns, the MSCs can express some pluripotency-related genes (markers). Gonzalez et al. [13] showed that MSCs can express pluripotency-related genes, and even more interesting these MSCs were isolated from adult human testes, which could indicate that these cells and PO-MSCs originate from the same cells in the developmental stage of indifferent gonads. Moreover, there are more and more studies confirming the trans-differentiation potential of MSCs [49–55] which makes them still more interesting to be applied in regenerative medicine and cellular therapies in the future. In the case of cells cultured in this study, we would particularly suggest using them in the frame of restoring ovarian function in cases of its impairment or of premature ovarian failure. It has already been shown in animal models that MSCs from different sources (e.g. adipose) can improve or even restore ovarian function [56–59]. To determine the ideal age at which PO-MSCs could be used to regenerate ovaries, the ovarian tissue of more patients at different ages would need to be included. This is difficult because it is not easy to get ovarian tissue, especially in younger women. We can only speculate on an ideal age based on the literature; this age could be under 30 years-old [34]. Moreover, our data showed that the female age did not affect the characteristics of PO-MSCs and that even in the oldest patient, aged 75 years, PO-MSCs expressed a high potential to differentiate into adipogenic in osteogenic cells.
The analysis of HDFs which were used in our study shows that the isolated population of PO-MSCs was a distinct population of cells and additionally, HDFs did not show any differentiation potential. In spite of non-differentiation potential, the HDFs shared the expression of a proportion of genes with PO-MSCs and BM-MSCs, although 8 genes were not expressed in HDFs but were expressed in PO-MSCs and BM-MSCs (M-CAM, VCAM1, LIF, EGF, IGF1, IL1B, NES, ABCB1). Moreover, more genes were differentially expressed when PO-MSCs were compared to HDFs than when compared to BM-MSCs. The functions of these differentially expressed genes vary, but only three of them are MSCs-specific genes (ANPEP, ALCAM and ITGAV). The gene ANPEP (known also as CD13), which was down-regulated in PO-MSCs in comparison with both BM-MSCs and HDFs, influences the MSCs’ adhesion, migration and vascular network formation, and its expression is important for the normal behaviour of MSCs [60]. On the other hand, the expression of ANPEP/CD13 could be related to pathogenesis, since its expression is connected with the invasion of cancer cells, including human ovarian cancers [61, 62]. Two other differentially expressed genes (ALCAM, ITGAV) were up-regulated in PO-MSCs when compared to HDFs. ALCAM (CD166) is a common MSCs marker detected in MSCs isolated from various sources [63], including granulosa cells [64]. It works as a cell adhesion molecule and is involved in immunological processes as well as in tumor growth and metastasis [65, 66]. The gene ITGAV (also known as CD51) encodes the molecule (integrin αv), which is involved in cell adhesion and is important for controlling the stem cell niche [67]. Other differentially expressed genes are mostly involved in the differentiation processes, which indicate the presence of a heterogeneous population of cells, as previously discussed.
An important question arises: why are cells showing MSCs characteristics resident in adult human ovaries? They are probably the residue from the period of fetal gonadal development and therefore retain some stemness that allows them to regulate the ovarian function, particularly (to some extent) regeneration. This is important, since during ovulation the oocytes are released monthly from the ovaries and the ovarian surface is damaged. The MSCs could also have some influence on the follicular development with the production of active molecules or in some other way, considering that they are most likely located in the vicinity of follicles. Moreover, it is not excluded that they could include a subpopulation of granulosa cells showing the characteristics of MSCs [64].
In conclusion, the cortex of healthy adult human ovaries can be a source of cells showing typical MSCs characteristics in conditions in vitro and for this reason we named these cells PO-MSCs. These cells express genes related to MSCs, such as CD105, CD44, CD90, M-CAM, CD73, VCAM1, and have an interesting differential potential. When exposed to differentiation media PO-MSCs are able to differentiate into cells of adipogenic, osteogenic, neural and pancreatic lineage. Based on these results, we propose PO-MSCs as a novel type of MSCs which share some similarities with BM-MSCs but nevertheless show distinct and specific characteristics.
Electronic supplementary material
(DOC 42 kb)
Acknowledgments
The authors would like to thank Dr. Branko Cvjeticanin, Department of Obstetrics and Gynecology, University Medical Centre Ljubljana, for providing the ovarian tissue biopsies, to all patients who donated ovarian tissue for this research, to Dr. Elvira Malicev and Prof. Primoz Rozman, the Blood Transfusion Centre of Ljubljana for providing flow-cytometry analysis, to all the colleagues at the Reproductive Unit of our department and to the Slovenian Research Agency (grant J3-4195 to Dr. Irma Virant-Klun) for financial support.
Competing interests
The authors declare that they have no conflict of interests.
Footnotes
Capsule We propose putative ovarian mesenchymal stem cells (PO-MSCs) as a novel type of MSCs which share some similarities with bone marrow-derived MSCs but nevertheless show distinct and specific characteristics.
References
- 1.Zuk PA, Zhu M, Mizuno H, Huang J, Futrell JW, Katz AJ, et al. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7(2):211–28. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 2.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 3.Uezumi A, Ojima K, Fukada S, Ikemoto M, Masuda S, Miyagoe-Suzuki Y, et al. Functional heterogeneity of side population cells in skeletal muscle. Biochem Biophys Res Commun. 2006;341(3):864–73. doi: 10.1016/j.bbrc.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 4.Laino G, Graziano A, D’Aquino R, Pirozzi G, Lanza V, Valiante S, et al. An approachable human adult stem cell source for hard-tissue engineering. J Cell Physiol. 2006;206(3):693–701. doi: 10.1002/jcp.20526. [DOI] [PubMed] [Google Scholar]
- 5.De Bari C, Dell’Accio F, Tylzanowski P, Luyten FP. Multipotent mesenchymal stem cells from adult human synovial membrane. Arthritis Rheum. 2001;44(8):1928–42. doi: 10.1002/1529-0131(200108)44:8<1928::AID-ART331>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 6.Wang HS, Hung SC, Peng ST, Huang CC, Wei HM, Guo YJ, et al. Mesenchymal stem cells in the Wharton’s jelly of the human umbilical cord. Stem Cells. 2004;22(7):1330–7. doi: 10.1634/stemcells.2004-0013. [DOI] [PubMed] [Google Scholar]
- 7.Romanov YA, Svintsitskaya VA, Smirnov VN. Searching for alternative sources of postnatal human mesenchymal stem cells: candidate MSC-like cells from umbilical cord. Stem Cells. 2003;21(1):105–10. doi: 10.1634/stemcells.21-1-105. [DOI] [PubMed] [Google Scholar]
- 8.Erices A, Conget P, Minguell JJ. Mesenchymal progenitor cells in human umbilical cord blood. Br J Haematol. 2000;109(1):235–42. doi: 10.1046/j.1365-2141.2000.01986.x. [DOI] [PubMed] [Google Scholar]
- 9.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8(4):315–7. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 10.da Silva ML, Chagastelles PC, Nardi NB. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci. 2006;119(Pt 11):2204–13. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 11.Tang KH, Dai YD, Tong M, Chan YP, Kwan PS, Fu L, et al. A CD90(+) tumor-initiating cell population with an aggressive signature and metastatic capacity in esophageal cancer. Cancer Res. 2013;73(7):2322–32. doi: 10.1158/0008-5472.CAN-12-2991. [DOI] [PubMed] [Google Scholar]
- 12.He Z, Kokkinaki M, Jiang J, Zeng W, Dobrinski I, Dym M. Isolation of human male germ-line stem cells using enzymatic digestion and magnetic-activated cell sorting. Methods Mol Biol. 2012;825:45–57. doi: 10.1007/978-1-61779-436-0_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez R, Griparic L, Vargas V, Burgee K, Santacruz P, Anderson R, et al. A putative mesenchymal stem cells population isolated from adult human testes. Biochem Biophys Res Commun. 2009;385(4):570–5. doi: 10.1016/j.bbrc.2009.05.103. [DOI] [PubMed] [Google Scholar]
- 14.Riekstina U, Cakstina I, Parfejevs V, Hoogduijn M, Jankovskis G, Muiznieks I, et al. Embryonic stem cell marker expression pattern in human mesenchymal stem cells derived from bone marrow, adipose tissue, heart and dermis. Stem Cell Rev. 2009;5(4):378–86. doi: 10.1007/s12015-009-9094-9. [DOI] [PubMed] [Google Scholar]
- 15.Kuroda Y, Kitada M, Wakao S, Nishikawa K, Tanimura Y, Makinoshima H, et al. Unique multipotent cells in adult human mesenchymal cell populations. Proc Natl Acad Sci U S A. 2010;107(19):8639–43. doi: 10.1073/pnas.0911647107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trubiani O, Zalzal SF, Paganelli R, Marchisio M, Giancola R, Pizzicannella J, et al. Expression profile of the embryonic markers nanog, OCT-4, SSEA-1, SSEA-4, and frizzled-9 receptor in human periodontal ligament mesenchymal stem cells. J Cell Physiol. 2010;225(1):123–31. doi: 10.1002/jcp.22203. [DOI] [PubMed] [Google Scholar]
- 17.Kopen GC, Prockop DJ, Phinney DG. Marrow stromal cells migrate throughout forebrain and cerebellum, and they differentiate into astrocytes after injection into neonatal mouse brains. Proc Natl Acad Sci U S A. 1999;96(19):10711–6. doi: 10.1073/pnas.96.19.10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Y, Chen L, Hou XG, Hou WK, Dong JJ, Sun L, et al. Differentiation of bone marrow-derived mesenchymal stem cells from diabetic patients into insulin-producing cells in vitro. Chin Med J (Engl) 2007;120(9):771–6. [PubMed] [Google Scholar]
- 19.Keating A. Mesenchymal stromal cells: new directions. Cell Stem Cell. 2012;10(6):709–16. doi: 10.1016/j.stem.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 20.McLean K, Gong Y, Choi Y, Deng N, Yang K, Bai S, et al. Human ovarian carcinoma–associated mesenchymal stem cells regulate cancer stem cells and tumorigenesis via altered BMP production. J Clin Invest. 2011;121(8):3206–19. doi: 10.1172/JCI45273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stimpfel M, Skutella T, Cvjeticanin B, Meznaric M, Dovc P, Novakovic S, et al. Isolation, characterization and differentiation of cells expressing pluripotent/multipotent markers from adult human ovaries. Cell Tissue Res. 2013;354(2):593–607. doi: 10.1007/s00441-013-1677-8. [DOI] [PubMed] [Google Scholar]
- 22.Stimpfel M, Skutella T, Kubista M, Malicev E, Conrad S, Virant-Klun I. Potential stemness of frozen-thawed testicular biopsies without sperm in infertile men included into the in vitro fertilization programme. J Biomed Biotechnol. 2012;2012:291038. doi: 10.1155/2012/291038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandra VGS, Phadnis S, Nair PD, Bhonde RR. Generation of pancreatic hormone-expressing islet-like cell aggregates from murine adipose tissue-derived stem cells. Stem Cells. 2013;27(8):1941–53. doi: 10.1002/stem.117. [DOI] [PubMed] [Google Scholar]
- 24.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 25.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19(3):368–75. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 28.Haniffa MA, Collin MP, Buckley CD, Dazzi F. Mesenchymal stem cells: the fibroblasts’ new clothes? Haematologica. 2009;94(2):258–63. doi: 10.3324/haematol.13699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hematti P. Mesenchymal stromal cells and fibroblasts: a case of mistaken identity? Cytotherapy. 2002;14(5):516–21. doi: 10.3109/14653249.2012.677822. [DOI] [PubMed] [Google Scholar]
- 30.Wagner W, Wein F, Seckinger A, Frankhauser M, Wirkner U, Krause U, et al. Comparative characteristics of mesenchymal stem cells from human bone marrow, adipose tissue, and umbilical cord blood. Exp Hematol. 2005;33(11):1402–16. doi: 10.1016/j.exphem.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Kern S, Eichler H, Stoeve J, Klüter H, Bieback K. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells. 2006;24(5):1294–301. doi: 10.1634/stemcells.2005-0342. [DOI] [PubMed] [Google Scholar]
- 32.Wagner W, Roderburg C, Wein F, Diehlmann A, Frankhauser M, Schubert R, et al. Molecular and secretory profiles of human mesenchymal stromal cells and their abilities to maintain primitive hematopoietic progenitors. Stem Cells. 2007;25(10):2638–47. doi: 10.1634/stemcells.2007-0280. [DOI] [PubMed] [Google Scholar]
- 33.Alves H, van Ginkel J, Groen N, Hulsman M, Mentink A, Reinders M, et al. A mesenchymal stromal cell gene signature for donor age. PLoS One. 2012;7(8):e42908. doi: 10.1371/journal.pone.0042908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choudhery MS, Badowski M, Muise A, Pierce J, Harris DT. Donor age negatively impacts adipose tissue-derived mesenchymal stem cell expansion and differentiation. J Transl Med. 2014;12:8. doi: 10.1186/1479-5876-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaim M, Karaman S, Cetin G, Isik S. Donor age and long-term culture affect differentiation and proliferation of human bone marrow mesenchymal stem cells. Ann Hematol. 2012;91(8):1175–86. doi: 10.1007/s00277-012-1438-x. [DOI] [PubMed] [Google Scholar]
- 36.Alt EU, Senst C, Murthy SN, Slakey DP, Dupin CL, Chaffin AE, et al. Aging alters tissue resident mesenchymal stem cell properties. Stem Cell Res. 2012;8(2):215–25. doi: 10.1016/j.scr.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Geißler S, Textor M, Kühnisch J, Könnig D, Klein O, Ode A, et al. Functional comparison of chronological and in vitro aging: differential role of the cytoskeleton and mitochondria in mesenchymal stromal cells. PLoS One. 2012;7(12):e52700. doi: 10.1371/journal.pone.0052700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baddoo M, Hill K, Wilkinson R, Gaupp D, Hughes C, Kopen GC, et al. Characterization of mesenchymal stem cells isolated from murine bone marrow by negative selection. J Cell Biochem. 2003;89(6):1235–49. doi: 10.1002/jcb.10594. [DOI] [PubMed] [Google Scholar]
- 39.Muraglia A, Cancedda R, Quarto R. Clonal mesenchymal progenitors from human bone marrow differentiate in vitro according to a hierarchical model. J Cell Sci. 2000;113(Pt 7):1161–6. doi: 10.1242/jcs.113.7.1161. [DOI] [PubMed] [Google Scholar]
- 40.Lee CC, Christensen JE, Yoder MC, Tarantal AF. Clonal analysis and hierarchy of human bone marrow mesenchymal stem and progenitor cells. Exp Hematol. 2010;38(1):46–54. doi: 10.1016/j.exphem.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell KC, Phinney DG, Lacey MR, Barrilleaux BL, Meyertholen KE, O’Connor KC. In vitro high-capacity assay to quantify the clonal heterogeneity in trilineage potential of mesenchymal stem cells reveals a complex hierarchy of lineage commitment. Stem Cells. 2010;28(4):788–98. doi: 10.1002/stem.312. [DOI] [PubMed] [Google Scholar]
- 42.Okamoto T, Aoyama T, Nakayama T, Nakamata T, Hosaka T, Nishijo K, et al. Clonal heterogeneity in differentiation potential of immortalized human mesenchymal stem cells. Biochem Biophys Res Commun. 2002;295(2):354–61. doi: 10.1016/s0006-291x(02)00661-7. [DOI] [PubMed] [Google Scholar]
- 43.Guilak F, Lott KE, Awad HA, Cao Q, Hicok KC, Fermor B, et al. Clonal analysis of the differentiation potential of human adipose-derived adult stem cells. J Cell Physiol. 2006;206(1):229–37. doi: 10.1002/jcp.20463. [DOI] [PubMed] [Google Scholar]
- 44.Wang X, Dai J. Concise review: isoforms of OCT4 contribute to the confusing diversity in stem cell biology. Stem Cells. 2010;28(5):885–93. doi: 10.1002/stem.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sy J, Xt X, Jiang H, Zhou JJ, Li F, Cao P. Low expression of basic fibroblastic growth factor in mesenchymal stem cells and bone marrow of children with aplastic anemia. Pediatr Hematol Oncol. 2014;31(1):11–9. doi: 10.3109/08880018.2013.792402. [DOI] [PubMed] [Google Scholar]
- 46.Lu P, Blesch A, Tuszynski MH. Induction of bone marrow stromal cells to neurons: differentiation, transdifferentiation, or artifact? J Neurosci Res. 2004;77(2):174–91. doi: 10.1002/jnr.20148. [DOI] [PubMed] [Google Scholar]
- 47.Croft AP, Przyborski SA. Formation of neurons by non-neural adult stem cells: potential mechanism implicates an artifact of growth in culture. Stem Cells. 2006;24(8):1841–51. doi: 10.1634/stemcells.2005-0609. [DOI] [PubMed] [Google Scholar]
- 48.Wenisch S, Trinkaus K, Hild A, Hose D, Heiss C, Alt V, et al. Immunochemical, ultrastructural and electrophysiological investigations of bone-derived stem cells in the course of neuronal differentiation. Bone. 2006;38(6):911–21. doi: 10.1016/j.bone.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 49.Chen G, Wang Y, Xu Z, Fang F, Xu R, Wang Y, et al. Neural stem cell-like cells derived from autologous bone mesenchymal stem cells for the treatment of patients with cerebral palsy. J Transl Med. 2013;11:21. doi: 10.1186/1479-5876-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Razavi S, Ahmadi N, Kazemi M, Mardani M, Esfandiari E. Efficient transdifferentiation of human adipose-derived stem cells into Schwann-like cells: a promise for treatment of demyelinating diseases. Adv Biomed Res. 2012;1:12. doi: 10.4103/2277-9175.96067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paldino E, Cenciarelli C, Giampaolo A, Milazzo L, Pescatori M, Hassan HJ, et al. Induction of dopaminergic neurons from human wharton’s jelly mesenchymal stem cell by Forskolin. J Cell Physiol. 2014;229(2):232–44. doi: 10.1002/jcp.24442. [DOI] [PubMed] [Google Scholar]
- 52.Boroujeni Z, Aleyasin A. Human Umbilical Cord Derived Mesenchymal Stem Cells Can Secret Insulin In Vitro and In Vivo. Biotechnol Appl Biochem. May 31 2013; doi:10.1002/bab.1127 [Epub ahead of print]. [DOI] [PubMed]
- 53.Dave SD, Vanikar AV, Trivedi HL. Extrinsic factors promoting in vitro differentiation of insulin-secreting cells from human adipose tissue-derived mesenchymal stem cells. Appl Biochem Biotechnol. 2013;170(4):962–71. doi: 10.1007/s12010-013-0250-y. [DOI] [PubMed] [Google Scholar]
- 54.Karaoz E, Okcu A, Ünal ZS, Subasi C, Saglam O, Duruksu G. Adipose tissue-derived mesenchymal stromal cells efficiently differentiate into insulin-producing cells in pancreatic islet microenvironment both in vitro and in vivo. Cytotherapy. 2013;15(5):557–70. doi: 10.1016/j.jcyt.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 55.Prasajak P, Leeanansaksiri W. Developing a new two-step protocol to generate functional hepatocytes from Wharton’s jelly-derived mesenchymal stem cells under hypoxic condition. Stem Cells Int. 2013;2013:762196. doi: 10.1155/2013/762196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang S, Yu L, Sun M, Mu S, Wang C, Wang D, et al. The therapeutic potential of umbilical cord mesenchymal stem cells in mice premature ovarian failure. Biomed Res Int. 2013;2013:690491. doi: 10.1155/2013/690491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abd-Allah SH, Shalaby SM, Pasha HF, El-Shal AS, Raafat N, Shabrawy SM, et al. Mechanistic action of mesenchymal stem cell injection in the treatment of chemically induced ovarian failure in rabbits. Cytotherapy. 2013;15(1):64–75. doi: 10.1016/j.jcyt.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 58.Takehara Y, Yabuuchi A, Ezoe K, Kuroda T, Yamadera R, Sano C, et al. The restorative effects of adipose-derived mesenchymal stem cells on damaged ovarian function. Lab Invest. 2013;93(2):181–93. doi: 10.1038/labinvest.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu T, Huang Y, Guo L, Cheng W, Zou G. CD44+/CD105+ human amniotic fluid mesenchymal stem cells survive and proliferate in the ovary long-term in a mouse model of chemotherapy-induced premature ovarian failure. Int J Med Sci. 2012;9(7):592–602. doi: 10.7150/ijms.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rahman MM, Subramani J, Ghosh M, Denninger JK, Takeda K, Fong GH, et al. CD13 promotes mesenchymal stem cell-mediated regeneration of ischemic muscle. Front Physiol. 2014;4:402. doi: 10.3389/fphys.2013.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Surowiak P, Drag M, Materna V, Suchocki S, Grzywa R, Spaczyński M, et al. Expression of aminopeptidase N/CD13 in human ovarian cancers. Int J Gynecol Cancer. 2006;16(5):1783–8. doi: 10.1111/j.1525-1438.2006.00657.x. [DOI] [PubMed] [Google Scholar]
- 62.Terauchi M, Kajiyama H, Shibata K, Ino K, Nawa A, Mizutani S, et al. Inhibition of APN/CD13 leads to suppressed progressive potential in ovarian carcinoma cells. BMC Cancer. 2007;7:140. doi: 10.1186/1471-2407-7-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roche S, Delorme B, Oostendorp RA, Barbet R, Caton D, Noel D, et al. Comparative proteomic analysis of human mesenchymal and embryonic stem cells: towards the definition of a mesenchymal stem cell proteomic signature. Proteomics. 2009;9(2):223–32. doi: 10.1002/pmic.200800035. [DOI] [PubMed] [Google Scholar]
- 64.Kossowska-Tomaszczuk K, De Geyter C, De Geyter M, Martin I, Holzgreve W, Scherberich A, et al. The multipotency of luteinizing granulosa cells collected from mature ovarian follicles. Stem Cells. 2009;27(1):210–9. doi: 10.1634/stemcells.2008-0233. [DOI] [PubMed] [Google Scholar]
- 65.Lunter PC, van Kilsdonk JW, van Beek H, Cornelissen IM, Bergers M, Willems PH, et al. Activated leukocyte cell adhesion molecule (ALCAM/CD166/MEMD), a novel actor in invasive growth, controls matrix metalloproteinase activity. Cancer Res. 2005;65(19):8801–8. doi: 10.1158/0008-5472.CAN-05-0378. [DOI] [PubMed] [Google Scholar]
- 66.Weidle UH, Eggle D, Klostermann S, Swart GW. ALCAM/CD166: cancer-related issues. Cancer Genomics Proteomics. 2010;7(5):231–43. [PubMed] [Google Scholar]
- 67.Chen S, Lewallen M, Xie T. Adhesion in the stem cell niche: biological roles and regulation. Development. 2013;140(2):255–65. doi: 10.1242/dev.083139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 42 kb)