Abstract
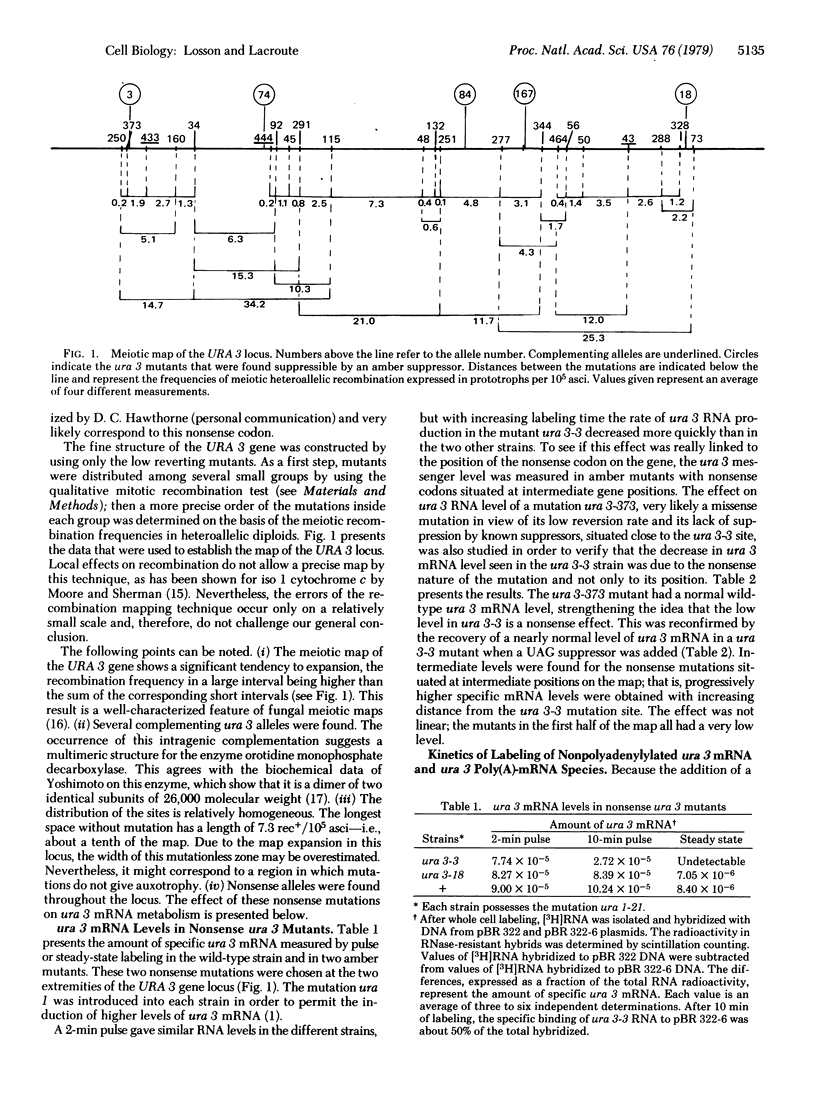
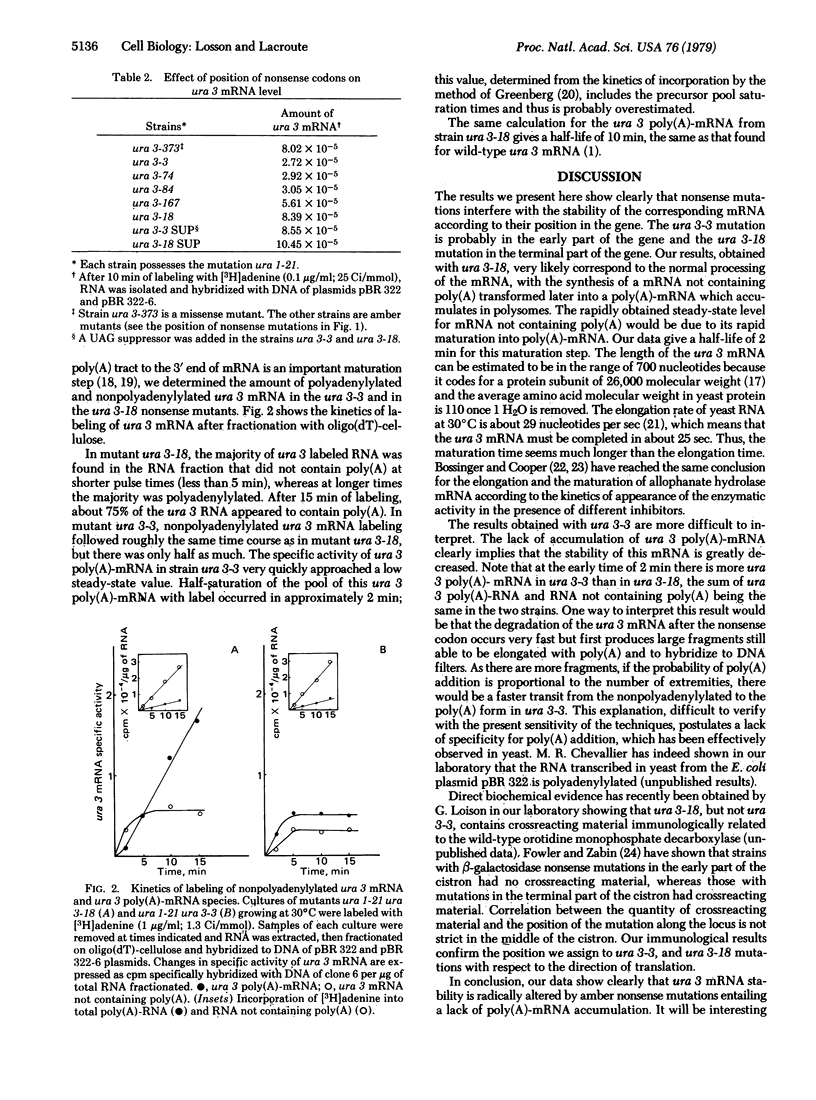
The fine structure map of the yeast URA 3 gene was established by meiotic recombination, and amber nonsense mutations were located at different points on the map. The effect of the length of the labeling time on the specific radioactivity of ura 3 messenger RNA and on its repartition between poly(A)-RNA and RNA not containing poly(A) has been followed in nonsense mutants. Nonsense mutations reduce the messenger level without lowering its instantaneous rate of synthesis. The strength of the reduction depends on the position of the nonsense codon within the locus and concerns essentially the accumulation of polyadenylylated ura 3 mRNA.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bach M. L., Lacroute F., Botstein D. Evidence for transcriptional regulation of orotidine-5'-phosphate decarboxylase in yeast by hybridization of mRNA to the yeast structural gene cloned in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Jan;76(1):386–390. doi: 10.1073/pnas.76.1.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach M. L., Lacroute F. Direct selective techniques for the isolation of pyrimidine auxotrophs in yeast. Mol Gen Genet. 1972;115(2):126–130. doi: 10.1007/BF00277292. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bossinger J., Cooper T. G. Execution times of macromolecular synthetic processes involved in the induction of allophanate hydrolase at 15 degrees C. J Bacteriol. 1976 Oct;128(1):498–501. doi: 10.1128/jb.128.1.498-501.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossinger J., Cooper T. G. Sequence of molecular events involved in induction of allophanate hydrolase. J Bacteriol. 1976 Apr;126(1):198–204. doi: 10.1128/jb.126.1.198-204.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CANNON M., KRUG R., GILBERT W. THE BINDING OF S-RNA BY ESCHERICHIA COLI RIBOSOMES. J Mol Biol. 1963 Oct;7:360–378. doi: 10.1016/s0022-2836(63)80030-3. [DOI] [PubMed] [Google Scholar]
- Clewell D. B. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972 May;110(2):667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincham J. R., Holliday R. An explanation of fine structure map expansion in terms of excision repair. Mol Gen Genet. 1970;109(4):309–322. doi: 10.1007/BF00267701. [DOI] [PubMed] [Google Scholar]
- Fowler A. V., Zabin I. Beta-galactosidase: immunological studies of nonsense, missense and deletion mutants. J Mol Biol. 1968 Apr 14;33(1):35–47. doi: 10.1016/0022-2836(68)90279-9. [DOI] [PubMed] [Google Scholar]
- Fraser R. S. Turnover of polyadenylated messenger RNA in fission yeast. Evidence for the control of protein synthesis at the translational level. Eur J Biochem. 1975 Dec 15;60(2):477–486. doi: 10.1111/j.1432-1033.1975.tb21026.x. [DOI] [PubMed] [Google Scholar]
- Gillespie D., Spiegelman S. A quantitative assay for DNA-RNA hybrids with DNA immobilized on a membrane. J Mol Biol. 1965 Jul;12(3):829–842. doi: 10.1016/s0022-2836(65)80331-x. [DOI] [PubMed] [Google Scholar]
- Greenberg J. R. High stability of messenger RNA in growing cultured cells. Nature. 1972 Nov 10;240(5376):102–104. doi: 10.1038/240102a0. [DOI] [PubMed] [Google Scholar]
- Gupta R. S., Schlessinger D. Coupling of rates of transcription, translation, and messenger ribonucleic acid degradation in streptomycin-dependent mutants of Escherichia coli. J Bacteriol. 1976 Jan;125(1):84–93. doi: 10.1128/jb.125.1.84-93.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S., Yanofsky C. Hyper-labile messenger RNA in polar mutants of the tryptophan operon of Escherichia coli. J Mol Biol. 1972 Dec 14;72(1):103–110. doi: 10.1016/0022-2836(72)90072-1. [DOI] [PubMed] [Google Scholar]
- Korch C. T., Lacroute F., Exinger F. A regulatory interaction between pyrimidine and purine biosyntheses via ureidosuccinic acid. Mol Gen Genet. 1974;133(1):63–75. doi: 10.1007/BF00268678. [DOI] [PubMed] [Google Scholar]
- Lacroute F. RNA and protein elongation rates in Saccharomyces cerevisiae. Mol Gen Genet. 1973 Sep 27;125(4):319–327. doi: 10.1007/BF00276587. [DOI] [PubMed] [Google Scholar]
- Moore C. W., Sherman F. Role of DNA sequences in genetic recombination in the iso-1-cytochrome c gene of yeast. I. Discrepancies between physical distances and genetic distances determined by five mapping procedures. Genetics. 1975 Mar;79(3):397–418. doi: 10.1093/genetics/79.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Hawthorne D. C. Genetic mapping in Saccharomyces. Genetics. 1966 Jan;53(1):165–173. doi: 10.1093/genetics/53.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. P. Processing of RNA. Annu Rev Biochem. 1976;45:605–629. doi: 10.1146/annurev.bi.45.070176.003133. [DOI] [PubMed] [Google Scholar]
- Schneider E., Blundell M., Kennell D. Translation and mRNA decay. Mol Gen Genet. 1978 Apr 6;160(2):121–129. doi: 10.1007/BF00267473. [DOI] [PubMed] [Google Scholar]
- Waldron C., Lacroute F. Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J Bacteriol. 1975 Jun;122(3):855–865. doi: 10.1128/jb.122.3.855-865.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto A., Umezu K., Kobayashi K., Tomita K. Orotidylate decarboxylase (yeast). Methods Enzymol. 1978;51:74–79. doi: 10.1016/s0076-6879(78)51013-6. [DOI] [PubMed] [Google Scholar]



