Abstract
The checkpoint between the life and death of macrophages is crucial for the host's frontline immune defense during acute phase infection. However, the mechanism as to how the immune cell equilibrates between apoptosis and immune response is unclear. Using in vitro and ex vivo approaches, we showed that macrophage survival is synchronized by SAG (sensitive to apoptosis gene), which is a key member of the ubiquitin–proteasome system (UPS). When challenged by pathogen-associated molecular patterns (PAMPs), we observed a reciprocal expression profile of pro- and antiapoptotic factors in macrophages. However, SAG knockdown disrupted this balance. Further analysis revealed that ubiquitination of Bax and SARM (sterile α- and HEAT/armadillo-motif-containing protein) by SAG-UPS confers survival advantage to infected macrophages. SAG knockdown caused the accumulation of proapoptotic Bax and SARM, imbalance of Bcl-2/Bax in the mitochondria, induction of cytosolic cytochrome c and activation of caspase-9 and -3, all of which led to disequilibrium between life and death of macrophages. In contrast, SAG-overexpressing macrophages challenged with PAMPs exhibited upregulation of protumorigenic cytokines (IL-1β, IL-6 and TNF-α), and downregulation of antitumorigenic cytokine (IL-12p40) and anti-inflammatory cytokine (IL-10). This suggests that SAG-dependent UPS is a key switch between immune defense and apoptosis or immune overactivation and tumorigenesis. Altogether, our results indicate that SAG-UPS facilitates a timely and appropriate level of immune response, prompting future development of potential immunomodulators of SAG-UPS.
In an infection, the pattern recognition receptors (PRRs) of the macrophages recognize pathogen pattern-associated molecular patterns (PAMPs), leading to phagocytosis of the pathogen, release of cytokines and secretion of antimicrobial peptides. When overwhelmed by pathogens, macrophages may undergo apoptosis, which produces microbicidal reactive oxygen species.1 Apoptotic death of macrophages is a strategic sacrifice, representing a severe terminal stage of cellular defense against microbial invasion.
The mitochondria has a decisive role in cell death or survival by controlling apoptosis signals via recruitment of pro- and antiapoptosis factors.2 Although it is known that many pathogens regulate apoptosis in the host,3 the mechanisms underlying how the host immune cell equilibrates its own death and survival to elicit an optimal immune response is poorly understood. This prompted us to investigate how signaling proteins might regulate the checkpoint between apoptosis or immune response. In this regard, we noted several lines of evidence indicating a hitherto undiscovered phenomenon on the control of host cell death or survival versus immune defense. Firstly, it is proposed that the ubiquitin–proteasome system (UPS)-mediated degradation of Bcl-2 family proteins regulates apoptotic cell death.4 Secondly, the sensitive to apoptosis gene (SAG), a key component of UPS, is strongly induced during early infection,5 suggesting its role in frontline defense. Thirdly, SARM (sterile α- and HEAT/armadillo-motif-containing protein), an evolutionarily conserved mitochondria-associated protein,6, 7 which downregulates TLR-TRIF signaling,8 exerts a strong proapoptotic killing of infection-activated T cells during the pathogen-clearance phase.9 As SAG confers survival to cancer cells,10, 11 we hypothesize that SAG and SARM have opposing roles in modulating apoptosis and immune response. Thus, we investigated the dynamic expression profiles of SAG and Bcl-2 (antiapoptotic) and Bax and SARM (proapoptotic) in macrophages challenged with bacterial and viral PAMPs.
We demonstrated that SAG responds dynamically to PAMP stimulation. SAG knockdown abrogates ubiquitination and stabilizes the proapoptotic Bax and SARM proteins, leading to their accumulation in the mitochondria and resulting in intrinsic apoptosis. SAG overexpression in macrophages downregulated the antitumorigenic cytokine (IL-12p40) and anti-inflammatory cytokine (IL-10), but upregulated the protumorigenic cytokines (IL-1β, IL-6 and TNF-α), indicating cellular sensitization to SAG activation. Taken together, we propose that the crossroad between macrophage survival/death and immune response is synchronized to a large extent by SAG-UPS.
Results
SAG is upregulated in macrophages during early infection
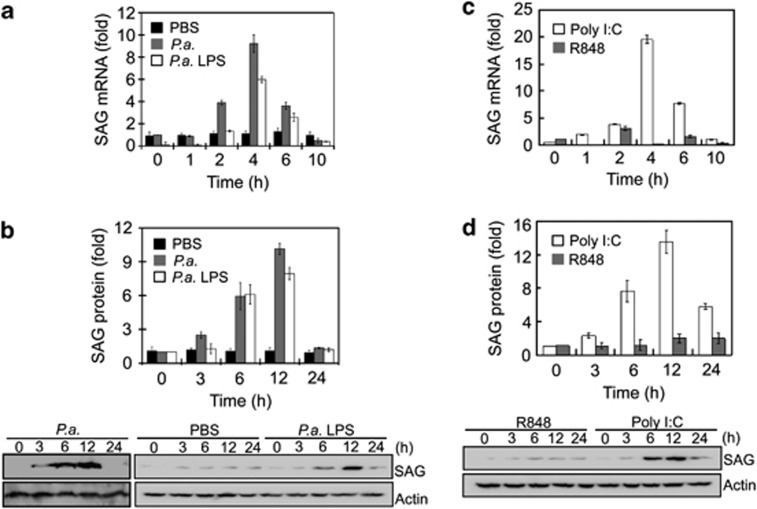
Our previous study with Limulus (horseshoe crab), an invertebrate species, which harbors a very powerful innate immune system, showed that SAG transcription was induced as early as 3 h after P. aeruginosa infection.5 Since heat-inactivated P. aeruginosa is known to induce immune response in mice,12 we used it to determine whether mammalian SAG is triggered in a pseudomonas infection. Macrophage J774 cells challenged with 1 × 106 CFU/ml of heat-inactivated P. aeruginosa showed a 10-fold rise in both the SAG mRNA within 4 h (Figure 1a) and SAG protein level at 12 h (Figure 1b). Since P. aeruginosa LPS was sufficient to trigger an inflammatory response, we further studied its effect on SAG expression in a dose-responsive manner in macrophages. SAG protein was upregulated by 5–9-fold at 12 h (Figure 1b and Supplementary Figure S1A). The profiles of SAG mRNA and protein expression were reproducible with Escherichia coli LPS (Supplementary Figure S1B).
Figure 1.
SAG expression is increased in J774 cells at the early stage of challenge with bacterial/viral PAMPs. (a and b) Cells were treated with heat-inactivated P. aeruginosa (P.a., PAO-Iglewski, 1 × 106 CFU/ml), P. aeruginosa LPS (P.a. LPS, 10 ng/ml) or phosphate-buffered saline (PBS). (c and d) Cells were treated with viral mimics; polyinosinic-polycytidylic acid (poly I:C) (10 μg/ml) and R848 (25 ng/ml), representing double-stranded RNA (dsRNA) and single-stranded RNA (ssRNA), respectively. (a and c) mRNA and (b and d) protein levels of SAG were induced upon treatment by P.a. LPS and poly (I:C), but not by R848. Data are presented as means±S.D. (n=3)
To further understand how SAG is involved in the signaling pathway(s), we tested its expression profile under stimulation by (i) poly I:C, a dsRNA analog, which triggers the TRIF pathway and (ii) R848, an ssRNA analog, which signals via MyD88. We found that poly I:C (10 μg/ml) induced SAG mRNA and protein by 20- and 14-fold, respectively, at 4 and 12 h, whereas R848 (25 ng/ml) showed minimal effect throughout the challenge (Figures 1c and d). As SAG activity appears unresponsive to R848, henceforth, we challenged macrophages with LPS and poly I:C.
The early induction of SAG by P. aeruginosa, LPS and poly I:C prompted us to hypothesize that SAG exerts its antiapoptotic activity at the initial phase of infection, to facilitate host innate immune defense rather than early death of the immune-responsive cell.
SAG-knockdown macrophages undergo intrinsic apoptosis during PAMP challenge
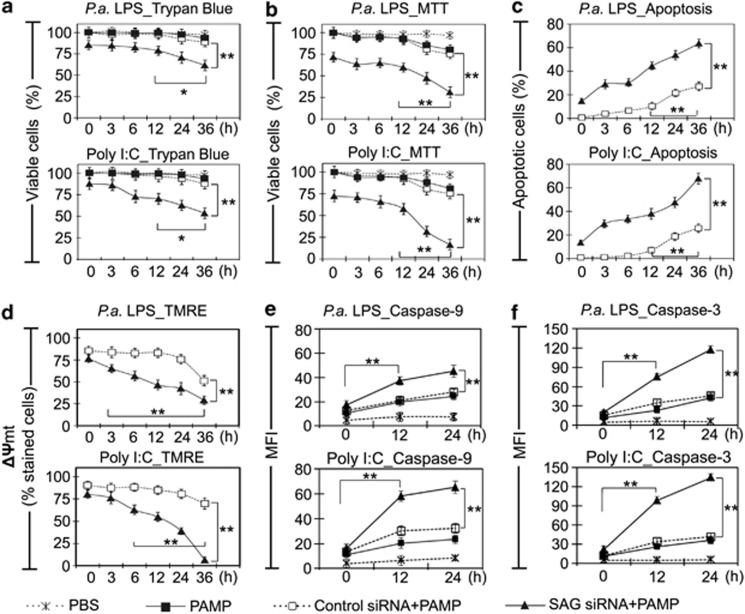
To investigate how SAG mediates death or survival of infected macrophages, SAG-specific RNAi was conducted in J774 cells, achieving 84% knockdown of SAG (Supplementary Figure S2). Cells treated with scrambled siRNA (control) or SAG-specific siRNA were challenged with LPS (10 ng/ml) or poly I:C (10 μg/ml) over a time course. The cell viability was evaluated using trypan blue dye exclusion and MTT assays. In SAG-expressing control macrophages, a significant reduction of 20% cell viability was indicated by MTT assay (Figures 2a and b). This is plausibly important for anti-microbial defense, albeit a mitochondrial respite. In contrast, SAG-knockdown macrophages treated with LPS and poly I:C exhibited significant (P<0.01) decreases in cell viability by 69.1% and 83.7%, respectively, at 36 h (Figure 2b). FACS analysis showed that SAG-knockdown cells challenged with LPS or poly I:C started to exhibit early apoptosis within 3 h, compared with control siRNA-treated cells, which only begun to show apoptosis after 12 h (Figure 2c). Control wild-type J774 cells (not treated with SAG-siRNA or scramble siRNA) challenged with PAMPs showed specific contribution of SAG to apoptosis of macrophages upon infection (Supplementary Figure S3A). SAG-knockdown cells without PAMP challenge showed minimal apoptosis, suggesting that the increased apoptosis of SAG-knockdown cells is specifically induced by the infection condition (Supplementary Figure S3A).
Figure 2.
SAG knockdown reduces cell viability and upregulates intrinsic apoptosis in infected macrophages. (a) Trypan blue exclusion test and (b) MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay demonstrate that knockdown of SAG resulted in loss of cell viability. (c) Annexin V and 7-aminoactinomycin (7-AAD) double staining was conducted to examine the early apoptosis. Representative histograms of apoptosis assay are shown in Supplementary Figure S3A. PAMP-only controls and no-PAMP controls (only SAG small interfering RNA (siRNA) treatment) are shown in Supplementary Figure S3A. (d) TMRE assay show mitochondrial dysfunction when J774 cells were challenged with LPS (10 ng/ml) or polyinosinic-polycytidylic acid (poly I:C) (10 μg/ml). Quantitative results show % of cells stained with TMRE. No PAMP controls (only SAG siRNA or PBS) and fluorescence-activated cell sorting (FACS) histograms are shown in Supplementary Figure S3B. (e and f) Caspase-9 and -3 activities in cells exposed to PAMPs for 0, 12 and 24 h were determined based on the hydrolysis of caspase-specific substrate (see Materials and Methods). Representative histograms of caspase assay are shown in Supplementary Figure S3C. Results are expressed as mean fluorescence intensity (MFI)±S.D. (n=3). *P<0.05; **P<0.01
Next, we analyzed the mitochondrial membrane potential (ΔΨmt) of infected J774 cells. The formation of mitochondrial permeability transition pore (MPTP), which indicates the loss of ΔΨmt, was monitored by the mitochondrial membrane potential-driven uptake of fluorescent dye, tetramethylrhodamine ethyl ester (TMRE). SAG-knockdown cells or control siRNA-treated cells were challenged with LPS (10 ng/ml) or poly I:C (10 μg/ml) over time, followed by TMRE staining and FACS analysis. Supplementary Figure S3B shows a clear separation in fluorescence signal between unstained and TMRE-stained macrophages. Control siRNA-treated cells challenged with PAMPs induced up to 50% mitochondria-mediated cell death at 36 h. On the contrary, SAG-knockdown cells showed a significant drop in ΔΨmt at 36 h (70% with LPS treatment and 93% with poly I:C treatment) (P<0.01) (Figure 2d). Furthermore, SAG knockdown elevated the activation of caspase-9 and -3 (Figures 2e and f). Representative histograms of FACS analysis are shown in Supplementary Figure S3C. Taken together, both the loss in ΔΨmt and the activation and cleavage of caspase-9 corroborate the importance of SAG in conferring mitochondria-specific antiapoptosis to macrophages during early infection.
The ratio of pro- and antiapoptotic factors is regulated by SAG, suggesting a kinetic switch between macrophage death/survival in response to infection
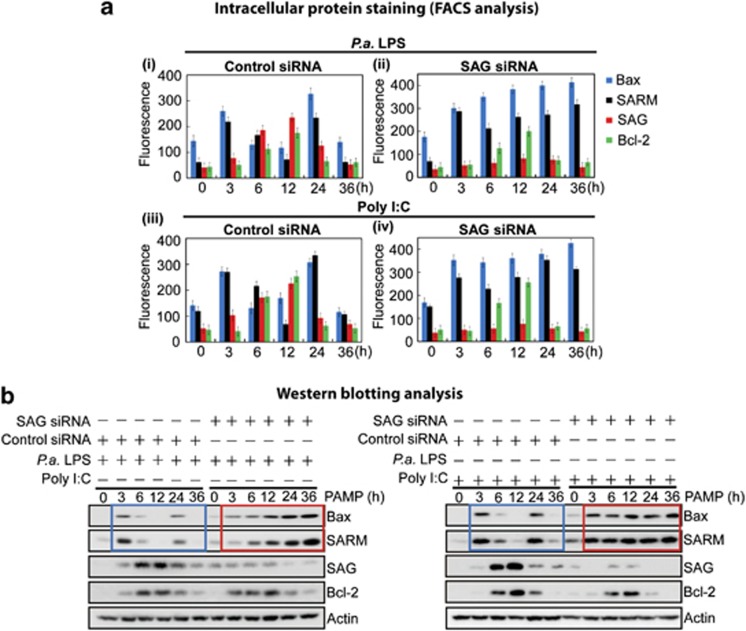
To identify critical regulator(s) of SAG-mediated survival of macrophages during early infection, we examined transcription profiles of selected apoptotic factors, including Bcl-2, Bax, SARM, Noxa and Bcl-xL, based on the literature.4, 6, 8, 9, 10 Supplementary Figures S4A and S4B show that neither Noxa (proapoptotic) nor Bcl-xL (antiapoptotic) responded to PAMP challenge, suggesting that these proteins may not directly participate in macrophage survival under the infection conditions studied. Subsequently, we focused on examining the profiles of antiapoptotic Bcl-2 and SAG and proapoptotic Bax and SARM to understand how SAG regulates macrophage survival during infection. Intracellular staining followed by FACS analysis of J774 cells that had been treated with LPS (10 ng/ml) or poly I:C (10 μg/ml) showed bimodal expression of the proapoptotic SARM and Bax as early as 3 h and again at 24 h. In contrast, both the antiapoptotic SAG and Bcl-2 appeared maximally at 12 h (Figure 3a (i and iii), left panels). SAG siRNA knockdown abolished these expression profiles (Figure 3a (ii and iv), right panels). Representative histograms of FACS analysis are shown in Supplementary Figure S5. These results are confirmed by immunoblot analysis (Figure 3b). The rate and timing of expression of the pro- and antiapoptotic proteins appear reciprocal. The balance is in favor of proapoptotic factors reaching a first peak as early as 3 h of infection and resurging at 24 h, which may explain how the infected cells switch to apoptosis beyond 24 h PAMP stimulation. SAG-knockdown cells maintained the expression of Bcl-2, but the balance of expression between the pro- and antiapoptosis factors was disrupted; from 3 h onwards, the expression of the proapoptotic Bax and SARM was sustained at high levels (Figure 3b, red box) instead of the bimodal peaks observed with control SAG-expressing cells (Figure 3b, blue box). This trend is consistent for both LPS and poly (I:C) treatments, suggesting that SAG may be a dynamic master regulator of macrophage death/survival during infection, without which apoptosis ensues.
Figure 3.
SAG regulates apoptosis of J774 macrophages by modulating the balance between anti- and proapoptotic factors in a timely manner. LPS (10 ng/ml) and polyinosinic-polycytidylic acid (poly I:C) (10 μg/ml) stimulation resulted in bimodal peaks (at 3 and 24 h) of proapoptotic Bax and SARM, and a single peak (over 6–12 h) each of antiapoptotic SAG and Bcl-2 (a (i and iii) and b, blue box). SAG-knockdown cells maintained expression of Bcl-2 but sustained the expression of the proapoptotic Bax and SARM at high levels (over 3–36 h; a (ii and iv) and red box in b). The PAMP-challenged cells were examined by both (a) intracellular staining (see FACS analysis data; Supplementary Figure S5) and (b) immunoblotting analysis. Data are presented as fluorescence intensity ±S.D. (n=3)
While the bimodal profile of Bax and SARM mRNAs was maintained during infection of SAG-knockdown cells (Supplementary Figure S4A), we observed a sustained rise in the Bax and SARM proteins (Figure 3a (ii and iv) and Figure 3b, red box), suggesting that the accumulation of Bax and SARM proteins was attributable to a posttranscriptional effect. SAG is known to elicit E3 ubiquitin ligase activity in the ubiquitination of specific target proteins leading to their proteasomal degradation.11 Therefore, it is conceivable that the balance of pro- and antiapoptotic factors in macrophages under infection might be regulated via SAG-dependent ubiquitination pathway.
SAG-dependent UPS regulates proapoptotic Bax and SARM
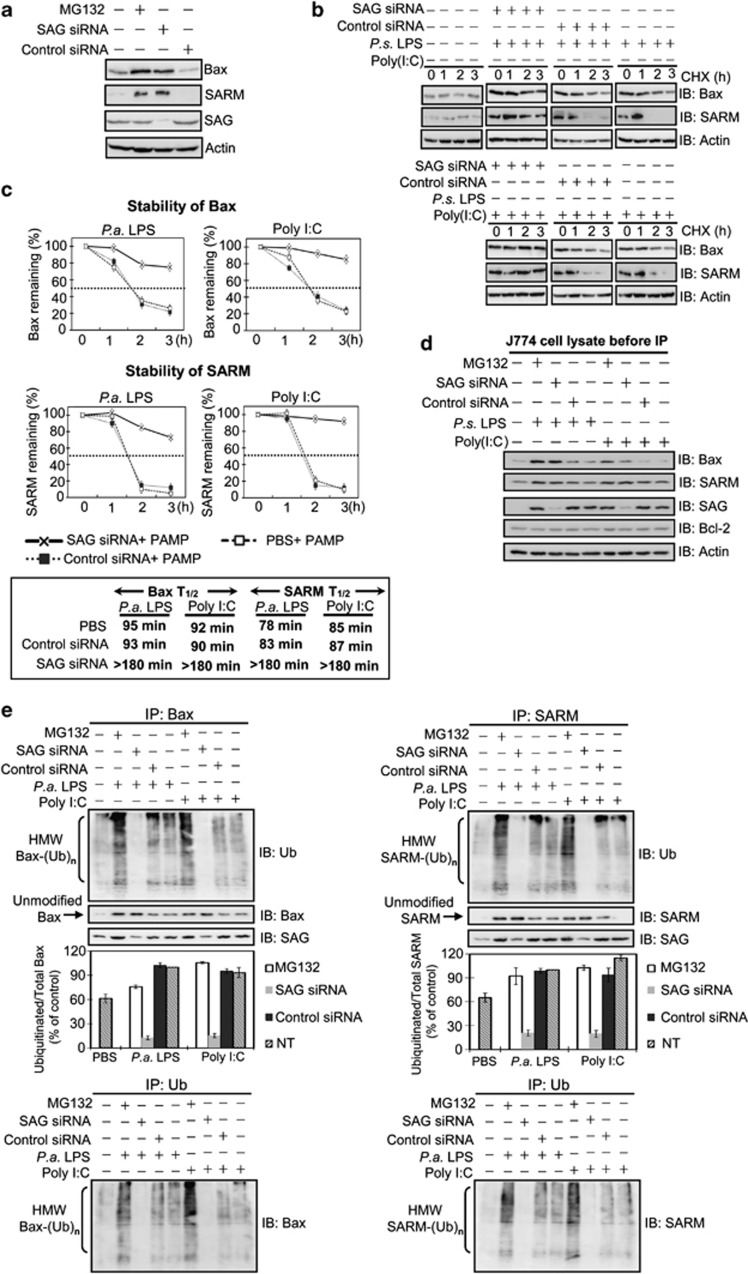
To test whether Bax and/or SARM may be a direct target of the UPS, a proteasome inhibitor, MG132, was administered into J774 cells. Figure 4a shows that Bax and SARM were accumulated under proteasome inhibition, indicating the involvement of UPS. In addition, SAG knockdown caused the accumulation of Bax and SARM proteins, suggesting that SAG regulates the UPS-mediated turnover of Bax and SARM. We then examined the degradation rate (half-life) of the proteins after blocking de novo protein synthesis with cycloheximide (CHX) while challenging the cells with LPS (10 ng/ml) or poly I:C (10 μg/ml). Results showed that SAG knockdown stabilized Bax and SARM proteins by up to 3 h of CHX treatment (Figure 4b). A twofold increase in the half-life of Bax and SARM was observed: from <100 min in control siRNA-treated cells to >180 min in SAG-knockdown cells (Figure 4c). Thus, the absence of the antiapoptotic SAG prolonged the lifespan of Bax and SARM proteins. Plausibly in an infection, SAG facilitates the degradation of proapoptotic Bax and SARM, to sustain cell survival.
Figure 4.
SAG knockdown blocks ubiquitination and degradation of Bax and SARM, which accumulate in PAMP-treated macrophage J774 cells. (a) Cells were treated with MG132 or SAG siRNA to determine the involvement of SAG–proteasome system in the regulation of proapoptotic Bax and SARM. (b) The stability of Bax and SARM was tested in J774 cells with CHX, a protein synthesis inhibitor, and the lifespan of the proteins was examined by western blotting. (c) The half-lives (T1/2) of Bax and SARM were quantified by densitometry analysis. (d) Western blot analysis of whole-cell lysates shows the presence of Bax, SARM, SAG and Bcl-2 in the J774 cells (before IP). Actin was used as a loading control. (e) To investigate the association between SAG and Bax (or SARM) in infected cells, total cell lysates were immunoprecipitated with anti-Bax (or anti-SARM) antibody, separated by 12% and 10% SDS-PAGE, respectively, immunoblotted and probed with SAG, ubiquitin, Bax or SARM antibodies. SAG-knockdown attenuated the high-molecular-weight (HMW) modified forms of Bax and SARM (but not that of Bcl-2; see Supplementary Figure S6A). Arrows indicate unmodified (un-ubiquitinated) Bax or SARM. A densitometric analysis of ubiquitinated Bax or SARM, relative to total Bax or SARM is plotted (mean ±S.D., n=3). To confirm that the observed ubiquitination is due to Bax (or SARM) proteins, IP with ubiquitin, followed by immunoblotting with Bax (or SARM) was performed. IP with control rabbit IgG demonstrates the specificity of the assay (Supplementary Figure S7A). IgG, immunoglobulin G; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis
If Bax and SARM are direct targets of SAG-dependent UPS, the inhibition of SAG activity should conceivably result in the attenuation of ubiquitinated forms of these proapoptotic proteins. To test this hypothesis, protein extracts of J774 cells, verified to contain Bax, SARM, SAG and Bcl-2 (Figure 4d), were treated with MG132 (or SAG siRNA or control siRNA). Then, the cell lysates were co-immunoprecipitated with Bax or SARM antibody, followed by immunodetection using anti-ubiquitin antibody. To confirm that observed ubiquitination was due to Bax (or SARM) proteins, the immunoprecipitation (IP) with ubiquitin followed by immunoblotting with Bax (or SARM) was performed. Co-IP shows that both Bax and SARM pulled down SAG (Figure 4e). Inhibition of proteasome increased the levels of ubiquitinated Bax and SARM. Consistently, we observed a decrease in the high-molecular-weight ubiquitinated Bax (HMW Bax-(Ub)n) and SARM (HMW SARM-(Ub)n) in SAG-silenced cells, compared with control siRNA-treated cells. Furthermore, Co-IP using Bcl-2 as bait showed that Bcl-2 ubiquitination was not regulated by SAG (Supplementary Figure S6). IP with control IgG demonstrated the specificity of the assay (Supplementary Figure S7). These data also corroborate that both Bax and SARM (but not Bcl-2) are specific substrates of SAG-dependent ubiquitination, which is enhanced under infection when the macrophage encounters an apoptosis stimulus. To show that our observations are not restricted to only one cell line, we performed key experiments in another macrophage cell line, RAW264.7. Results show consistent profile of SAG induction, cytochrome c release and ubiquitin activity upon PAMP stimulation (Supplementary Figure S8), suggesting that ubiquitination of apoptosis factors by SAG-UPS indeed regulates macrophage survival/death and its response to infection.
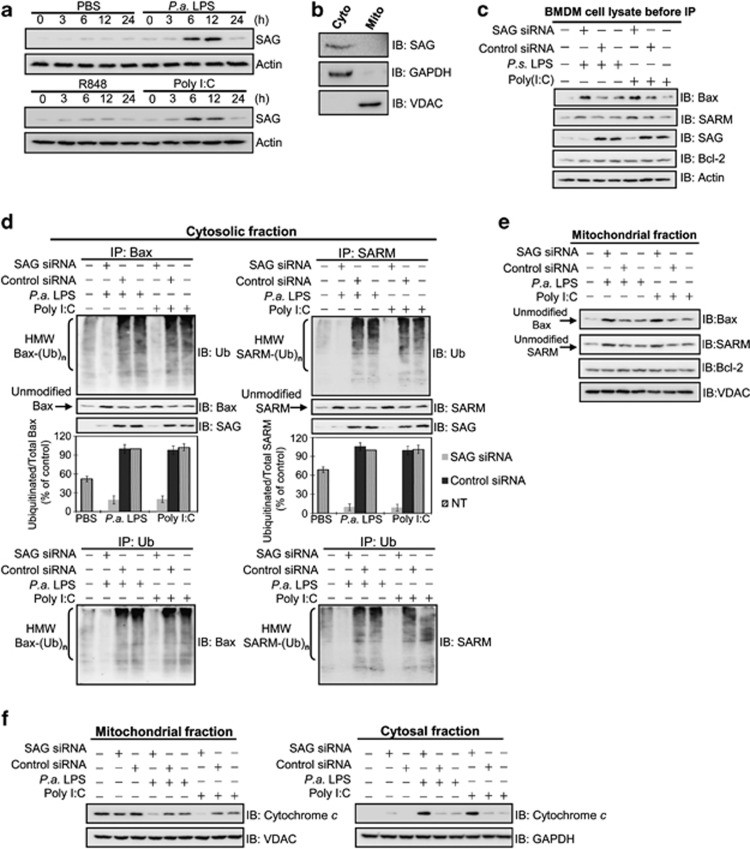
SAG hijacks proapoptotic cytoplasmic Bax and SARM for ubiquitination: observations in primary BMDM
To ensure that our findings in J774 and RAW cells are physiologically relevant, we performed ex vivo experiments using bone marrow-derived macrophages (BMDM) from BALB/c mice. Consistently, we found that SAG was upregulated in BMDM in response to LPS (10 ng/ml) and poly I:C (10 μg/ml), but not to R848 (Figure 5a). To further affirm that SAG mediates ubiquitination of Bax and SARM in primary cells, we examined their potential interactions and subcellular colocalization in BMDM challenged with PAMPs. As SAG is cytosolic (Figure 5b), we fractionated the cell lysate and used the cytosolic extract for Co-IP. Before IP experiments, immunoblotting analysis of whole-cell lysates shows the presence of Bax, SARM, SAG and Bcl-2 in primary BMDM cells (Figure 5c). Figure 5d shows that under either LPS (10 ng/ml) or poly I:C (10 μg/ml) challenge, there were interactions between both SAG-Bax and SAG-SARM in the cytosol. It is plausible that SAG binds the cytosolic Bax and SARM and directs these proapoptotic factors toward ubiquitination. Supportive of this speculation was that silencing SAG abrogated the ubiquitination activity, causing the accumulation of unubiquitinated (unmodified) Bax and SARM, but not Bcl-2, in the mitochondria (Figure 5e). To confirm the contribution by SAG in intrinsic apoptosis, we tested the release of cytochrome c by the infected BMDM. We found that SAG knockdown increased the cytosolic cytochrome c at the expense of mitochondrial cytochrome c (Figure 5f). Taken together, we have shown that infection causes SAG to interact with the cytosolic forms of proapoptotic Bax and SARM, hijacking them from the mitochondria and diverting these proteins toward ubiquitination and turnover. This maneuver exerts an influence on the balance of mitochondrial Bcl-2/Bax and subsequently sensitizes the macrophages to infection-induced intrinsic apoptosis.
Figure 5.
SAG knockdown blocks ubiquitination of Bax and SARM in the cytosol of primary BMDM cells, with increased cytochrome c release. (a) The induction of SAG in infected BMDM was immunodetected. (b) Cells were pretreated with , polyinosinic-polycytidylic acid (poly I:C) (10 μg/ml) for 12 h, followed by cellular fractionation and western blotting. SAG is primarily expressed in the cytosol. (c) Western blot analysis of whole-cell lysates shows the presence of Bax, SARM, SAG and Bcl-2 in primary BMDM cells (before IP). The role of SAG in the ubiquitination of proapoptotic Bax or SARM was determined by Co-IP of subcellular fractions. Cells were treated with SAG siRNA, small interfering RNA (siRNA) (or control siRNA) for 16 h, followed by PAMP stimulation for 24 h. (d) Cytosolic fraction and (e) mitochondrial fraction. Both SAG-Bax and SAG-SARM immunoprecipitates were found in the cytosolic fraction. A densitometric analysis of the ubiquitinated Bax and SARM relative to total Bax and SARM in the cytosol is provided. Data are representative of means±S.D. (n=3). To confirm that the observed ubiquitination is due to Bax (or SARM) proteins, the IP with ubiquitin, followed by immunoblotting with Bax (or SARM) was performed. IP with control rabbit IgG shows the specificity of the assay (Supplementary Figure S7B). (f) Cytochrome c release was examined to determine the levels of intrinsic apoptosis, which is increased in SAG-knockdown BMDM.
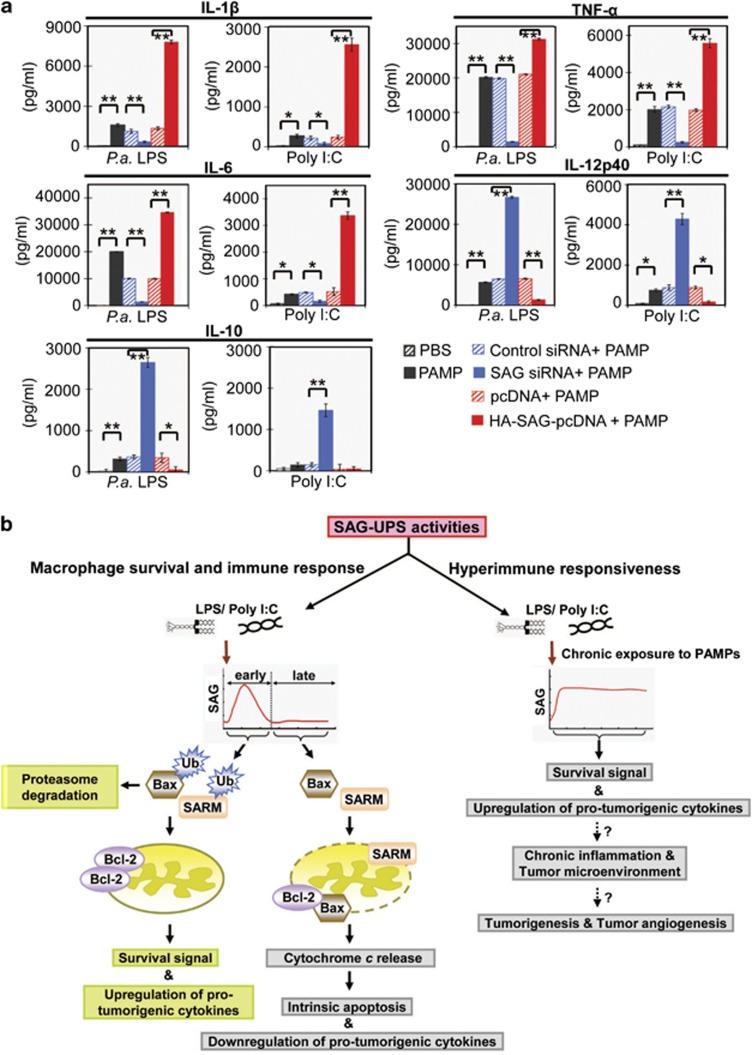
SAG enhances production of protumorigenic cytokines in primary BMDM
Upon infection, the key functions of macrophages are to release cytokines and attract secondary defense cells from the blood. Proinflammatory cytokines are key mediators of inflammatory responses.13 Figure 6a shows that BMDM cells that overexpress SAG significantly increased (P<0.01) the production of proinflammatory/protumorigenic cytokines (IL-1β, IL-6 and TNF-α), whereas SAG knockdown significantly reduced (P<0.01) the production of these cytokines. Concordantly, SAG overexpression downregulated IL-12p40, a proinflammatory/antitumorigenic cytokine.14, 15, 16 Thus, SAG appears to have a dual role in modulating both tumor growth and immune cell survival. We further measured IL-10, an anti-inflammatory cytokine, known to be regulated by LPS.17, 18, 19 Figure 6a shows that SAG-knockdown cells challenged with LPS produced high levels (P<0.01) of IL-10, whereas SAG overexpression significantly reduced (P<0.05) the production of IL-10. Thus in infected macrophages, SAG seems to regulate negatively both proinflammatory IL-12p40 and anti-inflammatory IL-10. Therefore, induction of the proinflammatory cytokines (IL-1, IL-6 and TNF-α) is unlikely attributable to a general increase in the survival advantage resulting from the overexpression of SAG. Rather, there appears to be a tight control of apoptosis in macrophages at early infection, during which SAG-UPS is induced to perform an intricate balance between apoptosis and immune response. Figure 6b depicts a model to illustrate the potential role of SAG-UPS linking immune defense and apoptosis or immune overactivation and tumorigenesis.
Figure 6.
SAG-UPS upregulates protumorigenic cytokines, balances apoptosis and infection–inflammation in macrophages. (a) The levels of protumorigenic cytokines (IL-1β, TNF-α and IL-6), antitumorigenic cytokine (IL-12p40) and anti-inflammatory cytokine (IL-10), in BMDM culture supernatants exposed to PAMP for 24 h, were examined using enzyme-linked immunosorbent assay (ELISA) (n=3). *P<0.05; **P<0.01. (b) A model showing a hypothetical mechanism of action of SAG-UPS during infection, which may lead to chronic inflammation and tumorigenesis. SAG-UPS maintains macrophage survival signal to facilitate immune response against infection at the early phase. SAG-mediated ubiquitination of cytosolic Bax and SARM is upregulated rapidly, thus sequestering these proapoptotic proteins away from the mitochondria. However, continuously overactivated SAG-UPS may induce hyperimmune responsiveness with uncontrolled survival signals in macrophages. Overactivated protumorigenic/proangiogenic cytokines may create a tumor microenvironment, which favors proliferation of overactivated immune cells, causing them to switch into a tumorigenic mode. IL, interleukin; LPS, lipopolysaccharide; PBS, phosphate-buffered saline; poly I:C, polyinosinic-polycytidylic acid; siRNA, small interfering RNA; TNF, tumor necrosis factor; Ub, ubiquitin
Discussion
At the frontline of immune defense, macrophages undergo apoptosis as an optimal strategy against microbial infection. However, the mechanism as to how macrophages equilibrate between apoptosis and immune response is unclear. Here, we show for the first time that under infection, SAG acts as a survival determinant for macrophages by interacting dynamically with the pro- and antiapoptotic factors, which coregulate the life or death of the cells. Coincidently, the SAG promoter harbors multiple transcription factor binding sites, including AP-1 and NF-κB, which are upregulated under TLR activation.20 Our findings reveal a novel mechanism by which SAG-UPS balances macrophage life or death.
The pivotal role of SAG to life is underscored by its evolutionary conservation from yeast to human (Supplementary Figure S9). SAG−/− mice are embryonically lethal, further indicating the fundamental significance of SAG to survival.21 With its ROS-scavenging antioxidant activity and E3 ubiquitin ligase activity, SAG was reported to exert antiapoptotic activity and stimulate cancer growth.10, 22 In agreement, our data demonstrate for the first time that during early infection, macrophages gained survival advantage owing to the antiapoptosis potency of SAG (Figures 1 and 2), which ubiquitinates proapoptosis factors, Bax and SARM (Figures 4 and 5 and Supplementary Figure S8). The SCF (Skp1-cullin-F-box proteins) is the largest family of E3 ubiquitin ligases, which is composed of four components: SKP1, a cullin, an F-box protein, and RBX1 or RBX2. The specificity of tetrameric SCF E3 ligase is determined by the F-box, which constitutes ∼70 members.23, 24 To better understand cellular responses during infection, future work will be required to delineate the key components contributing to distinct SCFs in a modular manner.
Not just apoptosis25 but timely regulation of apoptosis is an important defense mechanism against pathogens. In our study, we observed minimal apoptosis of macrophages at early infection (Figure 2c), consistent with the notion that pathogens inhibit or delay the host's cell death for their own intracellular replication and survival.2, 26 We provide compelling evidence for an apoptosis-escape mechanism of macrophages where apoptosis-related signaling pathways are inactivated via downregulation of proapoptotic proteins or upregulation of antiapoptotic proteins (Figures 2c and 3). However, a significant increase in apoptosis at the later stage (>24 h; Figure 2c) indicates that persistent infection induces apoptosis, possibly to minimize the host's inflammatory response.27 Coincidentally, SARM, which regulates immune deactivation by its proapoptotic activity,9 was also found to respond dynamically to infection (Figure 3), albeit the reciprocal to that of SAG. We showed that SAG-UPS mediates ubiquitination and turnover of SARM (Figures 4 and 5 and Supplementary Figure S8), which probably ensures that an appropriate level of immune response is achieved before SARM-mediated apoptosis ensues. Thus, SAG-UPS controls SARM activity, which in turn regulates immune response and cell death on a periodic basis.
Bax is a key proapoptosis member of the Bcl-2 family, which is regulated by UPS.28, 29 In cancer cells, Bax was reported to undergo proteasomal degradation, although the involvement of SAG-mediated ubiquitination was untested.29 The turnover of Bax in immune cells and how it responds to an infection is hitherto unclear, although it is known that during stress-induced apoptosis, the imbalance between the mitochondrial Bax and Bcl-2 compromises mitochondrial integrity.30 We found that consistent with the observation on SAG-mediated SARM turnover, Bax was susceptible to ubiquitin recognition and conjugation by SAG-UPS, thus reducing the half-life of the protein and disrupting the ratio of Bax and Bcl-2 (Figures 4 and 5 and Supplementary Figure S8). Furthermore, SAG opposes Bax translocation to the mitochondria (Figures 5d and e), thus downregulating apoptosis. This mechanism explains the underlying resistance of macrophages to apoptosis during early infection.
To ascertain the downstream effects of SAG-UPS on infected macrophages, we examined the cytokine expression. In cancer growth SAG is overexpressed persistently, but in an infection, SAG accumulates to a peak at 12 h (Figures 1 and 5a and Supplementary Figure S1). Here, our observation of SAG-mediated upregulation of protumorigenic IL-6, TNF-α and IL-1β (Figure 6a) is novel and indicates that SAG has a key role in inflammatory responses, facilitating macrophage survival during early infection. Supporting our finding are recent studies using MLN4924, an inhibitor of neddylation, which reduced the production of IL-6 and TNF-α by dendritic cells.31 Figure 6b (supported by experimental evidence of Figures 2, 3, 4, 5, 6a) depicts tightly controlled apoptosis of macrophages upon infection, showing that SAG-UPS is induced during early infection to counter apoptosis and to allow immune response. In contrast, SAG overexpression and SAG-mediated ubiquitination would continuously drive cell survival and infection-induced proinflammatory response (Figures 2 and 6a), which could conceivably create a chronic inflammatory state, attaining a tumor microenvironment. This may trigger immune overactivation and subsequently tumorigenesis. Recent studies using SAG deletion further highlighted the critical role of SAG in tumorigenesis32 and tumor angiogenesis.33 These interesting findings lend credence to our observation of the upregulation of protumorigenic cytokines (e.g. IL-6, TNF-α and IL-1β), when immune cells undergo SAG overexpression. Corroboratively, these cytokines have also been referred to as proangiogenic cytokines.34, 35 Recent evidence also suggests the correlation between persistent activation of TLRs at the sites of chronic inflammation and the development of cancer.36, 37 Therefore, understanding how TLR signaling contributes to immune overactivation and the development of chronic inflammation-associated tumors may provide insights to link immunology and cancer biology. Future work using animal models with transient/conditional SAG knockdown may help to evaluate the impact of SAG responses on infection, disease progression and its potential contribution to the recruitment of immune cells to a tumor microenvironment under chronic infection.
In conclusion, we have demonstrated that macrophage life or death is synchronized by SAG-mediated ubiquitination of proapoptotic factors, which strategically sustains immune defense during early infection. Our findings clearly established the power of SAG-UPS as a functional link between immune defense and apoptosis or immune overactivation with a potential towards tumorigenesis. We propose SAG-UPS to be an efficient target for potential future development of immunomodulators.
Materials and Methods
Experiments were performed according to Institutional Animal Care and Use Committee guidelines (NUS protocol, 012/09).
Bacteria
P. aeruginosa (PAO-Iglewski) was grown in Luria–Bertani broth at 37 °C to mid-log phase. After washing three times with PBS, the bacterial density and the colony-forming unit (CFU) were determined. The bacteria were heat inactivated at 56 °C for 1 h.38
Mouse J774, RAW264.7 cell lines, BMDM and reagents
Both J774 and RAW264.7 cells were maintained in DMEM supplemented with 10% fetal bovine serum. To verify the observations made with J774 and RAW264.7 cells, key experiments were performed with primary cells, using mouse BMDM. Briefly, bone marrow cells were isolated from the femur of 8- to 12-week-old BALB/c mice and incubated with 10 ng/ml mouse M-CSF (eBioscience, San Diego, CA, USA) for 7 days to allow differentiation. LPS from P. aeruginosa and E. coli (Sigma Chemical Co., St. Louis, MO, USA; L8643 and L4524, respectively) were used at 10 ng/ml. Poly (I:C) and R848 (InvivoGen, San Diego, CA, USA) were used at 10 μg/ml and 25 ng/ml, respectively. Proteasomes were effectively inhibited by 10 μM MG132 (Sigma-Aldrich, St. Louis, MO, USA) for 6 h.39 To evaluate the half-life of a protein, 80 μg/ml CHX was administered. The primary antibodies used in this study were: SAG polyclonal antibody (Abcam, Cambridge, UK), Bcl-2 monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), Bax polyclonal antibody (Cell Signaling Technology, Inc., Danvers, MA, USA), SARM polyclonal antibody (Santa Cruz Biotechnology), Noxa polyclonal antibody (Santa Cruz Biotechnology), Bcl-xL polyclonal antibody (Cell Signaling Technology, Inc.), ubiquitin monoclonal antibody (Santa Cruz Biotechnology), GAPDH monoclonal antibody (Santa Cruz Biotechnology), VDAC polyclonal antibody (Cell Signaling Technology, Inc.), cytochrome c monoclonal antibody (BD Biosciences, San Jose, CA, USA), control rabbit IgG (Santa Cruz Biotechnology), control mouse IgG (Santa Cruz Biotechnology) and a loading control, β-actin (Sigma Chemical Co.).
Real-time PCR
Total RNA was extracted with TRIzol reagent (Invitrogen) and the RNA was reverse transcribed using random hexamers and Superscript III (Invitrogen), according to the manufacturer's instructions. The synthesized cDNA was used for real-time PCR analysis using SYBR Green on a LightCycler 480II instrument (Roche, Mannheim, Germany). The primers were SAG (109 bp product): sense, 5′-CAGGCTCCAAGTCGGGAGGCG-3′, antisense, 5′-TGGACCCTGCAGATGGCACAGG-3′ Bcl-2 (350 bp product): sense, 5′-TACCGTCGTGACTTCGCAGAG-3′, antisense, 5′-GGCAGGCTGAGCAGGGTCTT-3′40 Bax (160 bp product): sense, 5′-CGGCGAATTGGAGATGAACTG-3′, antisense, 5′-GCAAAGTAGAAGAGGGCAACC-3′40 SARM (132 bp product): sense, 5′-CATCACCCGCAAGAGGTTC-3′, antisense, 5′-CCATAGGTGTACTGGCGGAA-3′ Noxa (130 bp product): sense, 5′-CCCAGATTGGGGACCTTAGT-3′, antisense, 5′-CTGCGAACTCAGGTGGTAGC-3′41 Bcl-xL (399 bp product): sense, 5′-AGGCAGGCGATGAGTTTGAAC-3′, antisense, 5′-GAACCACACCAGCCACAGTCA-3′40 β-actin (103 bp product): sense, 5′-GCTGACAGGATGCAGAAGGAG-3′, antisense, 5′-AGCCACCGATCCACACAGA-3′. All expression values were normalized based on β-actin as an endogenous control.
SAG knockdown, HA-SAG overexpression and PAMP stimulation in J774, RAW264.7 macrophages or primary BMDM cells
SAG siRNA (Silencer select predesigned siRNA) and control (scrambled) siRNA were purchased from Ambion (Carlsbad, CA, USA) and Invitrogen, respectively. siRNA transfection into J774, RAW264.7 or BMDM cells (at 8 × 105 cells per well of a 6-well plate) was conducted using 8 μl X-tremeGENE Transfection Reagent (Roche) with 90 pmol of siRNAs per well. HA-SAG-pcDNA3 and pcDNA3 plasmids were used for SAG overexpression study. For BMDM cells, DNA transfection was conducted with 8 × 105 cells per well of a 6-well plate using 4 μl X-tremeGENE Transfection Reagent (Roche) with 2 μg DNA plasmids. The transfection efficiency for siRNA knockdown was determined by immunodetection of SAG protein and real-time PCR of SAG mRNA (Supplementary Figure S2). The transfection efficiency for cDNA overexpression was determined by immunodetection of the HA-tag protein (Supplementary Figure S10). At 16 h after transfection, cells were treated with PAMP(s) for different time periods.
TMRE staining
Mitochondrial membrane potential was determined using TMRE (Sigma Chemical Co.), which stains intact mitochondria. Briefly, cells were loaded with 200 nM TMRE dye for 20 min at 37 °C to stain the mitochondrial matrix. The samples were then subjected to flow cytometric analysis for a minimum of 15 000 events.
Caspase-9 and -3 assays
The J774 cells were transfected with SAG-specific siRNA or control scrambled siRNA for 16 h, followed by PAMP stimulation at time points indicated. Apoptosis was confirmed by determining the caspase-specific cleavages of activated caspase-9 and -3 (OncoImmunin Inc., Gaithersburg, MD, USA). The fluorescence emission was measured at excitation (552 nm) and emission (580 nm) wavelengths, using a FACScan flow cytometer (Beckman Coulter, Inc., Brea, CA, USA).
Intracellular staining of representative apoptosis markers
The expression of SAG, Bcl-2, Bax and SARM was measured by staining intracellularly with specific antibodies. Briefly, cells were harvested at indicated time points and washed in PBS-TX (PBS containing 0.1% Triton X-100). The cells were incubated with primary antibody for 30 min, washed two times with PBS-TX and conjugated to secondary antibody, Alexa Fluor 488 (Invitrogen) or with phycoerythrin (Invitrogen) for 20 min. After washing two times with PBS-TX, the cells were analyzed on a FACScan flow cytometer.
Cellular apoptosis assay
Early-stage apoptosis was measured using annexin V (eBioscience) in conjunction with 7-AAD (eBioscience). Annexin V identifies surface-exposed phosphatidylserine, and 7-AAD is retained in late apoptotic cells. Apoptosis was then analyzed using FACScan flow cytometer for a minimum of 20 000 events.
Trypan Blue dye exclusion test
To assess total cell viability, cells were scraped and resuspended in equal volumes of culture medium and trypan blue dye (0.05% solution; GIBCO-Invitrogen, Paisley, UK) and counted using a Neubauer improved hemocytometer.
MTT assay
To estimate cell death due to loss of mitochondrial activity at the end of PAMP treatments, the cells were incubated with 100 μl MTT reagent (Sigma Chemical Co.) at 37 °C for 3 h. The metabolic activity of viable cells was measured at 570 nm using a BioTek microplate reader (Synergy Mx Monochromator, Winooski, VT, USA).
Subcellular fractionation, Co-IP and immunodetection
To prepare cytosolic and mitochondrial lysates, BMDM cells were homogenized and mitochondria were pelleted by a mitochondria isolation kit (Thermo Fisher Scientific, Rockford, IL, USA). The cytosolic fractions were subjected to Co-IP with anti-Bax or anti-SARM antibody. The mitochondrial fraction products were immunoblotted and probed sequentially with antibodies to Bax, SARM and VDAC. For Co-IP study with J774 or RAW264.7 total cell lysates, the cells were lysed in CHAPS lysis buffer containing 20 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 137 mM KCl, 1 mM EDTA, 1 mM EGTA, 1% CHAPS and 1 × protease inhibitors (complete EDTA-free cocktail, Roche). Then, the cell lysate was precleared by incubating with protein G Sepharose (GE Healthcare, Little Chalfont, UK) at 4 °C for 2 h. The supernatant was incubated overnight at 4 °C with Bax or SARM or Bcl-2 antibody, followed by a 3-h incubation with protein G Sepharose. The washed immunoprecipitates, resuspended in Laemmli buffer, was boiled at 95 °C for 5 min before immunodetection of SAG, Bax, SARM, Bcl-2 or ubiquitin.
ELISA
To determine the effects of SAG on cytokine release, BMDM cells were transiently transfected with SAG-specific siRNA, control siRNA, HA-SAG or empty vector for 16 h, followed by PAMP stimulation for 24 h. The levels of IL-6, TNF-α, IL-1β, IL-12p40 and IL-10, secreted by the cells, were quantified in 24-h supernatants by using OptEIA Mouse ELISA Set (BD Biosciences).
Statistical analysis
Data are presented as means±S.D. of three independent experiments, with three replicates per sample/condition tested. Differences between averages were analyzed by two-tailed Student's t-test. Significance was set at P-value of<0.05 (*P<0.05, **P<0.01). All target signals from western blots were quantified by Scion Image software (Frederick, MD, USA). The acquired data from FACS were analyzed with Summit software (Version 4.3.02, Fort Wayne, IN, USA).
Acknowledgments
We thank Dr. BH Iglewski (University of Rochester) for P. aeruginosa strain PAO-Iglewski; Dr. Yi Sun (University of Michigan, USA) for plasmids HA-SAG-pcDNA3 and pcDNA3, and Drs Zhu Yong and Laishram Pradeepkumar Singh for proofreading the manuscript. This work was supported by grants from the A*STAR Biomedical Research Council, Singapore (10/1/21/19/658) and the National Medical Research Council, Singapore (NMRC/CBRG/0055/2014).
Glossary
- PAMP
pathogen pattern-associated molecular pattern
- SAG
sensitive to apoptosis gene
- SARM
sterile α- and HEAT/armadillo-motif-containing protein
- UPS
ubiquitin–proteasome system
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
Supplementary Material
References
- Subramanian K, Du R, Tan NS, Ho B, Ding JL. CD163 and IgG codefend against cytotoxic hemoglobin via autocrine and paracrine mechanisms. J Immunol. 2013;190:5267–5278. doi: 10.4049/jimmunol.1202648. [DOI] [PubMed] [Google Scholar]
- Rudel T, Kepp O, Kozjak-Pavlovic V. Interactions between bacterial pathogens and mitochondrial cell death pathways. Nat Rev Microbiol. 2010;8:693–705. doi: 10.1038/nrmicro2421. [DOI] [PubMed] [Google Scholar]
- Hasnain SE, Begum R, Ramaiah KV, Sahdev S, Shajil EM, Taneja TK, et al. Host–pathogen interactions during apoptosis. J Biosci. 2003;28:349–358. doi: 10.1007/BF02970153. [DOI] [PubMed] [Google Scholar]
- Neutzner A, Li S, Xu S, Karbowski M. The ubiquitin/proteasome system-dependent control of mitochondrial steps in apoptosis. Semin Cell Dev Biol. 2012;23:499–508. doi: 10.1016/j.semcdb.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding JL, Tan KC, Thangamani S, Kusuma N, Seow WK, Bui THH, et al. Spatial and temporal coordination of expression of immune response genes during Pseudomonas infection of horseshoe crab, Carcinoscorpius rotundicauda. Genes Immun. 2005;6:557–574. doi: 10.1038/sj.gene.6364240. [DOI] [PubMed] [Google Scholar]
- Panneerselvam P, Singh LP, Ho B, Chen J, Ding JL. Targeting of pro-apoptotic TLR adaptor SARM to mitochondria: definition of the critical region and residues in the signal sequence. Biochem J. 2012;442:263–271. doi: 10.1042/BJ20111653. [DOI] [PubMed] [Google Scholar]
- Mink M, Fogelgren B, Olszewski K, Maroy P, Csiszar K. A novel human gene (SARM) at chromosome 17q11 encodes a protein with a SAM motif and structural similarity to Armadillo//beta-catenin that is conserved in mouse, Drosophila, and Caenorhabditis elegans. Genomics. 2001;74:234–244. doi: 10.1006/geno.2001.6548. [DOI] [PubMed] [Google Scholar]
- Carty M, Goodbody R, Schroder M, Stack J, Moynagh PN, Bowie AG. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat Immunol. 2006;7:1074–1081. doi: 10.1038/ni1382. [DOI] [PubMed] [Google Scholar]
- Panneerselvam P, Singh LP, Selvarajan V, Chng WJ, Ng SB, Tan NS, et al. T-cell death following immune activation is mediated by mitochondria-localized SARM. Cell Death Differ. 2013;20:478–489. doi: 10.1038/cdd.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Wang Y, Aviram M, Swaroop M, Loo JA, Bian J, et al. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol Cell Biol. 1999;19:3145–3155. doi: 10.1128/mcb.19.4.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Yang J, Hao X, Zheng M, He H, Xiong X, et al. Validation of SAG/RBX2/ROC2 E3 ubiquitin ligase as an anticancer and radiosensitizing target. Clin Cancer Res. 2010;16:814–824. doi: 10.1158/1078-0432.CCR-09-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratieva TK, Kobets NV, Khaidukov SV, Yeremeev VV, Lyadova IV, Apt AS, et al. Characterization of T cell clones derived from lymph nodes and lungs of Pseudomonas aeruginosa-susceptible and resistant mice following immunization with heat-killed bacteria. Clin Exp Immunol. 2000;121:275–282. doi: 10.1046/j.1365-2249.2000.01222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan SM, Rothwell NJ. Cytokines and acute neurodegeneration. Nat Rev Neurosci. 2001;2:734–744. doi: 10.1038/35094583. [DOI] [PubMed] [Google Scholar]
- Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J Pathol. 2002;196:254–265. doi: 10.1002/path.1027. [DOI] [PubMed] [Google Scholar]
- Etoh T, Shibuta K, Barnard GF, Kitano S, Mori M. Angiogenin expression in human colorectal cancer: the role of focal macrophage infiltration. Clin Cancer Res. 2000;6:3545–3551. [PubMed] [Google Scholar]
- Pinto A, Morello S, Sorrentino R. Lung cancer and Toll-like receptors. Cancer Immunol Immunother. 2011;60:1211–1220. doi: 10.1007/s00262-011-1057-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlato C, Cassatella MA, Kinjyo I, Gatto L, Yoshimura A, Bazzoni F. Involvement of suppressor of cytokine signaling-3 as a mediator of the inhibitory effects of IL-10 on lipopolysaccharide-induced macrophage activation. J Immunol. 2002;168:6404–6411. doi: 10.4049/jimmunol.168.12.6404. [DOI] [PubMed] [Google Scholar]
- Park P-h, McMullen MR, Huang H, Thakur V, Nagy LE. Short-term treatment of RAW264.7 macrophages with adiponectin increases tumor necrosis factor-α (TNF-α) expression via ERK1/2 activation and Egr-1 expression: role of TNF-α in adiponectin-stimulated interleukin-10 production. J Biol Chem. 2007;282:21695–21703. doi: 10.1074/jbc.M701419200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhao Y, Qian J, Sun L, Lu Y, Li H, et al. Toll-like receptor-4 signaling in mantle cell lymphoma. Cancer. 2013;119:782–791. doi: 10.1002/cncr.27792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q, Tan M, Sun Y. SAG/ROC2/Rbx2 is a novel activator protein-1 target that promotes c-Jun degradation and inhibits 12-O-tetradecanoylphorbol-13-acetate-induced neoplastic transformation. Cancer Res. 2007;67:3616–3625. doi: 10.1158/0008-5472.CAN-06-4020. [DOI] [PubMed] [Google Scholar]
- Tan M, Zhao Y, Kim S-J, Liu M, Jia L, Saunders T, et al. SAG/RBX2/ROC2 E3 ubiquitin ligase is essential for vascular and neural development by targeting NF1 for degradation. Dev Cell. 2011;21:1062–1076. doi: 10.1016/j.devcel.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Sun Y. Small RING finger proteins RBX1 and RBX2 of SCF E3 ubiquitin ligases: the role in cancer and as cancer targets. Genes Cancer. 2010;1:700–707. doi: 10.1177/1947601910382776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenciarelli C, Chiaur DS, Guardavaccaro D, Parks W, Vidal M, Pagano M. Identification of a family of human F-box proteins. Curr Biol. 1999;9:1177–S1173. doi: 10.1016/S0960-9822(00)80020-2. [DOI] [PubMed] [Google Scholar]
- Winston JT, Koepp DM, Zhu C, Elledge SJ, Harper JW. A family of mammalian F-box proteins. Curr Biol. 1999;9:1180–S1183. doi: 10.1016/S0960-9822(00)80021-4. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. Manipulation of host cell death pathways during microbial infections. Cell Host Microbe. 2010;8:44–54. doi: 10.1016/j.chom.2010.06.007. [DOI] [PubMed] [Google Scholar]
- Brune W. Inhibition of programmed cell death by cytomegaloviruses. Virus Res. 2011;157:144–150. doi: 10.1016/j.virusres.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Behar SM, Divangahi M, Remold HG. Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy. Nat Rev Microbiol. 2010;8:668–674. doi: 10.1038/nrmicro2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F-T, Agrawal SG, Gribben JG, Ye H, Du M-Q, Newland AC, et al. Bortezomib blocks Bax degradation in malignant B cells during treatment with TRAIL. Blood. 2008;111:2797–2805. doi: 10.1182/blood-2007-08-110445. [DOI] [PubMed] [Google Scholar]
- Li B, Dou QP. Bax degradation by the ubiquitin/proteasome-dependent pathway: involvement in tumor survival and progression. Proc Natl Acad Sci. 2000;97:3850–3855. doi: 10.1073/pnas.070047997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- Mathewson N, Toubai T, Kapeles S, Sun Y, Oravecz-Wilson K, Tamaki H, et al. Neddylation plays an important role in the regulation of murine and human dendritic cell function. Blood. 2013;122:2062–2073. doi: 10.1182/blood-2013-02-486373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Tan M, Jia L, Wei D, Zhao Y, Chen G, et al. Inactivation of SAG/RBX2 E3 ubiquitin ligase suppresses KrasG12D-driven lung tumorigenesis. J Clin Invest. 2014;124:835–846. doi: 10.1172/JCI70297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Li H, Sun Y.Endothelial deletion of Sag/Rbx2/Roc2 E3 ubiquitin ligase causes embryonic lethality and blocks tumor angiogenesis Oncogene 2013. e-pub ahead of print 11 November 2013; doi: 10.1038/onc.2013.473 [DOI] [PMC free article] [PubMed]
- Fan Y, Ye J, Shen F, Zhu Y, Yeghiazarians Y, Zhu W, et al. Interleukin-6 stimulates circulating blood-derived endothelial progenitor cell angiogenesis in vitro. J Cereb Blood Flow Metab. 2007;28:90–98. doi: 10.1038/sj.jcbfm.9600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini A, Carraro F. Role of inflammatory mediators in angiogenesis. Curr Drug Targets Inflamm Allergy. 2005;4:3–8. doi: 10.2174/1568010053622830. [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Medzhitov R. Toll-like receptors and cancer. Nat Rev Cancer. 2009;9:57–63. doi: 10.1038/nrc2541. [DOI] [PubMed] [Google Scholar]
- Chen R, Alvero AB, Silasi DA, Steffensen KD, Mor G. Cancers take their Toll—the function and regulation of Toll-like receptors in cancer cells. Oncogene. 2008;27:225–233. doi: 10.1038/sj.onc.1210907. [DOI] [PubMed] [Google Scholar]
- Tertilt C, Joh J, Krause A, Chou P, Schneeweiss K, Crystal RG, et al. Expression of B-cell activating factor enhances protective immunity of a vaccine against Pseudomonas aeruginosa. Infect Immun. 2009;77:3044–3055. doi: 10.1128/IAI.00927-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H-M, Wen H-C, Lin W-W. Proteasome inhibitors stimulate interleukin-8 expression via ras and apoptosis signal-regulating kinase-dependent extracellular signal-related kinase and c-Jun N-terminal kinase activation. Am J Resp Cell Mol Biol. 2002;27:234–243. doi: 10.1165/ajrcmb.27.2.4792. [DOI] [PubMed] [Google Scholar]
- Exley GE, Tang C, McElhinny AS, Warner CM. Expression of caspase and BCL-2 apoptotic family members in mouse preimplantation embryos. Biol Reprod. 1999;61:231–239. doi: 10.1095/biolreprod61.1.231. [DOI] [PubMed] [Google Scholar]
- Knowlton JJ, Dermody TS, Holm GH. Apoptosis induced by mammalian reovirus is beta interferon (IFN) independent and enhanced by IFN regulatory factor 3- and NF-κB-dependent expression of Noxa. J Virol. 2012;86:1650–1660. doi: 10.1128/JVI.05924-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.