Abstract
Proline-, glutamic acid- and leucine-rich protein-1 (PELP1) is a scaffolding oncogenic protein that functions as a coregulator for a number of nuclear receptors. p53 is an important transcription factor and tumor suppressor that has a critical role in DNA damage response (DDR) including cell cycle arrest, repair or apoptosis. In this study, we found an unexpected role for PELP1 in modulating p53-mediated DDR. PELP1 is phosphorylated at Serine1033 by various DDR kinases like ataxia-telangiectasia mutated, ataxia telangiectasia and Rad3-related or DNAPKc and this phosphorylation of PELP1 is important for p53 coactivation functions. PELP1-depleted p53 (wild-type) breast cancer cells were less sensitive to various genotoxic agents including etoposide, camptothecin or γ-radiation. PELP1 interacts with p53, functions as p53-coactivator and is required for optimal activation of p53 target genes under genomic stress. Overall, these studies established a new role of PELP1 in DDRs and these findings will have future implications in our understanding of PELP1's role in cancer progression.
p53 is considered as the guardian of genomic integrity and has an important role in initiating cellular response to various genomic stresses such as cell cycle arrest, senescence, DNA repair and apoptosis.1, 2 Loss of p53 or mutations in p53 is observed in >50% of the cases of all cancers.3, 4, 5 Stabilization of p53 upon genomic stress and activation of its transcription functions are vital for its central role in the DNA damage/genomic stress response and in its tumor-suppressive functions.6, 7 Upon genomic stress, p53 is stabilized and activated because of a decreased interaction with its E3-ligase MDM2.8 Activated p53 then upregulates expression of target genes, such as p21/WAF1, GADD45, PUMA and NOXA, all of which are important in the cellular decisions for cell cycle arrest or apoptosis.9
Post-translational modifications of p53, including phosphorylation, acetylation, ubiquitinylation and methylation,10, 11, 12 and interactions with several cofactors13, 14 have a critical role in the p53-mediated transcriptional response to the DNA damage response (DDR). Proline-, glutamic acid- and leucine-rich protein-1 (PELP1), a large multi-domain protein, modulates a number of biological processes and several pathways including estrogen signaling, and cancer progression.15, 16 PELP1 functions as a coregulator for several nuclear receptors such as estrogen receptor (ER), androgen receptor (AR) and progesterone receptor (PR), and transcription factors such as STAT3, AP1 and E2F.17, 18, 19 PELP1 associates with the chromatin20 and interacts with histone-modifying complexes17 including acetylases (CBP/P300),21 methylases (SETDB1 and MLL1),22, 23 demethylases (KDM1)24 and deacetylases (HDAC2 and MTA1).17 It promotes cell proliferation by enhancing G1 to S phase progression via its interactions with the pRb/E2F pathway.25 PELP1 localizes to the nucleolus and has an important role in ribosomal biogenesis.26 PELP1 signaling is also implicated in apoptosis and differentiation, and PELP1 functions as a coactivator of RXR homodimers and RXR-PPAR heterodimers.27 Although PELP1's role in both cell proliferation and differentiation is evident, it is not known how PELP1 would affect p53-mediated DDR functions and whether PELP1 status would affect sensitivity to various genomic stresses.
In this study, we show that PELP1 has an unexpected role in the p53-mediated DDR. Using p53 wild-type (WT) cells with or without PELP1 expression, we demonstrated that cells lacking PELP1 are less sensitive to various genotoxic agents. PELP1 interacts with p53 and functions as a coactivator of p53 and modulates expression of p53 target genes upon DNA damage. Our studies also identified PELP1 as a novel substrate of DDR kinases (including ataxia-telangiectasia mutated (ATM), ataxia telangiectasia and Rad3-related (ATR) or DNAPKc) and showed phosphorylation of PELP1 is important for its DDR functions. Collectively, our data established a new role for PELP1 in the DDR.
Results
PELP1 is phosphorylated upon various genotoxic stresses
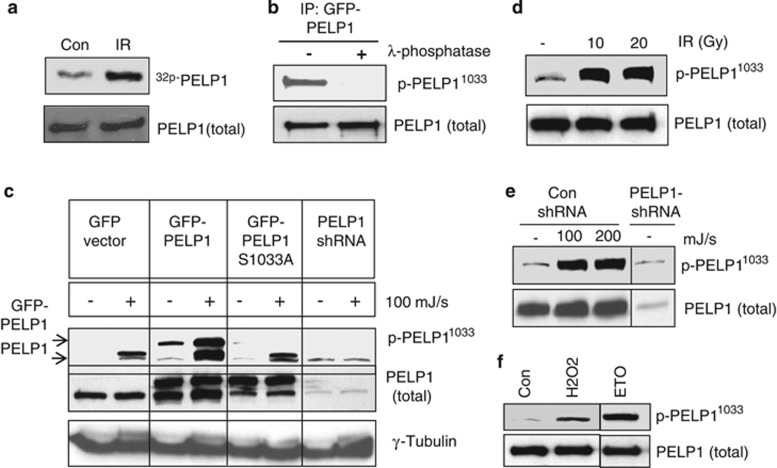
Upon DNA damage, ATM, ATR or DNAPKc kinases redundantly phosphorylate substrate proteins on S/T.Q motifs (serine/threonine followed by glutamine).28, 29 A large-scale mass spectrometric analysis identified ∼700 protein substrates with S/T.Q motifs that were phosphorylated upon γ-radiation.30 Interestingly, the C-terminus of PELP1 has an S/T.Q motif. To examine whether PELP1 indeed gets phosphorylated upon genotoxic stress, we performed an ortho-phosphate metabolic-labeling assay. MCF7 cells were metabolically labeled with 32P-orthophosphate and treated with or without γ-radiation. After 30 min of radiation treatment, PELP1 was immunoprecipitated and subjected to SDS-PAGE followed by autoradiography. Results indicated that PELP1 did indeed get phosphorylated upon DNA damage (Figure 1a). To confirm that PELP1 is phosphorylated at the Ser-1033 residue, we generated a novel affinity purified phospho-PELP1 antibody. The antibody-specific recognition of phosphorylated PELP1 was validated using a lambda phosphatase treatment. The λ-phosphatase was able to completely abolish the antibody detection of the phosphorylated form of PELP1 (Figure1b). To further characterize the antibody, we created the Ser1033Ala (S1033A) mutation in PELP1 using site-directed mutagenesis. ZR75 model cells stably expressing GFP, PELP1-WT, PELP1-mutant (MT) or PELP1-shRNA were treated with or without ultraviolet (UV) radiation, followed by western blotting with phospho-PELP1 antibody. The antibody efficiently recognized phosphorylated endogenous as well as GFP-tagged PELP1, and this antibody recognition was substantially reduced in ZR75-PELP1-shRNA cells (Figure 1c). The phospho-antibody failed to recognize the S1033A PELP1-MT, confirming the specificity of this antibody (Figure 1c). To further investigate the types of DNA damage that can induce PELP1 phosphorylation in vivo, we exposed cells to gamma radiation (Figure 1d), UV radiation (Figure 1e), genotoxic stress by H2O2 and etoposide (Figure 1f). Results showed that phosphorylation of PELP1 at the Ser-1033 residue occur in vivo in response to all DNA damaging agents tested.
Figure 1.
PELP1 is phosphorylated at Ser-1033 residue upon various genomic insults. (a) MCF7 cells were metabolically labeled with 32P-orthophosphate, and treated with radiation (10 Gy, 30 min). PELP1 phosphorylation was determined by immunoprecipitation followed by autoradiography. (b) MCF7 cells were treated with UV radiation (100 mJ/s), PELP1 was immunoprecipitated and treated with or without λ-phosphatase. Status of PELP1 phosphorylation was visualized by western blotting using PELP1 Ser1033 phospho-antibody. (c) ZR75 cells expressing GFP, GFP-PELP1, or GFP-PELP1-S1033A MT or PELP1-shRNA were treated with or without UV radiation, followed by western blotting with PELP1 Ser1033 phospho-antibody. (d) ZR75 cells were exposed to different doses of γ-radiation, followed by western blotting with PELP1 Ser1033 phospho-antibody. (e) ZR75 and ZR75-PELP1-shRNA were exposed to UV radiation, followed by western blotting with PELP1 Ser1033 phospho-antibody. (f) 293T cells were exposed to genotoxic stress by H2O2 (30 min) or etoposide (50 μM, 1 h) followed by western blot analysis with PELP1 Ser1033 phospho-antibody
PELP1 is phosphorylated at Ser-1033 by various DDR kinases upon DNA damage
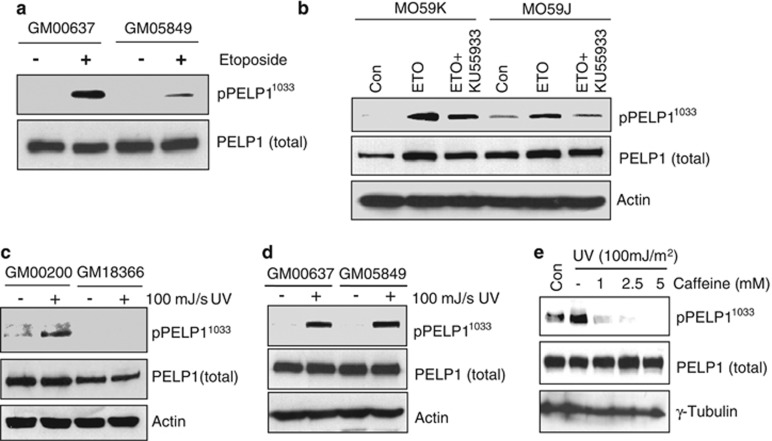
To identify the upstream DDR kinases that phosphorylate PELP1 upon DNA damage, we used various model cells that are either proficient or deficient in ATM, ATR or DNAPKc functions. We first treated the cells with 50 μM etoposide for 1 h to generate double-strand breaks (DSBs) in the human fibroblast cells, GM00637 (ATM proficient) and GM05849 (ATM function deficient). Western analysis using phospho-PELP1 antibody showed that ATM-deficient cells have reduced PELP1 phosphorylation at Ser1033 upon DNA damage (Figure 2a). Similarly, western analysis of lysates from etoposide-treated human glioma cell lines M059K and M059J that are either proficient or deficient in DNAPKc, respectively, revealed a defect in PELP1 phosphorylation in DNAPKc-deficient cells occur upon DNA damage (Figure 2b). Interestingly, pre-treatment of these cells with KU55933, a specific inhibitor of ATM further decreased the levels of phospho-PELP1 (Figure 2b), suggesting that while ATM and DNAPKc can both redundantly phosphorylate PELP1, ATM appears to be the major kinase in the context of DSBs. To identify the kinase that phosphorylates PELP1 upon single-strand breaks, we used UVC radiation as a damaging agent and GM18366 (ATR-deficient) and its matched control GM00200 fibroblasts as model cells. Western analysis revealed that phosphorylation of PELP1 at Ser-1033 was compromised in ATR-deficient cells (Figure 2c). Interestingly, ATM-deficient cells did not experience a decrease in PELP1 phosphorylation upon UVC treatment (Figure 2d). However, pre-treatment of ZR75 cells with increasing doses of caffeine abolished PELP1 phosphorylation (Figure 2e), suggesting involvement of ATR in UVC-mediated PELP1 phosphorylation.
Figure 2.
ATM, ATR and DNAPKc kinases can phosphorylate PELP1 at S1033 upon DNA damage. (a) ATM-proficient, human fibroblasts GM00637 and ATM-deficient fibroblast GM05849 cells were treated with or without etoposide (50 μM,1 h) and the status of PELP1 phosphorylation at S1033 was analyzed by western blotting. (b) Human glioma cells M059K and M059J, proficient and deficient in DNAPKc, respectively, were treated with or without etoposide (50 μM, 1 h) alone or in combination with KU55933 and the status of PELP1 phosphorylation at S1033 was analyzed by western blotting. (c and d) Human skin fibroblast GM00200 and GM18366, proficient and deficient in ATR functions, respectively, (c) or ATM proficient GM00637 and ATM-deficient cells GM05849 (d) were irradiated with UVC and the status of PELP1 phosphorylation at S1033 was analyzed by western blotting. (e) ZR75 cells were pre-treated with increasing doses of caffeine and irradiated with 100 mJ/s UVC. Status of PELP1 S1033 phosphorylation was analyzed by western blotting
Knockdown of PELP1 in p53-positive cancer cells reduces their sensitivity to genotoxic agents
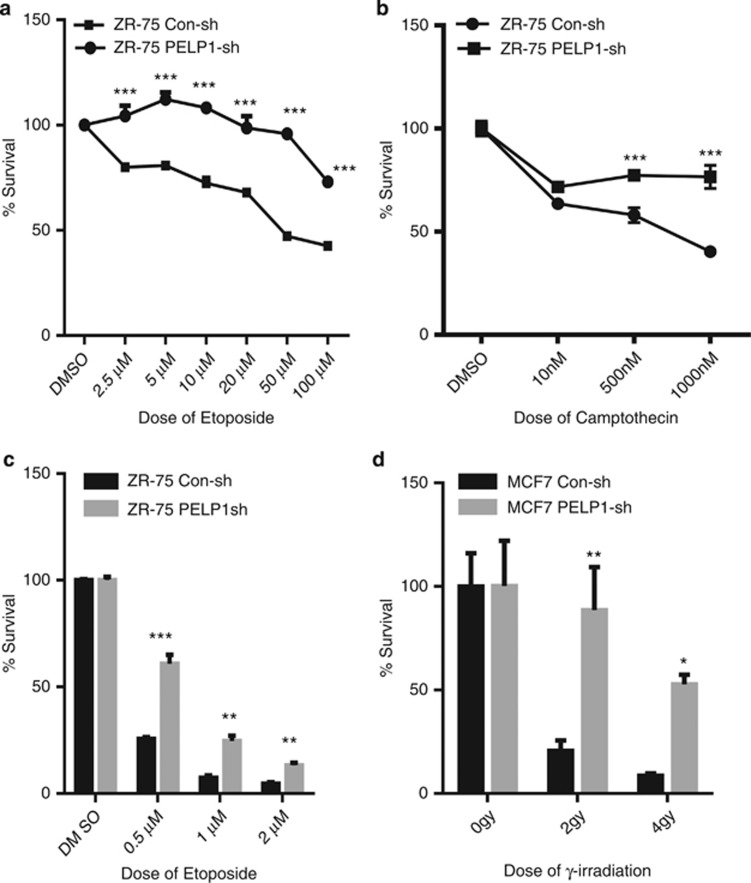
To examine whether PELP1 modulates sensitivity of breast cancer cells to various genotoxic agents, we used two p53 WT breast cancer cells (ZR75 and MCF7) that stably express the PELP1-shRNA. Western analysis showed that the PELP1 levels in PELP1-shRNA-transfected cells were about 70–80% less than the levels in control vector-transfected cells (Supplementary Figure S1a). ZR75 control and ZR75-PELP1-shRNA cells were treated with various doses of etoposide and camptothecin for 48 h, followed by analysis for cytotoxicity. PELP1-depleted cells exhibit reduced cytotoxicity to these drugs (Figures 3a and b). Similar results were obtained with MCF7 cells (Supplementary Figure 1b). Clonogenic survival assays after etoposide treatment confirmed that ZR75-PELP1-shRNA cells have greater survival ability after treatment with genotoxic agents (Figure 3c). We validated these findings using MCF7 model cells stably expressing PELP1-shRNA or transiently expressing PELP1-small interfering RNA (siRNA) in ZR75 cells and by using a different genotoxic agent (ionizing radiation). PELP1-depleted MCF7 and ZR75 cells survived better and were more clonogenic compared with control cells after genotoxic stress (Figure 3d and Supplementary Figure S1c). On the contrary, PELP1 knockdown in p53 non-functional cells such as 293T and IOMM-LEE further enhanced their sensitivity to DNA damage (Supplementary Figures S1d and e), suggesting that the less sensitivity to DNA damage seen in PELP1 knockdown breast cancer cells could depend on p53 status.
Figure 3.
PELP1-depleted cells exhibit decreased sensitivity to various genotoxic agents. ZR75 cells stably expressing PELP1-shRNA or the control shRNA were treated with different doses of chemotherapeutic drugs; etoposide (a) and camptothecin (b) and cytotoxicity was evaluated using MTT assay. Equal number of ZR75-Con shRNA and ZR75-PELP1shRNA (c) and MCF7-Con shRNA and MCF7-PELP1-shRNA cells (d) were subjected to different doses of etoposide or γ-radiation and clonogenic (survival) assays were performed after 14 days. The survival fraction for each dose of γ-radiation is expressed as percentage of colony formed in their respective untreated control group. Error bar depicts S.E. from triplicate experiment. *P<0.05; **P<0.01; ***P<0.001
PELP1-depleted cells are defective in p53-mediated DDR
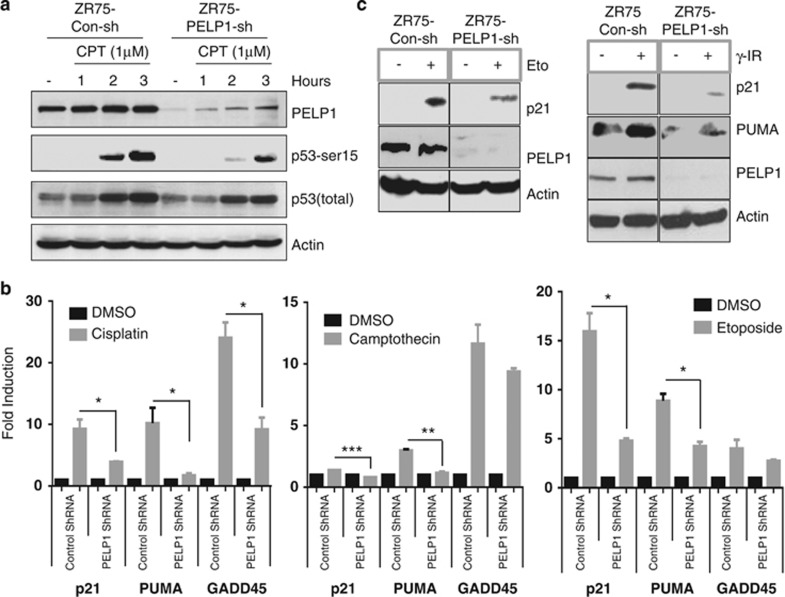
p53 has a crucial role in the apoptotic response to various genotoxic drugs. As PELP1-depleted cells were less sensitive to various genotoxic stresses, we examined the status of p53 activation. ZR75-conshRNA and ZR75-PELP1-shRNA cells were treated with camptothecin and total cellular lysates were analyzed for p53 activation by western blot using phospho-p53 (Ser-15) antibody. Levels of phospho-p53 were substantially less in PELP1-depleted cells than in controls cells (Figure 4a). Similarly, we observed decreased p53 acetylation upon DNA damage in PELP1 knockdown cells (Supplementary Figure S2a). PELP1 knockdown did not affect the expression of p53 as measured by RT-qPCR and by total p53 western analysis (Supplementary Figures S2a and b). To ascertain whether this reduction in p53 activation could translate into defective expression of its target genes, we analyzed the expression of p53 target genes by quantitative real-time reverse-transcriptase polymerase chain reaction (qRT-PCR) analysis. Expression of p53 target genes p21, PUMA and GADD45 was significantly less in PELP1-depleted cells than in the control cells (Figure 4b). Similar results were obtained in MCF7-PELP1-shRNA cells after treatment with etoposide or γ-radiation (Supplementary Figures S2c and d). Accordingly, western analysis revealed low levels of p21 and PUMA in PELP1-depleted cells upon genotoxic exposure (Figure 4c). Next, we determined whether PELP1 is essential for p53-mediated G1/S cell cycle checkpoint activation. Analysis of G1-S checkpoint revealed that PELP1 knockdown cells were defective in proper activation of G1-S checkpoint (Supplementary Figure S2e).
Figure 4.
Activation of p53 and its transcriptional functions are defective in PELP1-depleted cells. (a) ZR75 cells stably expressing PELP1-shRNA or the control shRNA vector were treated with 1 μM camptothecin and at different time periods, cell lysates were prepared and subjected to western blotting with indicated antibodies. (b) ZR75 cells stably expressing PELP1-shRNA or control shRNA vector were treated with either cisplatin (25 μM for 18 h), camptothecin (10 μM for 12h) or etoposide (50 μM for 12 h). RNA was isolated and the status of p53 target genes was analyzed by qRT-PCR analysis. *P<0.05; **P<0.01; ***P<0.001. (c) ZR75 cells stably expressing PELP1-shRNA or the control shRNA vector were treated with either 50 μM etoposide for 12 h, or γ-radiation (5 Gy). The status of p53 target genes was analyzed by western blotting of total lysates
PELP1 interacts with p53
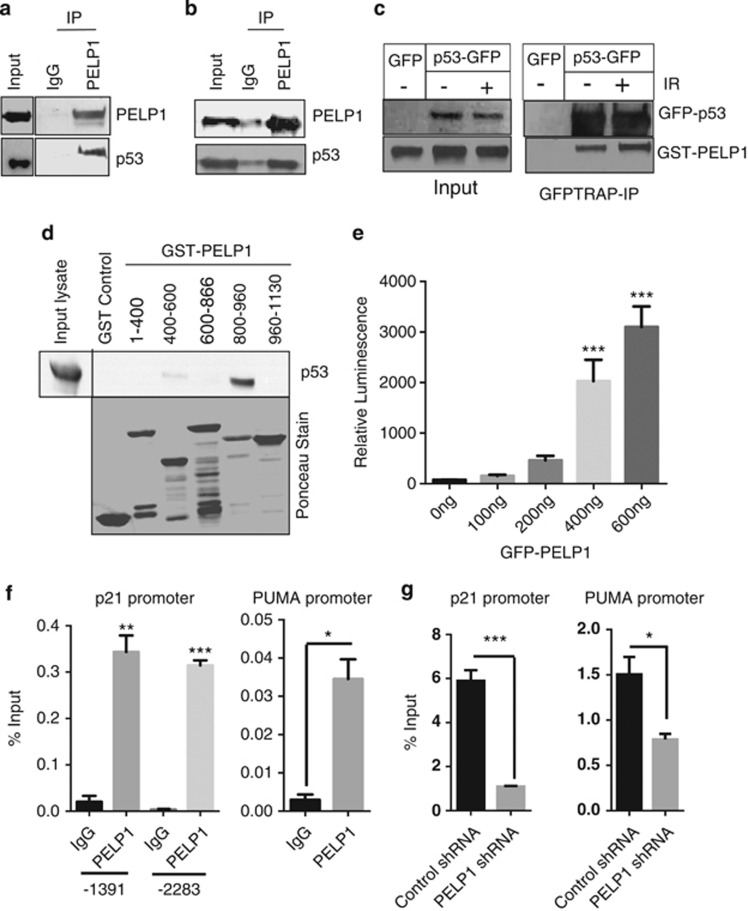
As PELP1 functions as a coregulator of many nuclear factors, and because reduced PELP1 expression compromised activation of p53 in response to various genotoxic agents, we investigated whether PELP1 functions as coregulator of p53. Western analysis of PELP1 immunoprecipitates from cells that were treated with IR and camptothecin revealed that endogenous PELP1 forms a complex with endogenous p53 (Figures 5a and b). Interaction of PELP1 and p53 was further confirmed using confocal analysis. Our results showed that endogenous PELP1 colocalizes with endogenous p53 upon camptothecin treatment, while no or little colocalization of PELP1 with p53 was observed in the absence of DNA damage (Supplementary figure S3). We also confirmed PELP1 and p53 interactions using model cells that transiently expressed epitope tagged GST-PELP1 and GFP-p53. Results showed GFP-p53 ability to interact with GST-PELP1 and increased interaction was observed under conditions of DNA damage (Figure 5c). GST pull-down assays identified PELP1 amino acids 800–960 contributed predominantly to the p53 interaction (Figure 5d). To assess whether PELP1 directly affects p53-mediated transactivation functions, we performed a p53-reporter assay in breast cancer cell lines by using p53-luc reporter plasmid that has p53-binding sites in its promoter. Cells were transfected with p53-luc reporter plasmid along with increasing doses of GFP-tagged PELP1 (PG) plasmids and luciferase activity was measured after 48 h. A dose-dependent increase in luciferase readouts was observed suggesting that PELP1 has the ability to increase p53-mediated transactivation functions in ZR75 (Figure 5e) and MCF7 cells (Supplementary Figure S4a). Similarly, luciferase readouts in PELP1 knockdown cells with p53-reporter plasmids were clearly compromised but could be rescued with transfection of shRNA-resistant, GFP-tagged PELP1 constructs (Supplementary Figure S4b). Chromatin immunoprecipitation (ChIP) analysis showed that PELP1 is recruited to the p53-binding sites on the promoter of p21 and PUMA (Figure 5f). More importantly, we found that the recruitment of p53 to these promoter sites upon genotoxic stress was reduced when PELP1 was knockdown in ZR75 cells (Figure 5g). Studies in the past have indicated the importance of c-terminal acetylation in the recruitment of p53 to target gene promoters.31 As PELP1 knockdown cells have reduced acetylation modification of p53, such reduction could contribute to reduced recruitment of p53 to the response elements.
Figure 5.
PELP1 interacts with p53 and modulates its transcriptional functions as a coactivator. (a) Total lysates from ZR75 cells treated with or without 10 Gy IR were subjected to immunoprecipitation using the PELP1 antibody and endogenous PELP1–p53 interaction was verified by western analysis. (b) Total lysates from MCF7 cells treated with or without camptothecin (1 μM) were subjected to immunoprecipitation using the PELP1 antibody and endogenous PELP1–p53 interaction was verified by western analysis. (c) Plasmids encoding GST-PELP1 and GFP-p53 were transiently transfected into HEK-293 cells. At 48 h after transfection, cells were exposed to IR (5 Gy). After 1.5 h, cells were harvested and lysed in NP-40 lysis buffer and subjected to GFP-trap IP followed by western analysis. (d) Nuclear extracts from ZR75 cells treated with γ-radiation were incubated with various deletions of PELP1 (GST-fusions) and the PELP1–p53 interaction site was determined using in vitro GST pull-down assays. (e) ZR75 cells were co-transfected with p53-reporter and β-gal reporter plasmids with increasing amount of GFP-PELP1 (WT). After 48 h, luciferase activity was measured and normalized with β-gal activity. Data shown are the means of ±S.E.M. performed in triplicate wells. (f) ChIP analysis was performed using IgG- or PELP1-specific antibodies and the status of PELP1 recruitment was analyzed by PCR using p21 and PUMA promoter-specific primers. (g) ZR75 cells expressing Con-shRNA or PELP1-shRNA1 were treated with γ-radiation and ChIP analysis was performed using p53-specific antibodies and the status of p53 recruitment was analyzed by using qRT-PCR with p21 and PUMA promoter-specific primers. *P<0.05; **P<0.01; ***P<0.001
PELP1 S1033A MT is defective in p53-mediated DDRs
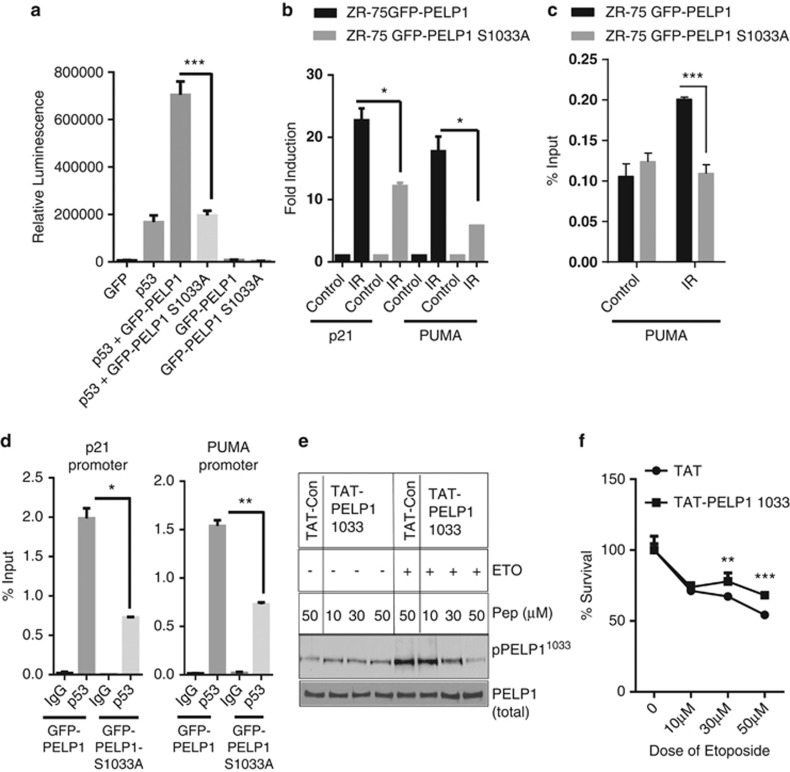
To further demonstrate the importance of PELP1 phosphorylation by DDR kinases, we tested the significance using p53 null cells (H1299, lung carcinoma cells). Cells were co-transfected either with PELP1-WT or PELP1-MT along with p53, and reporter gene assays were performed under conditions of genotoxic stress. PELP1-WT was able to activate p53-mediated transactivation function significantly more than the PELP1-S1033A-MT (Figure 6a). Next, we tested whether phosphorylation of PELP1 affects p53-mediated transactivation functions using a p53-reporter assay in ZR75 cells, with or without treatment of etoposide. PELP1 (WT) had the ability to increase p53-mediated transactivation functions upon genotoxic stress but PELP1S1033A mutation compromised PELP1 coactivator functions (Supplementary Figure S3c). Accordingly, expression of the p53 targets was significantly less in PELP1-MT-expressing cells than in PELP1-WT-expressing ZR75 cells (Figure 6b). To determine the mechanism, we examined whether PELP1 phosphorylation affects its ability to recruit to p53 target genes upon genotoxic stress. ChIP analysis revealed the ability of PELP1-MT to recruit to the p53 target gene PUMA was less than the ability of PELP1-WT upon genotoxic stress (Figure 6c). Similarly, recruitment of p53 to target gene promoter sites upon genotoxic stress was less in the PELP1S1033A-MT cells than in the PELP1-WT cells (Figure 6d). To demonstrate the in vivo significance of PELP1 phosphorylation, we have generated a cell permeable peptide inhibitor that efficiently inhibits PELP1 phosphorylation in vivo upon genotoxic stress. TAT-PELP1-Ser-1033 peptide contains 26-amino acids surrounding the PELP1Ser1033 phosphorylation site plus a 12-amino-acid TAT sequence that confers cell permeability. TAT-PELP1-1033 peptide efficiently reduced PELP1 phosphorylation upon genotoxic stress (Figure 6e). Cytotoxicity assays after etoposide treatment in the presence or absence of TAT-PELP1-1033 peptide inhibitor revealed that PELP1 inhibitor-treated cells have greater survival ability upon genotoxic agents compared with control TAT peptide (Figure 6f). Collectively, these results suggest that PELP1 phosphorylation at Ser-1033 by DDR kinases is needed for optimal DDR activity.
Figure 6.
Ser-1033 phosphorylation is needed for PELP1-mediated optimal p53 coactivation functions. (a) p53 null cells (H1299) were co-transfected with p53, p53-reporter and beta-galactosidase reporter plasmids with the GFP vector, GFP-PELP1 (WT) or GFP-PELP1S1033A (MT) plasmids. After 24 h of transfection, cells were treated with etoposide for 6 h and then luciferase activity was measured in the lysates and normalized with β-gal activity. Data shown are the means of ±S.E.M. performed in triplicate wells. ***P<0.001. (b) ZR75 cells expressing shRNA-resistant PELP1 WT or MT were transfected with PELP1-specific siRNA to downregulate endogenous PELP1. After 72 h, cells were treated with IR (10 Gy, 4 h), RNA was isolated and the status of p53 target genes was analyzed by RT-qPCR analysis. (c) ZR75 cells expressing GFP-PELP1-WT or GFP-PELP1-MT were treated with γ-radiation and ChIP analysis was performed using GFP epitope-specific antibodies and the status of PELP1 recruitment was analyzed by using RT-qPCR with PUMA promoter-specific primers. (d) ZR75 cells expressing GFP-PELP1-WT or GFP-PELP1-MT were treated with γ-radiation (10 Gy, 1 h) and ChIP analysis was performed using p53-specific antibodies and the status of p53 recruitment was analyzed by using RT-qPCR with p21 and PUMA promoter-specific primers. (e) ZR75 cells were pre-treated with indicated concentrations of TAT control or TAT-PELP1-1033 peptide. Cells were treated with or without etoposide and the PELP1 phosphorylation was analyzed via western blotting using the PELP1 Ser-1033 phospho antibody. (f) ZR75 cells were pre-treated with TAT control or TAT-PELP1-1033 peptide (50 μM) followed by treatment with different doses of etoposide for 48 h, and cytotoxicity was measured using MTT assays. Error bar depicts S.E. from triplicate experiments. *P<0.05; **P<0.01; ***P<0.001
Discussion
p53 status is an important determinant of cell fate when cells are exposed to chemotherapeutic drugs or genotoxic agents like γ-radiation or UV. Loss of p53 or its mutations are implicated both in the development and/or progression of cancer and in various types of cancers with the emergence of chemotherapy resistance.32, 33, 34 PELP1, a coregulator protein that interacts with various nuclear receptors, is a substrate of cyclin-dependent kinases and modulates the functions of cell cycle regulators to enhance G1/S cell cycle phase progression.25 In this study, we found that (a) PELP1 is phosphorylated at Ser-1033 after various genomic stresses including chemotherapeutic drugs and ionizing radiation; (b) PELP1 is redundantly phosphorylated by multiple DDR kinases and PELP1 phosphorylation at Ser-1033 is important for its p53-coactivation functions; (c) loss of PELP1 in p53 WT breast cancer cells decreases the apoptotic response to genomic stresses and causes a defect in the G1/S checkpoint; (d) PELP1 knockdown results in defective p53-activation and subsequently reduced expression of its key target genes; (e) PELP1 interacts with p53 and functions as a coactivator of p53 functions; and (f) PELP1 regulates the post-translational modification of p53 and thereby regulates its recruitment to the promoter of target genes. Collectively, these results suggest that PELP1 has diverse functions in activating p53-mediated transcriptional program.
Apoptotic response to various genotoxic agents is primarily mediated by p53, E2F, TGFβ or other apoptotic caspase cascades. All of the breast cancer model cells that we used in this study have WT p53. PELP1 knockdown increased the survivability of these cells when treated with various genotoxic agents including chemotherapeutic drugs and ionizing radiation. Mechanistic studies revealed that PELP1 functions as a coregulator of p53 and defective p53 activation in PELP1 knockdown cells is the primary reason for this phenotype. However, our studies did not eliminate the possibility of an impaired E2F-mediated apoptotic response in PELP1-knockdown cells. It will therefore be interesting to see whether PELP1 has a role in the apoptotic response of cells lacking p53 via E2F pathway and future studies are needed. PELP1 is known to modulate epigenetic changes at ER target gene promoters through histone modifiers like KDM1,24 and we speculate that PELP1 recruited to p53 target gene promoters may perform similar functions at p53 target genes upon genomic stress. The results from our study imply that PELP1 may function with p53 in safeguarding the genome from various types of stresses. As PELP1 has potential to act as a coregulator of p53 and augments cell death upon genomic stresses, it is most likely to be a protective mechanism that guarantees the genomic integrity.
The molecular mechanisms that underlie the seemingly disparate functions of PELP1 in cell cycle progression and DDR also remain elusive. However, the results from this study are reconcilable based on the notion that oncogene-mediated regulation of apoptosis is not completely unprecedented. For example, c-myc is an established oncogene in various types of cancer35, 36 however, overexpression of c-myc surprisingly sensitizes cells to apoptosis.37, 38, 39 E2F1 is another transcription factor that governs cell cycle progression and also regulates apoptosis under conditions of genotoxic stress.40 PELP1 overexpressing human breast cancer cells undergo significantly more apoptosis when treated with 9-cis-RA.27 Taken together, our data strongly support the idea that overexpression of PELP1 can sensitize cells to apoptosis via p53 pathway and that knocking down PELP1 in p53-WT cells may impair this process and increase survivability. As we observed increased sensitivity to genotoxic stress in p53 non-functional cells upon PELP1 knockdown, we speculate that during cancer development, an increase in PELP1's oncogenic functions is most likely to be associated with mutations in p53 and knocking down PELP1 functions in p53-deficient cancer is likely to have therapeutic benefits.
It is also well documented that overexpression of various oncogenes can result in an enhanced p53-mediated cellular response often termed as ‘Oncogene-Induced-Senescence (OIS)'. The DDR pathway has an essential role in the OIS through activation of ATM and other DDR kinases.41 It remains unknown whether PELP1 overexpression can similarly induce OIS and whether ATM-mediated phosphorylation has any role in the process. PELP1 proto-oncogene is overexpressed in multiple cancers, is an independent prognostic marker of reduced disease-free survival in breast cancer and is viewed as a viable therapeutic target against cancer.17 Our studies show that the cellular status of p53 represents a unique modifier to this qualification, and while this adds complexity, it also strengthens the concept of targeting PELP1. As recent studies have shown that p53 mutations or its inactivation is associated with invasive and metastatic cancers42 and because PELP1 also has a vital role in metastasis,17 it would be interesting to see how a MT p53-PELP1 axis governs the process of metastasis.
In summary, our study demonstrates for the first time that PELP1 functions as a p53-interacting protein and has a vital role in DDR pathway in breast cancer cells. Furthermore, we provided evidence that PELP1 phosphorylation by upstream DDR kinases, ATM, ATR or DNAPKc, are essential for optimal p53-mediated transcriptional program. These findings suggest that phosphorylation of PELP1 by upstream kinases constitutes an important step in ensuring genomic integrity after exposure to genotoxic agents.
Materials and Methods
Cell lines and reagents
Breast cancer cells MCF7, ZR75, and lung adenocarcinoma cell line H1299 were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained on either on RPMI or DMEM media with 10% fetal bovine serum (FBS) supplementation at 37°C and 5% carbon dioxide. Human fibroblast cells immortalized with SV40 antigen, GM00637 (ATM-proficient) and GM05849 (ATM function-deficient), human glioma cell lines M059K (DNAPKc-proficient) and M059J (DNAPKc-deficient) were purchased from ATCC and untransformed fibroblast from human Seckel syndrome patient GM018366 (ATR-deficient) and its matched control GM000200 were procured from Coriell Institute, Human Genetic Cell Repository (NIGMS, Camden, NJ, USA) and maintained on Eagle's minimum essential medium with Earle's salts, non-essential amino acids and 15% FBS as per the instructions from the repository. Details of GFP-tagged PELP1 plasmid was described.25 The PELP1 Ser1033Ala mutation on GFP-PELP1 backbone was generated using site-directed mutagenesis (Quick Change Mutagenesis Kit, Stratagene, La Jolla, CA, USA). p53-responsive pGL13-luc reporter construct with p53-binding sites was procured from Addgene (plasmid #16442; Cambridge, MA, USA). PELP1-specific shRNA (SureSilencing shRNA plasmids) and control shRNA vector with neomycin selection cassette were purchased from SA Biosciences (Qiagen, Germantown, MD, USA). ZR-75 controlshRNA and PELP1shRNA stable cell lines were generated through 500 μg/ ml G418 (neomycin) selection. MCF-7 controlshRNA and PELP1shRNA were generated using lentiviral transduction and the colonies were selected using puromycin. PELP1-phospho antibody was custom generated and affinity purified by Open Biosystems (Thermo-Fisher Scientifics, Huntsville, AL, USA) using phospho-PELP1 Serine-1033 (peptide sequence of PELP1: APTLAPEALP S(p)QGEVEREGES). TAT-control and TAT-PELP1-1033 peptides were custom synthesized by BioSynthesis (Lewisville, TX, USA). The sequences are: TAT-control-GRKKRRQRRRGG; TAT-PELP1-1033-GRKKRRQRRRGGRGADTAPTLAPEALPSQGEVEREGES. Etoposide, cisplatin (cis-diamineplatinum (II) dichloride), camptothecin and caffeine were purchased from Sigma Aldrich (St. Louis, MO, USA). Antibodies for Acetyl-p53, phospho-p53 and p53 (rabbit polyclonal) were purchased from Cell Signaling Technologies (Beverly, MA, USA). The antibody for PELP1 was purchased from Bethyl Laboratories (Montgomery, TX, USA). p53 (mouse monoclonal) antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Gamma-radiation
For radiation treatment, the exponentially growing cells were exposed to various doses of 137Cs – rays at a dose rate of 1.28 Gy/min at room temperature using a Gamma Cell-40 irradiator (Atomic Energy of Canada Ltd, Montreal, Canada). After the exposure, cells were returned to the 37 °C incubator and were either allowed to grow (for clonogenic assays) or harvested at different time periods of post-radiation for western blot analysis. Control cells were mock-irradiated (0 Gy) in a similar manner.
Cell lysis, immunoprecipitation and western blot analysis
For western analysis, lysates were prepared using a modified RIPA buffer (150 mM NaCl, 50 mM Tris-HCl, 50 mM NaF, 5 mM EDTA, 0.5% (wt/vol) sodium deoxycholate and 1% Triton X-100) containing phosphatase and protease inhibitors cocktails. For immunoprecipitation, lysates were prepared using a lysis buffer containing 50mM Tris-Hcl-pH7.5, 0.2% Triton X-100, 0.3% NP-40, 150 mM NaCl, 25 mM NaF, 0.1 mM sodium orthovanadate, and along with phosphatase and protease inhibitor cocktail and immunoprecipitation was done using PELP1 and p53 antibodies (Bethyl Lab, Montgomery, TX, USA). GFP Trap beads were purchased from Chromotek (Munich, Germany).
MTT and clonogenic assays
Model cells (2 × 103) were plated in six-replicates in 96-well tissue culture plate and allowed to attach overnight, and then treated with different doses of drugs for 48 h. Cell viability was measured using MTT assay. Toxicity was expressed as a percentage of control by dividing the average absorbance of each treated group by the average of the vehicle-treated controls. For clonogenic assays, cells were plated in six-well plates at a density of 1 × 103 cells per well in triplicates and treated with different doses of γ-radiation and allowed to grow for another 14 days. After 2 weeks, the cells were fixed with ice-cold methanol, and stained with 0.5% crystal violet. Image acquisition was done using a digital camera and clonogenic quantitation was performed by manual counting of colonies.
Quantitative RT-PCR analysis
Model cells with or without PELP1 expression were plated in 100 mm plates and treated with or without genotoxic drugs, etoposide (50 μM for 12 h), cisplatin (25 μM for 18 h) and camptothecin (10 μM for 12 h). Total RNA was isolated using Trizol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. cDNA synthesis was performed using Superscript III RT-PCR kit (Invitrogen). Real-time PCR was done using a Cepheid Smartcycler II (Sunnyvale, CA, USA) with specific real-time PCR primers for p21, GADD45, PUMA and actin. Results were normalized to actin transcript levels and the difference in fold expression was calculated using delta-delta-CT method. Primer sequences: p21F, 5′-CTGCCCAAGCTCTACCTTCC-3′ P21R, 5′-CAGGTCCACATGGTCTTCCT-3′ PumaF, 5′-ATGCCTGCCTCACCTTCATC-3′ PumaR, 5′-TCACACGTCGCTCTCTCTAAACC-3′ Gadd45AF, 5′-CTCAACGTCGACCCCGATAA-3′ GADD45AR, 5′-GCCTGGATCAGGGTGAAGTG-3′ p53F, 5′-TAACAGTTCCTGCATGGGCGGC-3′ p53R, 5′-AGGACAGGCACAAACACGCACC-3′ actin F, 5′-GTGGGCATGGGTCAGAAG-3′ actin R, 5′-TCCATCACGATGCCAGTG-3′.
Reporter gene and confocal microscopy assays
Reporter gene assays were performed using transient transfection of p53-responsive pGL3-luc construct with p53-binding sites, in six-well plates in triplicate using FuGENE6 method (Roche, Indianapolis, IN, USA) as described previously.25 Briefly, cells were transfected using 500 ng of pGL13-p53-luc, 10 ng pSV-β-galactosidase plasmid, with different dose of PELP1 WT or PELP1-S1033A (MT) expression plasmids. Cells were lysed in passive lysis buffer after 24 h of transfection, and the luciferase assay was performed using a luciferase assay kit (Promega, Madison, WI, USA). β-Galactosidase activity was analyzed by Galacto-light system (Applied Biosystems, Foster city, CA, USA). Data were normalized with β-gal activity for any variation in transfection. For confocal analysis, ZR75 breast cancer cells cultured on glass coverslips were treated with DMSO or camptothecin for 2 h followed by fixation, permeabilization and co-stained with antibodies against PELP1 (red) and p53/pS15-p53 (green) and colocalization was analyzed by confocal microscopy as described previously.24
Chromatin immunoprecipitation
The ChIP analysis was performed as described previously.24 In brief, model cells were grown in five 150 mm plates and after attaining a confluence of ∼75%, cells were irradiated with 10 Gy ionizing radiation and allowed to incubate at 37°C for 1–2 h. Cells were then cross-linked using formaldehyde and quenched by glycine. The chromatin was isolated and subjected to immunoprecipitation using the indicated antibodies. Isotype-specific IgG was used as a control. DNA was eluted and re-suspended in 50 μl of TE buffer and used for PCR amplification using the specific primers and primer sequence were taken from published reference.43, 44, 45
Acknowledgments
We thank all members of Vadlamudi lab for their critical reading and comments for the manuscript. This study was supported by the NIH/NCI grant CA095681 (RKV); NIH grants R01CA133093 and R01ES016354 (BX); NASA Grant NNX12-AC32G (MN); NIH/NINDS grant NS050730 (DB) and the Cancer Therapy and Research Center at the University of Texas Health Science Center at San Antonio through the NCI Cancer Center Support Grant P30CA054174-17.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by M Oren
Supplementary Material
References
- Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- Soussi T, Wiman KG. Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell. 2007;12:303–312. doi: 10.1016/j.ccr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Chehab NH, Malikzay A, Stavridi ES, Halazonetis TD. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci USA. 1999;96:13777–13782. doi: 10.1073/pnas.96.24.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Appella E, Anderson CW. Signaling to p53: breaking the posttranslational modification code. Pathol Biol (Paris) 2000;48:227–245. [PubMed] [Google Scholar]
- Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek DW, Anderson CW. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb Perspect Biol. 2009;1:a000950. doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- Coutts AS, La Thangue NB. The p53 response: emerging levels of co-factor complexity. Biochem Biophys Res Commun. 2005;331:778–785. doi: 10.1016/j.bbrc.2005.03.150. [DOI] [PubMed] [Google Scholar]
- Mann M, Zou Y, Chen Y, Brann D, Vadlamudi R. PELP1 oncogenic functions involve alternative splicing via PRMT6. Mol Oncol. 2013;13:10. doi: 10.1016/j.molonc.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard BJ, Daniel AR, Lange CA, Ostrander JH. PELP1: a review of PELP1 interactions, signaling, and biology. Mol Cell Endocrinol. 2014;382:642–651. doi: 10.1016/j.mce.2013.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadlamudi RK, Kumar R. Functional and biological properties of the nuclear receptor coregulator PELP1/MNAR. Nucl Recept Signal. 2007;5:e004. doi: 10.1621/nrs.05004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel AR, Gaviglio AL, Knutson TP, Ostrander JH, D'Assoro AB, Ravindranathan P, et al. Progesterone receptor-B enhances estrogen responsiveness of breast cancer cells via scaffolding PELP1- and estrogen receptor-containing transcription complexes Oncogene 2014. e-pub ahead of print 27 January 2014doi: 10.1038/onc.2013.579 [DOI] [PMC free article] [PubMed]
- Ravindranathan P, Lee TK, Yang L, Centenera MM, Butler L, Tilley WD, et al. Peptidomimetic targeting of critical androgen receptor-coregulator interactions in prostate cancer. Nat Commun. 2013;4:1923. doi: 10.1038/ncomms2912. [DOI] [PubMed] [Google Scholar]
- Nair SS, Mishra SK, Yang Z, Balasenthil S, Kumar R, Vadlamudi RK. Potential role of a novel transcriptional coactivator PELP1 in histone H1 displacement in cancer cells. Cancer Res. 2004;64:6416–6423. doi: 10.1158/0008-5472.CAN-04-1786. [DOI] [PubMed] [Google Scholar]
- Vadlamudi RK, Wang RA, Mazumdar A, Kim Y, Shin J, Sahin A, et al. Molecular cloning and characterization of PELP1, a novel human coregulator of estrogen receptor alpha. J Biol Chem. 2001;276:38272–38279. doi: 10.1074/jbc.M103783200. [DOI] [PubMed] [Google Scholar]
- Rosendorff A, Sakakibara S, Lu S, Kieff E, Xuan Y, DiBacco A, et al. NXP-2 association with SUMO-2 depends on lysines required for transcriptional repression. Proc Natl Acad Sci USA. 2006;103:5308–5313. doi: 10.1073/pnas.0601066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Nair SS, Nair BC, Cortez V, Chakravarty D, Metzger E, Schule R, et al. PELP1 is a reader of histone H3 methylation that facilitates oestrogen receptor-alpha target gene activation by regulating lysine demethylase 1 specificity. EMBO Rep. 2010;11:438–444. doi: 10.1038/embor.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair BC, Nair SS, Chakravarty D, Challa R, Manavathi B, Yew PR, et al. Cyclin-dependent kinase-mediated phosphorylation plays a critical role in the oncogenic functions of PELP1. Cancer Res. 2010;70:7166–7175. doi: 10.1158/0008-5472.CAN-10-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner E, Haindl M, Muller S. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 2011;30:1067–1078. doi: 10.1038/emboj.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RR, Gururaj AE, Vadlamudi RK, Kumar R. 9-Cis-retinoic acid up-regulates expression of transcriptional coregulator PELP1, a novel coactivator of the retinoid X receptor alpha pathway. J Biol Chem. 2006;281:15394–15404. doi: 10.1074/jbc.M601593200. [DOI] [PubMed] [Google Scholar]
- Lees-Miller SP, Sakaguchi K, Ullrich SJ, Appella E, Anderson CW. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven A, Heierhorst J. SQ/TQ cluster domains: concentrated ATM/ATR kinase phosphorylation site regions in DNA-damage-response proteins. Bioessays. 2005;27:397–407. doi: 10.1002/bies.20204. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, III, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Giannakakou P, Poy G, Zhan Z, Knutsen T, Blagosklonny MV, Fojo T. Paclitaxel selects for mutant or pseudo-null p53 in drug resistance associated with tubulin mutations in human cancer. Oncogene. 2000;19:3078–3085. doi: 10.1038/sj.onc.1203642. [DOI] [PubMed] [Google Scholar]
- Scata KA, El-Deiry WS. p53, BRCA1 and breast Cancer chemoresistance. Adv Exp Med Biol. 2007;608:70–86. doi: 10.1007/978-0-387-74039-3_5. [DOI] [PubMed] [Google Scholar]
- Aas T, Borresen AL, Geisler S, Smith-Sorensen B, Johnsen H, Varhaug JE, et al. Specific P53 mutations are associated with de novo resistance to doxorubicin in breast cancer patients. Nat Med. 1996;2:811–814. doi: 10.1038/nm0796-811. [DOI] [PubMed] [Google Scholar]
- Ponzielli R, Katz S, Barsyte-Lovejoy D, Penn LZ. Cancer therapeutics: targeting the dark side of Myc. Eur J Cancer. 2005;41:2485–2501. doi: 10.1016/j.ejca.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16:318–330. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Shi Y, Glynn JM, Guilbert LJ, Cotter TG, Bissonnette RP, Green DR. Role for c-myc in activation-induced apoptotic cell death in T cell hybridomas. Science. 1992;257:212–214. doi: 10.1126/science.1378649. [DOI] [PubMed] [Google Scholar]
- Albihn A, Loven J, Ohlsson J, Osorio LM, Henriksson M. c-Myc-dependent etoposide-induced apoptosis involves activation of Bax and caspases, and PKCdelta signaling. J Cell Biochem. 2006;98:1597–1614. doi: 10.1002/jcb.20816. [DOI] [PubMed] [Google Scholar]
- Guerra L, Albihn A, Tronnersjo S, Yan Q, Guidi R, Stenerlow B, et al. Myc is required for activation of the ATM-dependent checkpoints in response to DNA damage. PLoS One. 2010;5:e8924. doi: 10.1371/journal.pone.0008924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Herr W. E2F1 mediates DNA damage and apoptosis through HCF-1 and the MLL family of histone methyltransferases. EMBO J. 2009;28:3185–3195. doi: 10.1038/emboj.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- Hwang CI, Matoso A, Corney DC, Flesken-Nikitin A, Korner S, Wang W, et al. Wild-type p53 controls cell motility and invasion by dual regulation of MET expression. Proc Natl Acad Sci USA. 2011;108:14240–14245. doi: 10.1073/pnas.1017536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller PA, Vousden KH, Norman JC. p53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192:209–218. doi: 10.1083/jcb.201009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhang L, Jones KA. SKIP counteracts p53-mediated apoptosis via selective regulation of p21Cip1 mRNA splicing. Genes Dev. 2011;25:701–716. doi: 10.1101/gad.2002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.