SUMMARY
Reprogramming of somatic cells to induced pluripotent stem cells (iPSCs) holds enormous promise for regenerative medicine, but the underlying mechanisms remain poorly understood. We report a systematic dissection of human cellular reprogramming by combining a genome-wide RNAi screen, innovative computational methods, extensive single-hit validation and mechanistic dissection of relevant pathways and networks. We identify novel reprogramming barriers, including genes involved in transcription, chromatin regulation, ubiquitination, dephosphorylation, vesicular transport and cell adhesion. Specific a disintegrin and metalloproteinase (ADAM) proteins are inhibitors of reprogramming, and the disintegrin domain of ADAM29 is necessary and sufficient for this function. Clathrin-mediated endocytosis can be targeted with small molecules and opposes reprogramming by positively regulating TGFβ signaling. Genetic interaction studies of endocytosis or ubiquitination reveal that barrier pathways can act in linear, parallel or feed-forward loop architectures to antagonize reprogramming. Our online resource summarizing these results provides a global view of barriers to human cellular reprogramming.
Keywords: Pluripotency, induced Pluripotent Stem cells (iPSCs), genome-wide RNAi screen, barriers to reprogramming, ADAMs, endocytosis, RNF40, pathway interaction
INTRODUCTION
Cells generally become committed to increasingly differentiated fates during normal development, but experimental paradigms for cellular reprogramming have shown that differentiation is reversible. Initial methods for reprogramming included somatic cell nuclear transfer to enucleated oocytes (Gurdon et al., 1958) or fusion with pluripotent stem cells (Tada, 1997), both of which were technically difficult to dissect at the mechanistic level. The finding that overexpression of a key transcription factor leads to fibroblast-to-muscle transdifferentiation (Davis et al., 1987) suggested that perhaps other cell-fate transitions could similarly be induced. The recent discovery that combinations of transcription factors can reprogram somatic cells to induced pluripotent stem cells (iPSCs) has transformed stem cell research and holds enormous promise in regenerative medicine (Takahashi and Yamanaka, 2006; Takahashi et al., 2007; Yu et al., 2007). Understanding the process of reprogramming may also shed light on events that take place during cellular transformation in cancer (Daley, 2008; Ramalho-Santos, 2009). However, due to the low efficiency and protracted nature of the process, the mechanisms that underlie the generation of iPSCs remain poorly understood.
Recent studies have revealed that reprogramming to the iPSC state is a multi-step process involving large-scale changes in the transcriptional and epigenetic state of somatic cells along the path to pluripotency (Buganim et al., 2012; Polo et al., 2012). Importantly, the somatic cell is not a tabula rasa and expresses genes that antagonize reprogramming, as has been shown for tumor suppressors (p53, INK4a/ARF, LATS2) (Kawamura et al., 2009; Qin et al., 2012; Zhao et al., 2008) and H3K9 methyltransferases (SETDB1, SUV39H, EHMT2) (Chen et al., 2013). In addition, focused RNAi screens have revealed other pathways that act as barriers to reprogramming, such as TGFβ signaling (Samavarchi-Tehrani et al., 2010), H3K79 methylation by DOT1L (Onder et al., 2012), or protein ubiquitination (Buckley et al., 2012). These findings suggest that other critical barriers to reprogramming are likely to exist, but no genome-wide functional screen has yet been carried out in mouse or human iPSC generation.
RNAi provides a powerful technique for exploiting a cell’s endogenous machinery for mRNA degradation to obtain selective gene knockdown. Well-based genome-wide RNAi screens, where cells are transfected in separate wells with small pools or individual siRNAs, have been carried out successfully, including for the identification of genes that regulate human embryonic stem cell (ESC) self-renewal and pluripotency (Chia et al., 2010). However, the throughput of this approach is limited, particularly in the context of iPSC generation, because of the low reprogramming efficiency. An alternative to well-based screens yielding much higher throughput is a pooled short hairpin RNA (shRNA)-based screen combined with next generation sequencing (NGS). This approach has a significantly larger dynamic range and has enabled genome-wide screens at an unprecedented scale (Bassik et al., 2013; 2009). However, the extraction of robust biological information from genome-wide screen data is still challenging: the problems of false-positive hits caused by off-target effects, false-negative hits caused by ineffective RNAi, and variance in sequencing depth can limit reliability.
We use ultracomplex EXPANDed pooled shRNA libraries to report a genome-wide screen for barriers to human cellular reprogramming. We introduce a multi-objective optimization technique for analyzing NGS-based shRNA screen data, and combine our method with systems-level meta-analyses and in vitro experiments to discover critical barriers to reprogramming genome wide. Our integrative approach identifies 956 genes predicted to act as barriers to reprogramming, including genes involved in transcription, chromatin regulation, ubiquitination, dephosphorylation, vesicular transport and cell adhesion. We mechanistically dissect the roles of disintegrin proteins and clathrin-mediated endocytosis as reprogramming barriers, and show that barriers from different pathways interact and can have combinatorial effects to antagonize reprogramming. The results are compiled into an online resource (http://song.igb.illinois.edu/ipsScreen/), allowing researchers to browse, query, and visualize the analysis.
RESULTS AND DISCUSSION
Genome-Wide shRNA Libraries and NGS Enable an Unbiased Screen for Barriers to Reprogramming
We sought to implement a robust and unbiased screen for barriers to human iPSC generation. We used a recently described method (Bassik et al., 2009) to perform a genome-wide shRNA library screen targeting 19,527 human genes with an average coverage of 30 independent shRNAs per gene. Human BJ fibroblasts were co-infected with lentivirus expressing these shRNAs along with OCT4, SOX2, KLF4, cMYC (4F) and p53 RNAi (p53i). We chose to add p53i because it has been shown to enhance reprogramming efficiency (Kawamura et al., 2009; Zhao et al., 2008). Moreover, data from a pilot screen demonstrates that p53i increases the sampling rate of fully reprogrammed cells and, hence, improves sensitivity in the detection of reprogramming barriers (Figure S1A). Importantly, all of our downstream hit validation was done in the absence of p53i (see below). Following the appearance of colonies with iPSC characteristics on day 28, we Fluorescence-activated cell sorting (FACS) purified the transduced cells for TRA-1-81, a marker of fully reprogrammed human iPSCs (International Stem Cell Initiative et al., 2007). Integrated shRNAs were then recovered and identified by PCR amplification from genomic DNA of both the TRA-1-81+ and TRA-1-81-cell populations and quantified by NGS (Figure 1A).
Figure 1. A Genome-Wide RNAi Screen Identifies Known and Novel Regulators of Human iPSC Generation.
(A) Design of the genome-wide RNAi screen in human iPSC generation. (B) Positive control genes consisting of known barriers identified from the literature and the experimentally validated targets of miR-17, miR-200, and miR-372 (miRNAs known to enhance reprogramming efficiency) are enriched in TRA-1-81+. Shown are the medians of the distributions of collective shRNA activity levels for these genes. The statistical significance was measured by a hypergeometric test. (C) Each dot on the graph is a gene. The x-axis shows the number of distinct active shRNAs targeting the gene. The y-axis represents the collective shRNA activity level δ on a log scale; this summarizes the read-count odds ratios of all active shRNAs targeting a given gene. Genes with greater statistical significance are color-coded towards red. The top 1% most significant genes are denoted by stars. Examples of known barriers or enhancers of reprogramming are indicated.
The relative frequency of reads mapping to a given shRNA in TRA-1-81+ compared to TRA-1-81-, expressed as an odds ratio θ, estimates the positive effect size of that shRNA on reprogramming and, thus by inference, the negative effect size of its targeted gene as a potential reprogramming barrier. We call an shRNA active if it has greater odd of being sequenced in TRA-1-81+ compared to TRA-1-81-, i.e. if θ > 1 with sufficient coverage. To assess gene-wise collective shRNA activity levels and to quantify the negative effect size of a given gene on reprogramming, we combined the log-transformed odds ratios for all active shRNAs targeting a gene into a single statistic using a random-effects model. The combined statistic, δ, controls for variance in knockdown efficiency and variance in sequencing depth-of-coverage by estimating them separately from the data. We then developed a gene ranking algorithm based on multi-objective optimization (Handl et al., 2007). This technique simultaneously maximizes collective shRNA activity and the number of distinct active shRNAs. The result is a gene ranking which controls for off-target effects, since highly ranked genes are precisely the ones that demonstrate a reproducible effect across multiple shRNAs enriched in the TRA-1-81+ population. To assign statistical significance to gene ranks, we computed p-values using a permutation test with 5,000 permutations of the read counts. False discovery rates (FDRs) were computed based on library swap to adjust for multiple-hypothesis testing. We compared this multi-objective optimization algorithm, which we call HitSelect, with existing methods, such as RIGER (Luo et al., 2008) and RSA (König et al., 2007), and found that HitSelect has greater sensitivity and specificity, and is less prone to off-target effects (Figure S1B). This approach generated 956 TRA-1-81+ screen hits at the 5% significance level, corresponding to a FDR of 7% (Extended Experimental Procedures and File S1).
A Genome-Wide Screen Confirms Known Barriers to Reprogramming
To validate our screen, we used as positive controls previously identified gene barriers, as well as the experimentally validated targets of micro-RNAs (miRNAs) known to enhance reprogramming efficiency. Known barrier genes show significant activity levels in TRA-1-81+ compared to TRA-1-81-(Figure 1B, C). Specifically, we identified 26 barrier genes whose knockdown has been previously shown to enhance the efficiency of reprogramming, including genes associated with tumor suppression (CDKN2B), epithelial-to-mesenchymal transition (TGFBR2), heterochromatin (EHMT2, MECP2, RBL2, SMARCC2), and extracellular matrix and adhesion (ARHGAP26, RHOC, CD44) (Figure 1C and Table S1). These positive controls are significantly enriched in the TRA-1-81+ population (Figure 1B). Targets of miRNAs known to enhance human reprogramming efficiency, such as miR-17, miR-200 and miR-372 (Gregory et al., 2008; Li et al., 2011; Subramanyam et al., 2011), also show significant enrichments in the TRA-1-81+ population (Figure 1B). As negative controls, we used control coding sequences represented in the library (GFP, ampicillin resistance or luciferase), a random gene list, or validated targets of miR-218, a known tumor suppressor not linked to reprogramming. These controls all show weak median activity (below δ=1) and statistically insignificant enrichment in the TRA-1-81+ population (Figure 1B). Overall, these data indicate that our approach is well validated by the successful identification of previously reported barriers to reprogramming. This work focuses on genes that act as barriers to reprogramming for two main reasons: a) they can be easily knocked down to improve the efficiency of iPSC generation, providing straightforward single hit validation; b) a search for shRNAs enriched in the TRA-1-81-population is complicated by the high heterogeneity of that population, which is not the case in the TRA-1-81+ population. Nevertheless, we also identified shRNAs for multiple known positive regulators of reprogramming enriched in the TRA-1-81-population, including OCT4, SOX2, NANOG, PRDM14, CHD1, PARP1, KDM2A, KDM2B and KDM3B (Figure 1C).
Analysis of Screen Hits Identifies Ubiquitination, Cell Adhesion/Motility and Endocytosis Pathways as Putative Barriers to Reprogramming
We took an integrated approach to identifying barrier pathways and prioritizing genes for downstream validation. We clustered the 956 TRA-1-81+ screen hits by Interpro protein domain, Gene Ontology (GO) cellular component and GO biological process annotations via DAVID (Huang et al., 2009). The most frequently annotated protein domain is “WD40 repeat conserved site” (Figure 2A). This domain often serves as a substrate for protein-protein interactions, and the WD40 annotation cluster contains multiple ubiquitin conjugating enzymes and ligases. The importance of the ubiquitination pathway is highlighted by the recent finding that the E3 ligase F-box and WD-40 domain protein 7 (FBXW7) functions as a barrier to reprogramming in mouse (Buckley et al., 2012). FBXW7 is also a hit in our screen, and is part of the WD40 annotation cluster. Using GeneMANIA (Zuberi et al., 2013) to aggregate published genetic interaction data, we identified multiple screen hits documented to interact with FBXW7 (Figure 2B). Thus, the role of ubiquitination as a barrier to reprogramming may be conserved (see below, Figure S3).
Figure 2. An Integrated Resource Identifies Barrier Pathways to Reprogramming.
(A) Screen hits, significant at the 5% level, were clustered by over-represented GO biological process, GO cellular component, and Interpro protein domain annotations using the DAVID functional analysis tool. Shown are the clusters with the 5 highest enrichment scores. Each cluster is represented by the most frequently occurring annotation term. On the y-axis is the number of genes in each cluster. The p-value shown was obtained by using Fisher’s method to combine DAVID assigned p-values for individual annotation terms within a cluster. (B) Gene functional annotations, aggregated via DAVID, were compiled into an interactive tree-map, available online (http://song.igb.uiuc.edu/ipsScreen/docs/david_treemap.html). All genes shown are screen hits at the 5% significance level. Gene lists for subclusters were analyzed for network structure. This identified several gene networks and subclusters. (1) A genetic interaction network for the ubiquitin ligase FBXW7. This network documents that FBXW7 can directly and indirectly interact with multiple screen hits from a variety of pathways. (2) A dense network of protocadherins. Two genes are connected with an edge if their expression correlates across tissue types. (3) Genetic interaction networks for ADAM family proteins. (4) A subcluster enriched for endocytosis functional annotations.
Also enriched among screen hits are annotations associated with cell adhesion and secretion, as well as adhesion-related extracellular protein domains (Figure 2). The most statistically significant protein domain annotation cluster is “Proto-cadherin”. Protocadherins are the largest mammalian sub-family of cadherins and have been implicated in cell adhesion and signal transduction. Gene network analysis reveals that there is a dense co-localization sub-network of protocadherins among screen hits (Figure 2B). There are also a large number of proteins containing a “Fibronectin type III” domain, as well as a significant number of metallopeptidases. Several members of the metallopeptidase cluster are ADAM proteins. These molecular functions are likely related to the fact that the most enriched biological process term is “Cell adhesion”. Thus, pathways mediating cell adhesion and extracellular matrix interactions are likely to contain barriers to reprogramming (see below, Figure 4).
Figure 4. DAMs Are a Barrier to Reprogramming and Act Via their Desintegrin Loop Domain.
(A) Screen hits at the 5% significance level with GO biological process annotations related to cell motility and adhesion are highlighted. (B) 4F+ADAM29i induced iPSCs show strong positive immunostaining for pluripotency markers NANOG, SSEA3 and SSEA4. Scale bar, 300 m. (C) 4F+ADAM29i-induced iPSCs express endogenous pluripotency markers at similar levels to 4F iPSCs and ESCs. (D) Growth curves of fibroblasts infected with 4F, 4F+ non-sense (NS), and 4F+RNF40i, counted on D0, D4, D8, and D13 post-infection. RNF40i does not substantially alter total cell numbers during the first 13 days of reprogramming. Error bars represent standard deviations. (E) ADAM29 over-expression reduces reprogramming efficiency, and the disintegrin domain is critical for the barrier function of ADAM29 in reprogramming. BJ fibroblasts over-expressing wild-type (WD) or mutated ADAM29 (H333A in the active site of the metalloprotease domain; E461A and ECD461 deletion in the disintegrin domain) were induced to pluripotency by 4F. (F) Over-expression of wild-type or mutated ADAM29 was confirmed by Western Blotting. Tubulin was used as loading control. (G) ADAM29 disintegrin loop peptide reduces reprogramming efficiency. Human BJ fibroblasts were infected with 4F. Four days later, a synthetic ADAM29 disintegrin loop peptide (100 g/ml) was added. A peptide with substitutions of the three critical residues (ECD) was used as controls. (H) Human ESCs H9 express higher levels of Integrin α6, and lower levels of Integrin α5 and β1 compared with BJ fibroblasts.
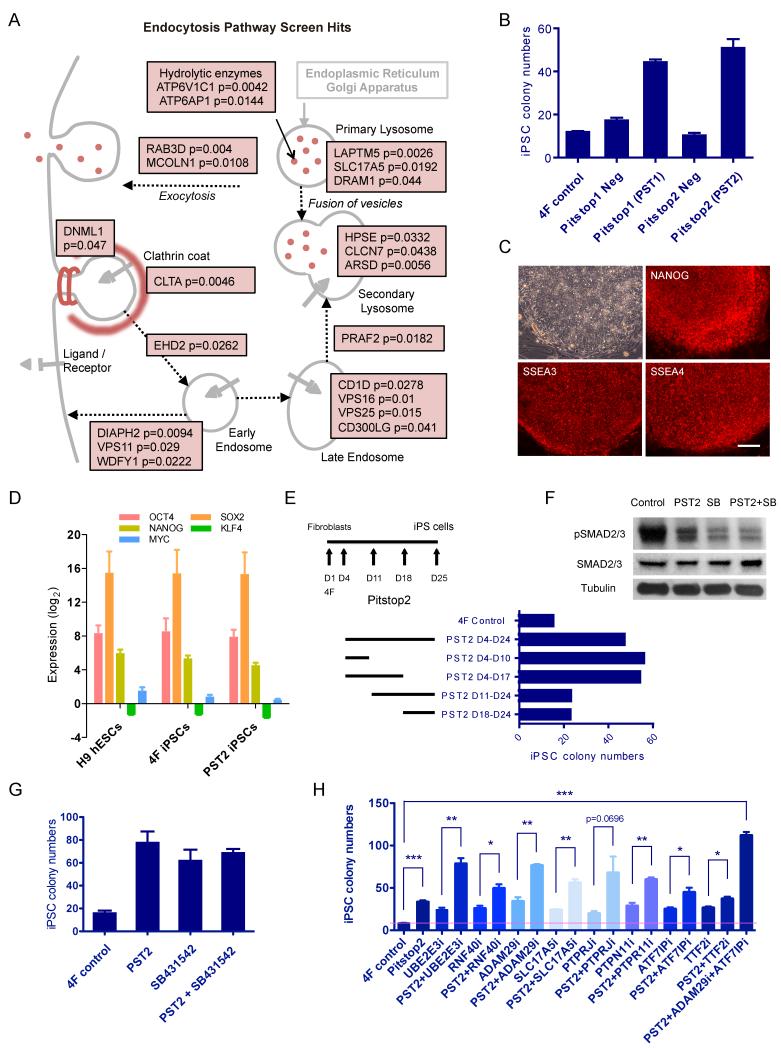
Lastly, screen hits showed a significant enrichment for functional annotations related to endocytosis (Figure 2). This enrichment occurs across all three annotation categories. The second most frequently annotated biological process is “Vesicle-mediated transport”. Moreover, the largest cellular component annotation clusters include multiple compartments of the endocytic pathway, such as “Cytoplasmic vesicle”, “Lysosome” and “Vacuole”. Together, this analysis suggests that endocytosis-mediated trafficking is a barrier to reprogramming (see below, Figure 5).
Figure 5. Endocytosis Is an Early-Acting Barrier to Reprogramming that Enhances TGF β Signaling.
(A) Screen hits at the 5% significance level with GO biological process annotations related to endocytosis and lysosome are highlighted. (B) Inhibition of clathrin-dependent endocytosis by small molecules Pitstop1 and Pitstop2 increases human iPSC generation efficiency. Human BJ fibroblasts were infected with 4F. Four days later, clathrin inhibitors (Abcam) were added to medium at a final concentration of 45 M (Pitstop1) and 30 M (Pitstop2) until colonies were counted. Pitstop1 Neg and Pitstop2 Neg, compounds structurally related to Pitstop1 and Pitstop2 respectively but lacking the ability to inhibit endocytosis (Kleist et al., 2011), were used as controls. (C) 4F+Pitstop2 iPSCs show strong immunostaining for pluripotency markers NANOG, SSEA3 and SSEA4. Scale bar, 300 m. (D) 4F+Pitstop2 iPSCs express endogenous pluripotency markers at similar levels to 4F iPSCs and ESCs. (E) Endocytosis acts as a barrier at an early stage of reprogramming. Human BJ fibroblasts were infected with 4F. Pitstop2 was added to the medium at the time points indicated. (F) Pitstop2 and TGFβ inhibitor SB431542 (SB, 2 μM) lead to a decrease in pSMAD2/3 levels by Western Blotting. Tubulin was used as loading control. (G) Both Pitstop2 and SB431542 (D2-D14) increase human iPSC generation efficiency. However, when combined together, no additional increase is observed. (H) Pitstop2 further enhances reprogramming efficiency upon knockdown of multiple other barriers.
We also examined published time-course data on gene expression and chromatin marks from reprogramming experiments in mouse. In particular, we considered two recent datasets where secondary mouse embryonic fibroblasts (MEFs) were reprogrammed by dox-inducible expression of 4F (Golipour et al., 2012; Polo et al., 2012). Both groups analyzed the gene expression profiles of intermediate populations during reprogramming, while Polo et al. additionally profiled H3K4me3 and H3K27me3 genome-wide. A key feature of both experimental designs was a strategy to enrich intermediate populations for cells with the potential to give rise to iPSCs. Interestingly, our re-analysis of these datasets reveals that functional annotations associated with cell adhesion, motility and extracellular matrix interactions are over-represented among genes down-regulated during the course of reprogramming in the Polo et al. data (Figure S2A). Furthermore, in the data of Golipour et al., there is significant enrichment for annotations associated with endocytosis in genes downregulated as reprogramming proceeds (Figure S2B). Thus, several of the pathways implicated as barriers by our functional analysis in human reprogramming have expression patterns consistent with such a role during the course of mouse reprogramming.
Taken together, our integrative analysis indicates that pathways associated with ubiquitination, cell adhesion/motility and endocytosis are putative reprogramming barriers. Other potential barriers include genes with roles in transcription, chromatin regulation, and dephosphorylation (Figure 2).
Single-Gene Analyses Validate Reprogramming Barriers from Multiple Pathways
To assess whether genes and pathways enriched in the 956 TRA-1-81+ hits truly represent reprogramming barriers, we subjected 23 genes to rigorous validation. In addition to genes related to ubiquitination (UBE2D3, UBE2E3, RNF40), ADAM family genes (ADAM7, ADAM21, ADAM29), and endocytosis (ARSD, SLC17A5, DRAM1), we also tested protein tyrosine phosphatases (PTPRJ, PTPRK, PTPN11), chromatin regulators (ATF7IP, ARID4A, CENPB, MED19), transcription factors (TTF1, TTF2, TMF1, T) and miscellaneous others (RPL30, PLCE1, INPP5K). For each gene, we chose the two most highly active shRNAs from the screen and performed single-gene validation of its knockdown effect on reprogramming. RNAi for 20 out of 23 genes tested (a hit validation of 87%) leads to significant increases in iPSC colony count compared with non-sense (NS) shRNA control (Figure 3A). The fold increase in reprogramming efficiency ranges from 1.5 to 4.2, which is comparable with previous studies of knockdown of reprogramming barriers, including p53/p21 (Kawamura et al., 2009; Zhao et al., 2008), LATS2 (Ohi et al., 2011), DOT1L and SUV39H1 (Onder et al., 2012). Knockdown of genes by shRNAs was confirmed by RT-PCR (Figure 3B).
Figure 3. Single-Gene Validation of Reprogramming Barriers.
(A) Individual knockdown of screen hits in different pathways increases the efficiency of human iPSC generation. The number of iPSC colonies was counted 25 days after infection of human BJ fibroblasts with 4F control, 4F with non-sense (NS) shRNA control, or 4F with the two most highly active shRNAs targeting the indicated gene. Error bars represent standard deviation compared to 4F+NS. * p<0.05; **, p<0.01; ***, p<0.001. (B) Reduction in the levels of gene expression achieved by shRNA constructs was confirmed by qRT-PCR.
We next present further validation and mechanistic dissection of the ubiquitin, cell adhesion and motility, and endocytosis pathways. To facilitate exploration of the entire screen data, we implemented our integrative analysis into a publicly available website. This resource combines the RNAi screen data from this study with a large collection of published genetic and epigenetic data, functional annotation analysis, gene interaction network analysis, and gene kinetics during reprogramming (http://song.igb.illinois.edu/ipsScreen/). Researchers are able to query, browse, and visualize this data collection.
The Ubiquitin Pathway Is a Barrier to Reprogramming
Fbxw7, an E3 ubiquitin ligase, was recently described as a barrier to mouse iPSC generation (Buckley et al., 2012). Our systems approach confirms and extends these findings to a much broader network of genes in human reprogramming. A variety of conjugating enzymes (UBE2D3, UBE2E3), ubiquitin ligases (RNF40, FBXW7, NEDD4, MARCH3), and deubiquitination enzymes (USP9X, OTUB2) are targeted in the TRA-1-81+ population (Figure S3A and File S2). Single-gene validation shows that knocking down UBE2D3, UBE2E3, or RNF40 individually significantly increases reprogramming efficiency compared to non-sense (NS) shRNA control, validating their functional role in reprogramming (Figure 3). We confirmed that iPSCs generated with RNF40 shRNAs express, in addition to TRA-1-81, the pluripotency markers NANOG, SSEA3 an SSEA4 at the protein level (Figure S3B), and express mRNAs of endogenous pluripotency factors at levels similar to human ESCs (Figure S3C). We further show that RNF40i increases reprogramming efficiency without accelerating cell expansion rates (Figure S3D). Finally, we show that inhibition of multiple members of the ubiquitin pathway increases the protein levels of the reprogramming factor OCT4 (Figure S3E), in support of previous mouse studies (Buckley et al., 2012). These results reveal that the previously reported role of the ubiquitin pathway as a reprogramming barrier in mouse is conserved in human and demonstrate the power of our systems approach to expand upon the pathways identified.
Cell Adhesion and Motility in Reprogramming
Genes involved in the controlled turnover of actin filament needed for cell motility are enriched for screen hits (Figure 4A and File S2). Actin, coronin, and RHOC, factors needed for filament assembly, branching, and disassembly are all significant hits in the screen. Moreover, several of the screen hits have well-documented interactions with the extracellular matrix, including MMP14, ADAM7, ADAM21 and ADAM29. Knockdown of ADAM7, ADAM21 or ADAM29 individually significantly increases human iPSC generation efficiency (Figure 3), with the most notable effect seen for ADAM29. We confirmed by immunofluorescence and qRT-PCR that iPSCs generated with ADAM29 shRNAs express high levels of several pluripotency markers, comparable to levels in hESCs (Figures 4B and 4C). . In addition, ADAM29i does not increase iPSC generation efficiency by accelerating cell expansion rates (Figure 4D). ADAM29 knockdown at both mRNA and protein levels and the consequential enhancement of reprogramming efficiency were confirmed with two independent shRNAs (Figure S4A, B, C). Moreover, we found that over-expression of ADAM29 significantly reduces reprogramming efficiency (Figure 4E) and decreases human ESC colony formation efficiency (Figure S4D), indicating that ADAM29 impedes both establishment and maintenance of the pluripotent state. To study the mechanism of ADAM29 as a reprogramming barrier, we mutated the metalloprotease and disintegrin domains of ADAM29, and over-expressed the altered proteins during reprogramming. Mutation of the disintegrin domain, but not the metalloprotease domain, abolishes the adverse effect of ADAM29 on reprogramming efficiency (Figure 4E, F). Remarkably, we found that adding a short synthetic peptide corresponding to the disintegrin loop of ADAM29 to the culture medium, but not a mutated peptide, significantly reduces iPSC colony formation efficiency (Figure 4G).
These data demonstrate that specific ADAM proteins are potent reprogramming barriers and that the disintegrin domain is necessary and sufficient for their activity during reprogramming. Disintegrin was originally identified from snake venom metalloprotease, which inhibits blood coagulation and is known to regulate Integrin-dependent cell adhesion. Multiple reports document the interaction between the ADAM disintegrin domain and Integrins, and the specificity of this interaction is mediated by a tripeptide motif in the disintegrin domain (Figure S4E) (Calvete et al., 2005; Igarashi et al., 2007). Interestingly, the screen hits that belong to the ADAM family, ADAM7, ADAM21 and ADAM29, all have an ECD tripeptide motif (Figure S4F), which can inhibit specific Integrin dimers including those containing α6. During reprogramming, parental fibroblasts predominantly express Integrin α5, which is not targeted by the ECD motif (Calvete et al., 2005), while iPSCs/ESCs express high levels of Integrin α6 (Figure 4H). Integrin α6 is a component of the Laminin receptor, and in turn Laminin is important for maintaining self-renewal of human ESCs and iPSCs (Rodin et al., 2010). Together, these findings suggest that ECD-containing ADAM proteins may antagonize an Integrin switch that occurs during reprogramming to pluripotency, a model that deserves further exploration. Notably, both ADAM7 and ADAM29 are often found mutated in melanoma (Wei et al., 2011), suggesting that ADAM dysfunction may also contribute to cellular transformation in certain cancers.
Endocytosis Is a Barrier to Reprogramming
To obtain a global view of the negative effect size of the endocytosis pathway on reprogramming, we aggregated all screen-hit genes with endocytosis-related GO biological process annotations (File S2). Remarkably, multiple screen hits were associated with a variety of components of the endocytic pathway: receptor-targeting ubiquitin ligases (MARCH3, RNF40, NEDD4); the mediator of vesicle transport, clathrin (CLTA); plasma-membrane remodeling and early-endosome fusion genes (EHD2, RABEP1); and endosomal/lysosomal surface proteins (MCOLN1, VPS25, ARSD, HSPA8, SCARB2, SLC17A5, DRAM1) (Figure 5A and File S2). Interestingly, NEDD4 has been reported to ubiquitinate cell surface receptors for lysosomal degradation (Persaud et al., 2011), suggesting a potential crosstalk between the endocytosis and ubiquitin pathways during reprogramming.
As validation of these findings, we found that knocking down DRAM1, SLC17A5 or ARSD individually with shRNAs leads to a significant increase in iPSC generation relative to non-sense (NS) shRNA control (Figure 3). We also used small molecules to inhibit the endocytosis pathway. We found that two clathrin-specific inhibitors, Pitstop1 and Pitstop2, can dramatically increase reprogramming efficiency, whereas structurally related controls (Kleist et al., 2011) have no effect (Figure 5B). Furthermore, Pitstop2 can also increase iPSC generation efficiency when episomal vectors are used to reprogram human dermal fibroblasts (HDF) (Figure S5A). Therefore, the effect of Pitstop is independent of the retroviral infection used in standard reprogramming methods or specific fibroblast lines. We confirmed that iPSCs generated with Pitstop2 express mRNA and protein levels of pluripotency markers similar to human ESCs (Figure 5D). In addition, Pitstop2 does not affect cell expansion rates during iPSC generation (Figure S5B). These data demonstrate that clathrin-mediated endocytosis is a barrier to reprogramming. Inhibitors of endocytosis may thus be added to the panel of small molecules that enhance reprogramming efficiency.
Endocytosis functions as a master organizer of signaling circuits by modulating the abundance of receptors, their ligands, and downstream effectors at different membrane compartments. We sought to further probe the mechanism by which endocytosis acts as a reprogramming barrier. Treatment of cells with Pitstop2 at different time points post infection with 4F revealed that endocytosis acts at an early stage of reprogramming (Figure 5E). During that stage, reprogramming cells must undergo a mesenchymal-epithelial transition (MET) (Samavarchi-Tehrani et al., 2010; Subramanyam et al., 2011), and we found that Pitstop2 up-regulates the epithelial marker E-cadherin at day 12 of reprogramming (Figure S5C, D). Of note, it has been shown that clathrin-mediated endocytosis positively regulates TGFβ signaling by recycling the receptors (Scita and Di Fiore, 2010), and TGFβ signaling is a known barrier to reprogramming that inhibits MET (Samavarchi-Tehrani et al., 2010; Subramanyam et al., 2011). In support of a role for the endocytosis pathway in TGFβ signaling, we found that Pitstop2 inhibits phosphorylation and activation of SMAD2/3, in particular SMAD2, during reprogramming (Figure S5C). Using GeneMANIA, we found that the endocytosis and TGFβ signaling pathways can interact at multiple levels (Figure S5E). Members of the TGFβ pathway, such as MAPK3 and SMAD2, are also screen hits as reprogramming barriers (Figure S5E). Importantly, in the presence of the TGFβ inhibitor SB431542, Pitstop2 does not further decrease the level of phosphorylated SMAD2/3 (Figure 5F), nor does it further increase iPSC generation efficiency (Figure 5G). Taken together, these data indicate that clathrin-mediated endocytosis antagonizes reprogramming via a linear positive interaction with the TGFβ signaling pathway.
Given the observation that inhibition of endocytosis is unable to synergize with inhibition of TGFβ signaling to increase reprogramming efficiency, we tested the effect of combining Pitstop2 with RNAi for members of other barrier pathways. In contrast to TGFβ signaling, we found that Pitstop2 can further enhance the iPSC generation efficiency of cells treated with UBE2E3i, RNF40i, ADAM29i, SLC17A5i, PTPN11i, ATF7IPi or TTF2i (Figure 5H). The interaction of Pitstop2 with the lysosomal protein SLC17A5 (also known as SIALIN) suggests that the lysosomal sub-pathway could have additional effects on reprogramming unrelated to TGFβ signaling. Remarkably, when ADAM29i, ATF7IPi and Pitstop2 are combined together, reprogramming efficiency can be elevated by up to 15 fold (Figure 5H). These data indicate that endocytosis can act in parallel with other barriers, including the ubiquitination and ADAM pathways, to antagonize reprogramming.
Identification of Feed-Forward Loops Between Reprogramming Barriers
To further explore potential interactions between validated reprogramming barriers, we searched the literature for experiments where one of the barrier genes significantly changed at transcriptional level (t-test p-value < 0.05) after RNAi against a second barrier gene. Using GeneMANIA, we also aggregated evidence of tissue-specific co-expression of barrier genes, and identified common barrier-gene protein domains. In the resulting barrier gene interaction network (Figure 6A thin edges), RNF40 stands out as a hub (i.e., it has both the highest “betweenness” and “closeness” centralities). We verified these interactions in human BJ fibroblasts and found that RNF40i induces both positive and negative effects on the expression of other barrier genes. RNF40i leads to significant increases in the expression of ADAM29, PTPN11, TTF2, TMF1, and MED19. RNF40i also induces a significant down-regulation of SLC17A5, PTPRJ, and CENPB (Figure 6A thick arrows, B). Transcriptional effects of RNF40i likely result from its role in regulating the levels of transcription factors such as OCT4 (Figure S3E) and in ubiquitinating H2B, a histone mark involved in transcriptional elongation and stem cell differentiation (Karpiuk et al., 2012). Thus, inhibition of a reprogramming barrier can alter the expression of other barriers in unexpected ways. We next tested the effect of combining RNF40i with RNAi for other barriers with which RFN40 interacts at the expression level (Figure 6B). RNF40i together with PTPN11i, MED19i, SLC17A5i or PTPRJi produces combinatorial effects on reprogramming efficiency (Figure 6C). These data suggest that different barrier pathways interact in dynamic feed-forward loops (FFLs) that can be classified as either coherent or incoherent and are thought to provide noise filtering and response delay capabilities to stabilize cell types (Mangan and Alon, 2003). As examples of coherent FFLs with an OR-gate integration, RNF40 positively regulates the other barrier genes SLC17A5 and PTPRJ. This kind of network motif can stabilize cells against transient fluctuating signals and can delay the response to removing the source repressor, which corresponds to RNF40 in our case. Thus, even though knockdown of RNF40 attenuates the second barrier’s expression, the residual amount of the second barrier can still partially suppress reprogramming without RNF40; as a result, simultaneous RNAi against the second barrier itself drastically relieves the suppression of reprogramming by the FFL as a whole (Figure 6D). Incoherent FFLs with an OR-gate integration has not been studied extensively to date, but we found that such network motifs can also inhibit reprogramming. For example, RNF40 represses the other barrier genes PTPN11 and MED19, so that knockdown of RNF40 alone aberrantly up-regulates PTPN11 and MED19, acting as a built-in compensatory suppression mechanism against reprogramming. In this case, a persistent knockdown of RNF40 may only transiently lower the barrier against reprogramming, because the subsequent derepression of PTPN11 and MED19 can help raise back the barrier. However, the dampening of the increase in reprogramming efficiency that could result from this derepression of a second barrier can be relieved via knockdown of the second gene (Figure 6D). The FFLs with OR-gate integration thus provide partial redundancy in cell fate control, and removing the redundancy with multiple RNAi manifests itself in the observed combinatorial knockdown effect (Figure 6C). Barriers can of course interact in multiple ways, not just at the transcriptional level. Nevertheless, the FFLs identified here contribute to revealing some of the complex interactions between barrier pathways. Taken together, our genetic interaction studies reveal that barrier pathways can act in linear, parallel (Figure 5) or feed-forward loop (Figure 6) architectures to antagonize reprogramming, and show that iPSC generation efficiency can be greatly increased by simultaneous targeting of different reprogramming barriers.
Figure 6. Genetic Interactions Between RNA40 and Other Reprogramming Barriers.
(A) In this barrier-gene network, non-directional interactions (colored edges) were identified via a literature search, and genetic interactions were inferred from knockdown experiments. We consider gene A to interact with gene B if B’s expression changes significantly (t-test p-value < 0.05) after the knockdown of A (red edges). Genes sharing common protein domains (blue edges), and genes which are co-expressed in the same tissue type (purple edges), were aggregated via GeneMANIA. Directional interactions (thick black edges) were identified in human fibroblasts from knockdown experiments shown in (B). (B) RNF40i affects the expression of other barrier genes. qRT-PCR was performed in BJ fibroblasts infected with non-sense (NS) shRNA control or shRNAs against RNF40. (C) The net effects on reprogramming efficiency of simultaneous knockdown of RNA40 and each of the other barrier-genes shown exceed the effect of knocking down either gene alone. (D) RNF40 forms coherent and incoherent feed-forward loops in the barrier-gene network.
PERSPECTIVE
We use an ultracomplex EXPANDed shRNA library to report the application of a genome-wide screen to the process of human cellular reprogramming to pluripotency. Our integrative study reveals multiple pathways as reprogramming barriers, including ubiquitination, endocytosis, cell adhesion, interaction with extracellular matrix, dephosphorylation, and transcription. Moreover, we found that different barrier pathways interact in complex combinatorial ways to antagonize reprogramming to the iPSC state. The data are implemented as a fully searchable integrative online resource to facilitate other studies of reprogramming.
Our genome-wide RNAi screen provides a systematic approach to interrogate iPSC generation. Several key aspects underlie the successful identification of the reprogramming barriers reported here: 1) the high (30 shRNAs) per-gene coverage of the library reduces off-target effects in hit selection; 2) using NGS for screen readout provides a large dynamic range; 3) a multi-objective optimization algorithm with greater sensitivity and specificity, and less prone to off-target effects than existing methods, helps reduce false positive hits; 4) systems-level analyses combining large-scale integration of public data with independent functional validation assays elucidate functional pathways. Benefiting from these aspects, our screen has a scale of 600,000 shRNAs tested and a hit validation rate of 87%, whereas previous genome-wide shRNA/siRNA screens tend to have lower scales (average <40,000) and average <50% success rates (Table S2).
More generally, our work provides a model for combining a genome-wide screen with large-scale data mining in contexts other than iPSC generation. Our preliminary findings indicate that some of the pathways identified may represent barriers to the induction of stem-like phenotypes in other cellular and disease processes. We applied our de-convolution method to a genome-wide screen for barriers against the proliferation of ovarian carcinoma stem cells (Tan et al., 2013). Cross-referencing our iPSC screen hits with the hits in carcinoma stem cells found 35 overlapping genes at the 5% significance level, and DAVID functional analysis identified transcription factors, cell membrane proteins, and ubiquitin pathway genes to be enriched in these hits (Figure S6). Of note, RNF40 was detected in both screens. Also enriched in both screens is arginine methyltransferase PRMT1, previously discussed as part of the FBXW7 interaction network (Figure 2B). Thus, our platform provides an integrative approach for identifying pathways that may act as barriers beyond the setting of reprogramming to pluripotency, including in cancer. We further anticipate that this approach will be useful in the dissection of direct lineage reprogramming and may reveal shared and unique aspects of different reprogramming paradigms.
EXPERIMENTAL PROCEDURES
Pooled shRNA Libraries
The design method for generating ultracomplex EXPANDed shRNA libraries targeting the human genome has been previously described (Bassik et al., 2009; 2013). In brief, the library contains approximately 30 independent shRNAs per gene for all human protein-coding genes (19,527 in total), for a total of ~600,000 shRNAs, hence the term “ultracomplex”. The full list of shRNA sequences present in the library is available upon request. Pooled sequences coding for the shRNAs were cloned downstream of a U6 promoter in a modified pSicoR lentiviral vector containing a CMV-Puro-T2A-mCherry cassette. All vectors were pooled to generate one lentiviral library representing the 600,000 shRNAs. The entire pooled library was then used to generate lentiviruses at the UCSF ViraCore.
RNAi Screen in Human iPSC Generation
Human primary newborn foreskin (BJ) fibroblasts were seeded at 630,000 cells per 15cm-dish the day before infection. Sixteen 15-cm dishes were used in total for the whole library screen, to represent 50 cells per shRNA. Cells were infected with concentrated retroviruses (Harvard Gene Therapy Initiative) leading to the over-expression of OCT4, SOX2 and KLF4 (MOI=10) and c-MYC (MOI=1) (4F), in combination with a lentivirus expressing an shRNA against p53 (MOI=1) (Zhao et al., 2008) and the lentiviral shRNA library (MOI=3). On d28 after infection, cells were trypsinized, live TRA1-81 (MAB4381, Millipore) staining was performed, and TRA-1-81+ and TRA-1-81-populations were isolated by FACS (FACSAria II, BD).
Bioinformatic Analyses
TRA-1-81+/TRA-1-81-differential shRNA effect size was estimated by read-count odds-ratio. shRNA effect sizes were summarized gene wise using a random effects model. Gene ranking was performed using multi-objective optimization, with collective shRNA effect size and the number of distinct shRNA as criterion. Significance was assessed using a permutation test, and library swap was used to control for false discovery in multiple hypothesis testing (Extended Experimental Procedures). Functional annotations were analyzed via DAVID (Huang et al., 2009) and gene interaction networks constructed via GeneMANIA (Zuberi et al., 2013) and Cytoscape. Betweenness centrality is defined as the number of shortest-paths through the network that pass through a given node. Closeness centrality is defined as the inverse of the sum of all shortest-path distances, from a given node, to all other nodes in the network.
Single-Gene Validation
For gene knockdown, the 2 most enriched shRNAs against each gene from the screen data were cloned into the lentiviral vector pSicoR downstream of a U6 promoter. Each shRNA was cloned into a separate pSicoR-CMV-Puro-T2A-GFP lentiviral vector. Viruses inducing the expression of 2 shRNAs against the same gene were combined together with 4F before infecting cells, unless otherwise indicated. Table S3 includes the target sequences of all shRNAs used for follow-up experiments. For gene over-expression, cDNA was cloned into the lentiviral vector pGAMA downstream of an EF1α promoter. Mutations were generated using QuickChange (Agilent) according to the manufacturer’s instructions. On d20-d28 after infection, live Tra1-81 (MAB4381, Millipore) staining was performed, and iPSC colonies were counted. All reprogramming experiments were performed in triplicates, and error bars represent standard deviation. *, p<0.05; **, p<0.01; ***, p<0.001.
Supplementary Material
HIGHLIGHTS.
Genome-Wide RNAi Screen Reveals Known and Novel Barriers to Human Cell Reprogramming
ADAM29 Antagonizes Reprogramming via Its Disintegrin Domain
Clathrin-Mediated Endocytosis Antagonizes Reprogramming by Enhancing TGFβ Signaling
Different Barrier Pathways Have a Combined Effect on Reprogramming Efficiency
ACKNOWLEDGEMENTS
We are grateful to Robert Blelloch, Keith Mostov and Zena Werb for critical reading of the manuscript, Jonathan Weissman, Martin Kampann and Michael Bassick for discussions and help with library generation, Nikki Shariat for input on the experiment design, Xiaoyin Wang for virus production, Fong Ming Koh for providing the pGAMA vector, Charles Nicolet and Henriette O’Geen for support on NGS, and Tara Rambaldo and Michael Kissner for support with FACS. This work was supported by NWO Rubicon grant 825.06.030 and Veni grant 916.10.138 to R.J.L., the UCSF Program for Breakthrough Biomedical Research and NIH grants 1U01CA168370 and R01 GM80783 to M.T.M., NIH R01CA163336 and Sontag Foundation Distinguished Scientist Award to J.S.S., and CIRM grant RB4-06028 and NIH Director’s New Innovator Award DP2OD4698 to M.R-S. E.M.L. is an employee of Agilent Technologies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
M.R.-S. and J.S.S. directed the project. H. Q. and M.R-S. developed the experimental design with participation from R.J.L. and M.T.M. R.J.L., E.M.L. and M.T.M. developed and produced the RNAi libraries. H.Q., L.B. and P.T. carried out the bench experiments, and A.D. and J.S.S. developed and carried out the statistical and bioinformatic analyses, with initial input from W.P. H.Q., A.D., J.S.S. and M.R.-S. wrote the manuscript. The authors declare no competing financial interests.
REFERENCES
- Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, et al. A Systematic Mammalian Genetic Interaction Map Reveals Pathways Underlying Ricin Susceptibility. Cell. 2013:1–14. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassik MC, Lebbink RJ, Churchman LS, Ingolia NT, Patena W, LeProust EM, Schuldiner M, Weissman JS, McManus MT. Rapid creation and quantitative monitoring of high coverage shRNA libraries. Nature Methods. 2009;6:443–445. doi: 10.1038/nmeth.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, et al. Regulation of Pluripotency and Cellular Reprogramming by the Ubiquitin-Proteasome System. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-Cell Expression Analyses during Cellular Reprogramming Reveal an Early Stochastic and a Late Hierarchic Phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvete JJ, Marcinkiewicz C, Monleón D, Esteve V, Celda B, Juárez P, Sanz L. Snake venom disintegrins: evolution of structure and function. Toxicon. 2005;45:1063–1074. doi: 10.1016/j.toxicon.2005.02.024. [DOI] [PubMed] [Google Scholar]
- Chen J, Liu H, Liu J, Qi J, Wei B, Yang J, Liang H, Chen Y, Chen J, Wu Y, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nature Genetics. 2013;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- Chia N-Y, Chan Y-S, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau M-S, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Daley GQ. Common themes of dedifferentiation in somatic cell reprogramming and cancer. Cold Spring Harbor Symposia on Quantitative Biology. 2008;73:171–174. doi: 10.1101/sqb.2008.73.041. [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Golipour A, David L, Liu Y, Jayakumaran G, Hirsch CL, Trcka D, Wrana JL. A Late Transition in Somatic Cell Reprogramming Requires Regulators Distinct from the Pluripotency etwork. Stem Cell. 2012;11:769–782. doi: 10.1016/j.stem.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biology. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, ELSDALE TR, FISCHBERG M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182:64–65. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- Handl J, Kell DB, Knowles J. Multiobjective optimization in bioinformatics and computational biology. IEEE/ACM Trans Comput Biol Bioinform. 2007;4:279–292. doi: 10.1109/TCBB.2007.070203. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi T, Araki S, Mori H, Takeda S. Crystal structures of catrocollastatin/VAP2B reveal a dynamic, modular architecture of ADAM/adamalysin/reprolysin family proteins. FEBS Lett. 2007;581:2416–2422. doi: 10.1016/j.febslet.2007.04.057. [DOI] [PubMed] [Google Scholar]
- International Stem Cell Initiative. Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nature Biotechnology. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Karpiuk O, Najafova Z, Kramer F, Hennion M, Galonska C, König A, Snaidero N, Vogel T, Shchebet A, Begus-Nahrmann Y, et al. The histone H2B monoubiquitination regulatory pathway is required for differentiation of multipotent stem cells. Molecular Cell. 2012;46:705–713. doi: 10.1016/j.molcel.2012.05.022. [DOI] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya Á, Wahl GM, Izpisua-Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleist von, L., Stahlschmidt W, Bulut H, Gromova K, Puchkov D, Robertson MJ, MacGregor KA, Tomilin N, Pechstein A, Chau N, et al. Role of the Clathrin Terminal Domain in Regulating Coated Pit Dynamics Revealed by Small Molecule Inhibition. Cell. 2011;146:471–484. doi: 10.1016/j.cell.2011.06.025. [DOI] [PubMed] [Google Scholar]
- König R, Chiang C-Y, Tu BP, Yan SF, DeJesus PD, Romero A, Bergauer T, Orth A, Krueger U, Zhou Y, et al. A probability-based approach for the analysis of large-scale RNAi screens. Nature Methods. 2007;4:847–849. doi: 10.1038/nmeth1089. [DOI] [PubMed] [Google Scholar]
- Li Z, Yang C-S, Nakashima K, Rana TM. Small RNA-mediated regulation of iPSC generation. The EMBO Journal. 2011;30:823–834. doi: 10.1038/emboj.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, et al. Highly parallel identification of essential genes in cancer cells. Proceedings of the National Academy of Sciences. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. U.S.a. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, Qi Z, Downey SL, Manos PD, Rossi DJ, et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPSCs. Nature Cell Biology. 2011;13:1–11. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Mancarci OB, Unternaehrer J, Gupta PB, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persaud A, Alberts P, Hayes M, Guettler S, Clarke I, Sicheri F, Dirks P, Ciruna B, Rotin D. Nedd4-1 binds and ubiquitylates activated FGFR1 to control its endocytosis and function. The EMBO Journal. 2011;30:3259–3273. doi: 10.1038/emboj.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A Molecular Roadmap of Reprogramming Somatic Cells into iPSCs. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Blaschke K, Wei G, Ohi Y, Blouin L, Qi Z, Yu J, Yeh RF, Hebrok M, Ramalho-Santos M. Transcriptional analysis of pluripotency reveals the Hippo pathway as a barrier to reprogramming. Human Molecular Genetics. 2012;21:2054–2067. doi: 10.1093/hmg/dds023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Santos M. iPSCs: insights into basic biology. Cell. 2009;138:616–618. doi: 10.1016/j.cell.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin S, Domogatskaya A, Ström S, Hansson EM, Chien KR, Inzunza J, Hovatta O, Tryggvason K. long-term self-renewal of human pluripotent stem cells on human recombinant laminin-511. Nature Biotechnology. 2010;28:611–615. doi: 10.1038/nbt.1620. [DOI] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung H-K, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional Genomics Reveals a BMP-Driven Mesenchymal-to-Epithelial Transitionin the Initiation of Somatic Cell Reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Scita G, Di Fiore PP. The endocytic matrix. Nature. 2010;463:464–473. doi: 10.1038/nature08910. [DOI] [PubMed] [Google Scholar]
- Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nature Biotechnology. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. The EMBO Journal. 1997;16:6510–6520. doi: 10.1093/emboj/16.21.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tan TZ, Miow QH, Huang RY-J, Wong MK, Ye J, Lau JA, Wu MC, Bin Abdul Hadi LH, Soong R, Choolani M, et al. Functional genomics identifies five distinct molecular subtypes with clinical relevance and pathways for growth control in epithelial ovarian cancer. EMBO Mol Med. 2013;5:1051–1066. doi: 10.1002/emmm.201201823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Moncada-Pazos A, Cal S, Soria-Valles C, Gartner J, Rudloff U, Lin JC, NISC Comparative Sequencing Program. Rosenberg SA, López-Otín C, et al. Analysis of the disintegrin-metalloproteinases family reveals ADAM29 and ADAM7 are often mutated in melanoma. Hum. Mutat. 2011;32:E2148–E2175. doi: 10.1002/humu.21477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, Zhang Q, Xiang C, Hou P, Song Z, et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Zuberi K, Franz M, Rodriguez H, Montojo J, Lopes CT, Bader GD, Morris Q. GeneMANIA prediction server 2013 update. Nucleic Acids Research. 2013;41:W115–W122. doi: 10.1093/nar/gkt533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.