Abstract
The p53 family of proteins has an important role in determining cell fate in response to different types of stress, such as DNA damage, hypoxia, or oncogenic stress. In recent years, p53 has also been shown to respond to metabolic stress, and to be induced by the AMP-activated protein kinase (AMPK), a central cellular energy sensor. A bioinformatic analysis revealed three putative AMPK phopshorylation sites in p73, a p53 tumor suppressor paralog. In vitro and in vivo assays confirmed that AMPK phosphorylates p73 on a novel residue, S426. Following specific pharmacologic stimulation of AMPK in cells, p73 protein half-life was prolonged leading to p73 accumulation in the nucleus. We show that p73 escaped the E3 ligase Itch resulting in reduced p73 ubiquitination and proteasomal degradation. Furthermore, chronic activation of AMPK led to apoptosis that was p73 dependent, but only in p53-expressing cells. Surprisingly, we found that p73 was required for p53 stabilization and accumulation under AMPK activation, but was dispensable under DNA damage. Our findings couple p73 with p53 in determining cell fate under AMPK-induced metabolic stress.
The p73 transcription factor belongs to a small but important family of proteins that includes p53 and p63. These family members cooperate as molecular hubs in a signaling network that coordinates cell fates such as cell death, proliferation, and differentiation. The high degree of similarity in the DNA binding domain of all the members suggests that they share certain target genes. Indeed, in response to DNA damage, p73 induces many p53-target genes responsible for triggering cell death in a p53-independent manner.1, 2 Under this physiological setting, each of the p53 paralogs appears to function in parallel modes in potentiating tumor suppression functions.
The expression of p73 is tightly regulated to accommodate effective response to genomic insults.3 At the level of mRNA, the p73 gene encodes multiple proteins originating from alternative promoter usage and alternative splicing events.4 At the protein level, several posttranslational modifications stimulate the accumulation and activation of p73.5 A unique p73 modification that is not shared by p53 is phosphorylation by the non-receptor tyrosine kinase c-Abl. Following γ-radiation (IR) or treatment with the chemotherapeutic drug cisplatin, c-Abl interacts with and phosphorylates p73. Tyrosine-phosphorylated p73 accumulates and induces proapoptotic target genes.6, 7, 8 The accumulation and activation of p73 is also controlled by several other kinases, including ATM,9, 10 IKK,11 Chk1/2,12, 13 p38 MAPK,14 JNK,15 and PKC.16
An important regulator of p73 protein level is the E3-ligase Itch that binds and polyubiquinates p73, thereby triggering its proteasomal degradation.17 The Itch-p73 axis is regulated on DNA damage to ensure the accumulation of p73. At the transcriptional level, Itch is regulated by Runx–Yap complex and under DNA damage stress Itch level is downregulated via a mechanism involving Yap tyrosine phosphorylation by c-Abl.18 In addition, Yap regulates p73 accumulation by negating Itch association with p73.19, 20, 21 The protein steady-state level of p73 is also controlled by ubiquitin-independent degradation by the 20S proteasome.22 Interestingly, degradation of p73 by this pathway is inhibited by the NAD(P)H quinone oxidoreductase 1 (NQO1) in a manner that depends on the presence of the cofactor NADH. These observations may link p73 protein accumulation to the redox state of the cell. It remains, however, an open question whether and to what extent the metabolic state of the cell regulates p73 accumulation.
A well-known regulator of cellular energy and metabolism is the AMP-activated protein (AMPK), which is activated in response to energy crisis. During cellular energy depletion when the AMP/ATP ratio rises,23 AMPK is phosphorylated and activated by the upstream kinase LKB1 (also known as STK11). The LKB1-AMPK pathway serves as a metabolic checkpoint in the cell, favoring restoration of energy through upregulation of catabolic processes and shutdown of ATP-consuming processes, and arresting cell growth.24 Given that cellular transformation is accompanied by marked changes in energy metabolism, interest in the role of the LKB1-AMPK axis in cancer has grown substantially. In fact, LKB1 was identified as the tumor suppressor gene mutated in patients with the inherited cancer disorder Peutz–Jegher syndrome.25, 26 Loss of AMPK activation is, therefore, thought to support the development of malignancy.27 For instance, AMPK activation by the drugs metformin or AICAR suppresses naturally arising tumors in transgenic mice and in carcinogen-treated rodent cancer models, and inhibits the growth of a wide range of tumor cells in culture.24, 28 The ability of AMPK not only to reprogram energy metabolism but also to enforce a metabolic checkpoint on growing cells, relies partially on p53 induction. In response to AMPK activation, p53 accumulates and activates target genes involved in programming growth arrest, senescence, and apoptosis, depending on the cell type.29, 30, 31
Here we demonstrate that p73 is a direct substrate of AMPK. We found that AMPK supports p73 accumulation by inhibiting p73 proteasomal degradation. Remarkably, we further found that AMPK-dependent p53 accumulation is p73 dependent. This study demonstrates a tight interdependent linkage between p73 and p53 in determining cell fate decision under metabolic stress as opposed to the parallel activity of these proteins under DNA damage.
Results
p73α is an AMPK substrate
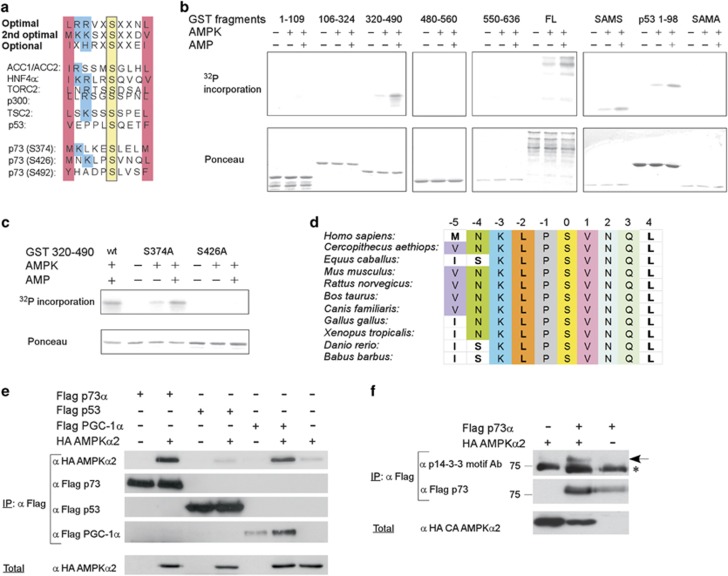
Biochemical and bioinformatics studies have identified the optimal motif for AMPK-mediated phosphorylation.32, 33 Inspection of the p73α sequence revealed that it contains three putative AMPK phosphorylation sites (Figure 1a). To examine these sites, we performed in vitro kinase reactions using purified AMPK and bacterially expressed recombinant full-length p73α and various p73 protein fragments. We obtained AMPK-induced phosphorylation of p73α, which was further increased by adenosine 5′-monophosphate sodium salt (AMP) (Figure 1b). The SAMS peptide, a synthetic peptide substrate specific for AMPK, and a fragment of p53 containing S15 were used as positive controls, and SAMA peptide that bears a mutation in the consensus site was used as a negative control. Only the full-length p73α and a 320–490-aa p73 fragment were phosphorylated. Two AMPK consensus sites were found inside this fragment and the suspected serine residues were mutated to alanine. Both the wild-type and the S374A fragments were phosphorylated to the same extent (Figure 1c). In contrast, the S426A fragment was not phosphorylated by AMPK, suggesting that S426 is a direct target of AMPK. Importantly, the residues flanking p73 S426, which constitute the consensus phospho-AMPK substrate motif, are highly conserved through different p73 orthologs, suggesting that p73 is an ancestral AMPK target (Figure 1d).
Figure 1.
AMPK phosphorylates p73α in vitro and in cells. (a) Optimal and optional AMPK phosphorylation substrate motifs. AMPK phosphorylation sites in several established AMPK substrates. Predicted AMPK phosphorylation sites in p73α. (b) Purified recombinant GST-p73α fragments and full length (FL) were incubated with purified AMPK and phosphorylation was determined by incorporation of (γ-32P)ATP. Ponceau-S staining is shown to visualize the recombinant fragments. (c) GST-p73 320–490 fragment contains two potential AMPK phosphorylation consensus sites in which their putative serines were individually mutated to alanine and subjected to an in vitro phosphorylation assay as in (b). (d) Multiple alignment of the AMPK phosphorylation consensus sites in p73 orthologs. (e) Co-expression of p73α, p53, and PGC-1α with AMPKα2 subunit in HEK-293 cells. Flag-tagged p73α, p53 or PGC-1α were IP using flag beads, and co-precipitated AMPKα2 was analyzed using anti-HA antibody. (f) Flag-tagged p73 was co-expressed with HA-tagged constitutively active (CA) AMPKα2 subunit in HEK-293. Flag beads were used for IP, and p73 phosphorylation was detected using the phospho-14-3-3 motif antibody. An asterisk marks nonspecific cross-reacting bands, and an arrow points to phospho-p73
To examine whether p73α is an AMPK substrate in cells, we first performed coimmunoprecipitation experiments to validate their interaction and used as controls PGC-1α and p53, as both are well-known AMPK substrates. We found that PGC-1α and AMPK coimmunoprecipitated as previously reported,34 and that p73α and AMPK also form a complex in cells (Figure 1e). Surprisingly, however, although p53 was reported to be a direct substrate of AMPK in-vitro,29 it did not interact with AMPK (Figure 1e). Next we asked whether p73α is phosphorylated by AMPK in cells. To this end we used a phospho-motif antibody approach to recognize phosphorylated residues within a specific sequence motif.35 It has been reported that a ‘14-3-3 motif' antibody, which was generated against peptides bearing R-X-X-(X)-pS sequences, is useful in detecting AMPK-phosphorylated substrates.32 We found that this antibody recognized p73α in HEK-293 cells when co-expressed with a constitutively active form AMPK (Figure 1f), and with the AMPK-upstream kinase LKB1 (Supplementary Figure S1). These data suggest that p73 is an AMPK substrate in vitro and in cells.
Active AMPK supports p73α accumulation
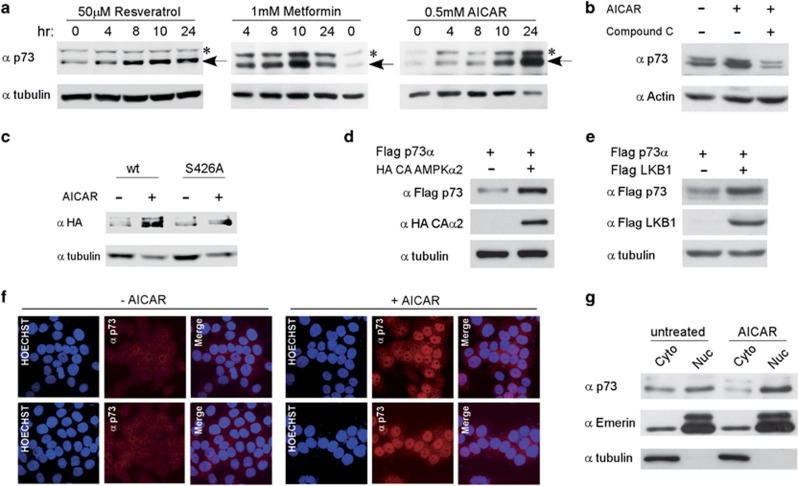
AMPK is activated by resveratrol, a natural polyphenolic compound,36 by the antidiabetic therapeutic drug metformin,37 and by AICAR, which is metabolized to AICAR-PO4 (ZMP), a 5′-AMP analog. Treating HCT116 cells with resveratrol, metformin, or AICAR resulted in a gradual increase in endogenous p73α protein levels (Figure 2a) the p73 alpha isoform was specifically increased by this treatment (Supplementary Figure S2). The mRNA levels of p73 did not increase in response to the treatments, but rather decreased, suggesting that the accumulation of p73α protein is not due to elevated transcription (Supplementary Figure S3). In subsequent experiments, we used only the AMP-mimetic AICAR, as it is considered to be a more direct activator of AMPK, and it also does not perturb the cellular levels of ATP, ADP, or AMP.38 AICAR-induced p73α accumulation was compromised when compound C, a partially selective pharmacological AMPK inhibitor,37 was added, validating the involvement of AMPK kinase activity in p73α accumulation (Figure 2b). Furthermore, wild-type p73α was increased to a greater extent than p73α S426A mutant, suggesting that direct phosphorylation by AMPK is responsible for p73α protein accumulation (Figure 2c). Co-expression of p73α and a constitutively active form of the α2 subunit of AMPK also resulted in the accumulation of p73 (Figure 2d), further validating the involvement of AMPK activity in p73 accumulation. LKB1 is an upstream kinase of AMPK.39, 40 Remarkably, LKB1 overexpression supported p73α accumulation (Figure 2e), suggesting that p73α is a target of the LKB1-AMPK pathway.
Figure 2.
Activation of AMPK results in p73 accumulation and localization in the nucleus. (a) HCT116 cells were treated with the indicated drugs for different time points. Cells were lysed together and subjected to immunoblot analysis for endogenous p73 protein. Asterisks mark nonspecific cross-reacting bands. (b) AMPK inhibitor, compound C (10 μM), was added an hour before the addition of 0.5 mM AICAR. Cells were lysed and analyzed by immunoblotting after 24 h. (c) HCT116 cells transiently expressing either HA p73α (wt) or HA p73α S426A were treated with 0.5 mM AICAR for 24 h, and analyzed by immunoblotting. (d) Co-expression of Flag-tagged p73α with HA-tagged constitutively active AMPKα2 in HEK-293 cells. Cell lysates were analyzed by immunoblotting 24 h post transfection. (e) Co-expression of Flag-tagged p73α with Flag-tagged LKB1 in HEK-293 cells. Cell lysates were analyzed by immunoblotting 24 h post transfection. (f) Immunofluorescence analysis of endogenous p73 protein in HCT116 cells that were either untreated (−AICAR), or treated with 0.5 mM AICAR for 24 h. Rhodamine red-X-conjugated secondary antibody was used to detect the primary anti-p73 antibody (red channel), and Hoechst was used to visualize the nuclei (blue channel). The cells were visualized by confocal laser microscope, and representative optical fields are shown. (g) HCT116 cells were either untreated or incubated with 0.5 mM AICAR for 24 h followed by subcellular fractionation. Equivalent cell lysates from cytoplasmic and nuclear fractions were analyzed by western blotting using the indicated antibodies. Tubulin and emerin were analyzed to indicate the purity of the cytoplasmic and nuclear fractions, respectively
p73α contains both a nuclear localization signal and a nuclear export signal,41 suggesting that p73α is regulated at the level of subcellular localization. In response to DNA damage, p73α accumulates in the nuclei in targeting growth-arrest and apoptotic genes.42, 43, 44, 45 To examine p73α cellular localization under AMPK activation, we utilized both microscopic and biochemical approaches. Microscopically, endogenous p73 was scarcely detected in untreated cells, but after AICAR treatment the accumulated p73 was predominantly nuclear (Figure 2f). Similar results were obtained by biochemical subcellular fractionation assays (Figure 2g), suggesting that p73α under metabolic stress accumulates in the nucleus.
AMPK inhibited p73α ubiquitination by Itch
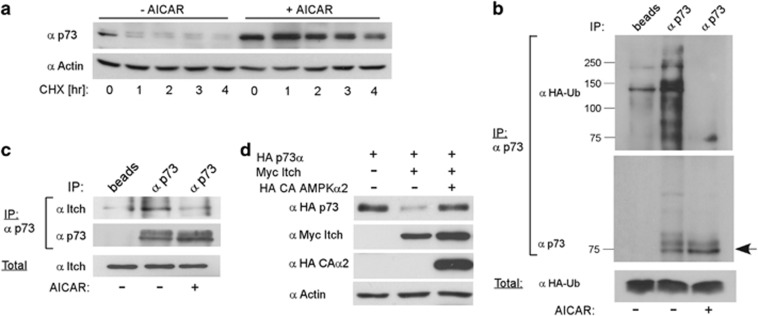
To examine whether p73α accumulation is the result of protein stabilization, we measured p73α decay with and without AMPK induction. HCT116 cells were treated with AICAR for 24 h, then cycloheximide (CHX) was added to inhibit protein synthesis, and cells were collected at the indicated time points (Figure 3a). Remarkably, AICAR treatment resulted in a marked increase in p73α protein half-life, as compared with the untreated cells. Ubiquitinated p73α accumulated in the presence of MG132, a proteasome inhibitor, suggesting ubiquitin-dependent degradation of the endogenous p73α. However, in the presence of AICAR, no ubiquitinated p73α accumulated (Figure 3b), suggesting that AICAR inhibited p73α ubiquitin-dependent degradation.
Figure 3.
AMPK stabilizes p73 by protecting it from Itch-mediated proteasomal degradation. (a) HCT116 cells were treated with 0.5 mM AICAR for 24 h and then 20 μg/ml CHX was added. Cells were collected at different time points and analyzed by immunoblotting to measure p73 protein half-life. (b) HCT116 cells were transiently transfected with a plasmid encoding HA-Ubiquitin (HA-Ub), 24 h later the cells were treated with 0.5 mM AICAR for another 24 h, followed by addition of MG132 (25 μM, 2 h). Lysates were subjected to IP with anti-p73 antibody for 3 h and collected on protein A/G beads overnight (untreated cell lysate was incubated only with the beads, and used as a control). The immunoprecipitants were analyzed by western blot using the indicated antibodies. An arrow indicates the migration size of the non-modified p73. (c) HCT116 cells were either untreated or treated with 0.5 mM AICAR for 24 h. Subsequently, 25 μM MG132 was added for 2 h to all plates. IP and analysis was carried out as in (b). (d) HA-tagged p73α was overexpressed in HEK-293 cells alone, together with Myc-tagged Itch, or with addition of HA-tagged constitutively active AMPKα2. Lysates were prepared 24 h post transfection and analyzed by immunoblotting
The Itch E3 ligase binds and ubiquitinates p73α and induces its rapid proteasomal degradation.17 Therefore, we examined the association between p73α and Itch under AMPK activation. Remarkably, p73α association with Itch was markedly reduced following AICAR treatment (Figure 3c). To validate the involvement of AMPK activity, we co-expressed p73α and Itch in the presence or absence of the constitutively active AMPKα2 subunit, and analyzed p73α steady-state protein levels. We found that activated AMPK markedly inhibited Itch-mediated p73α degradation (Figure 3d), suggesting that active AMPK negated the p73-Itch axis.
p73 supports AICAR-induced apoptosis
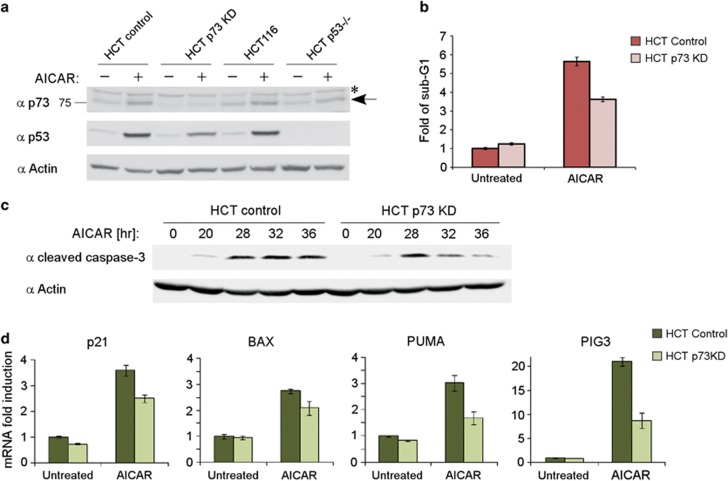
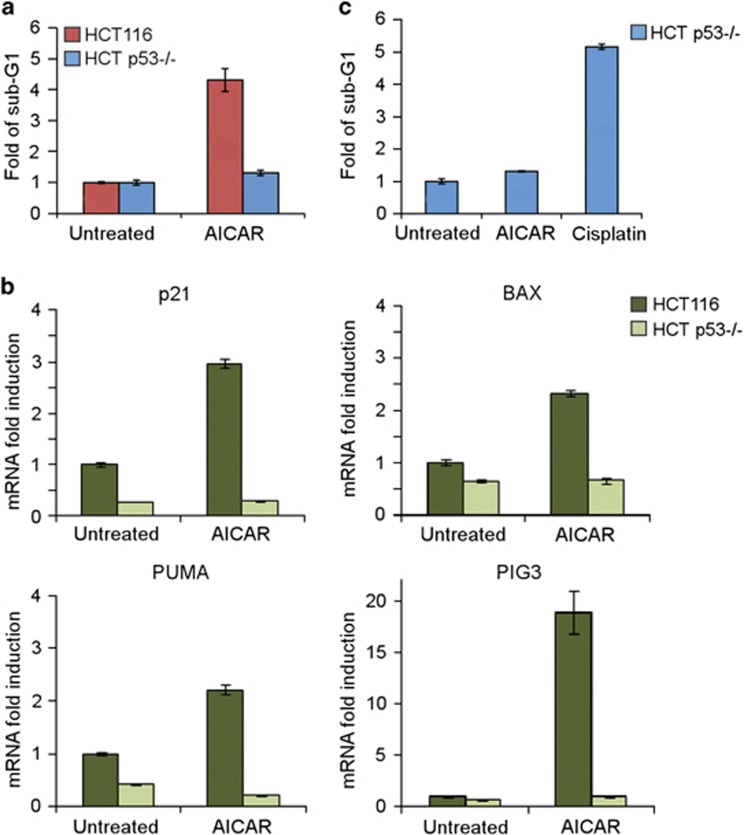
Chronic activation of AMPK in fast-growing and cancer cells often leads to apoptosis.46 We asked whether p73α is involved in the apoptotic response induced by AICAR-mediated activation of AMPK. To this end, we used a stable cell line constitutively expressing a p73 knockdown shRNA cassette, and compared it with a control non-silencing shRNA-expressing cell line. A marked decrease in the level of p73α was obtained in p73 knockdown cells both under basal conditions and under AICAR treatment (Figure 4a). In agreement with previous publications, the level of endogenous p53 was elevated following AMPK activation by AICAR.29, 30 Unexpectedly, in cells stably expressing p73-specific shRNA, p53 accumulation was attenuated (see below). AICAR-induced apoptosis, as detected by sub-G1 fraction, was compromised in p73 knockdown cells (Figure 4b). Similar results were obtained using another experimental setup in which double staining of annexin V and propidium iodide was analyzed (Supplementary Figure S4). As another means to analyze apoptosis, we probed for the cleaved caspase-3 as an indicator of caspase-3 activation. A time course experiment revealed that caspase-3 was activated 20 h after the addition of AICAR and in the control cells remained highly active up to 36 h (Figure 4c). In contrast, in the p73 knockdown cells, the process was transient and the level of active caspase-3 dropped after 28 h. Similar results were obtained in a cell line constitutively expressing a different p73 shRNA cassette (Supplementary Figure S5). Next, we quantified the RNA level of certain key p53/p73 target genes involved in cell cycle arrest and apoptosis, such as p21, Bax, Puma, and PIG3. The mRNA levels were induced by AICAR treatment, suggesting that the accumulated p73α and p53 proteins are functional (Figure 4d). In the p73 knockdown cells, however, the expression of the target genes was attenuated. These data suggest that p73α is required to potentiate and sustain apoptosis in response to AICAR treatment.
Figure 4.
p73 supports the apoptotic response induced by AMPK activation. (a) HCT116, HCT p53−/−, and HCT116 cells stably expressing a non-silencing cassette (control) or an shRNA directed against p73 (p73 KD) were either untreated or incubated with 0.5 mM AICAR for 24 h. Lysates were then analyzed by immunoblotting. An asterisk marks a nonspecific cross-reacting band, and an arrow points to the endogenous p73 band. (b) Apoptotic sub-G1 population analysis by flow cytometry. HCT116 cells stably expressing a non-silencing cassette (control) or an shRNA directed against p73 (p73 KD) were either untreated or incubated with 0.5 mM AICAR for 25 h. Represented is fold induction of the sub-G1 population over the untreated HCT control cells. Data are represented as mean±S.D (n=3). (c) HCT116 cells stably expressing a non-silencing cassette (control) or an shRNA directed against p73 (p73 KD) were incubated with 0.5 mM AICAR and collected at different time points, as indicated. Cell lysates were analyzed by immunoblotting, and actin served as a loading control. (d) Cells were treated the same as in (b). The expression levels of the indicated mRNA were analyzed by real-time PCR. Data are represented as mean±S.D. (n=3)
p53 is essential for AICAR-mediated apoptosis
We next wanted to examine the contribution of p73 to AICAR-induced apoptosis in a p53-null background. To this end we used HCT116 p53−/− cells. Surprisingly, however, no apoptosis induction was detected in HCT116 p53−/− cells, compared with HCT116 cells that exhibited about fivefold induction of apoptosis (Figure 5a). The critical p53 role was also evident at the RNA level of p53 target genes. These genes were induced by AICAR only in HCT116 cells, but not in HCT p53−/− cells (Figure 5b). The high dependency of apoptosis on p53 was not observed under DNA damage stress induced by cisplatin. HCT116 p53−/− cells treated with cisplatin underwent apoptosis rather efficiently compared with HCT p53−/− cells treated with AICAR (Figure 5c). This was also evident in the p53 null cell line H1299 (Supplementary Figure S6). These data suggest that under AMPK activation, p53 is essential in stimulating apoptosis.
Figure 5.
AICAR treatment results in p53-dependent apoptosis. (a) Apoptotic sub-G1 population analysis by flow cytometry of HCT116 or HCT p53−/− cells treated with 0.5 mM AICAR for 48 h. Represented is fold induction relative to the sub-G1 population of the untreated state in each cell line. Data are represented as mean±S.D. (n=3). (b) HCT116 and HCT p53−/− cells were either untreated or incubated with 0.5 mM AICAR for 24 h. Expression levels of the indicated mRNAs were quantified by real time PCR. (c) Apoptotic sub-G1 population analysis by flow cytometry of HCT p53−/− cells treated with either 0.5 mM AICAR or 25 μM cisplatin for 48 h. Represented is fold induction of the sub-G1 population over the untreated cells. Data are represented as mean±S.D. (n=3)
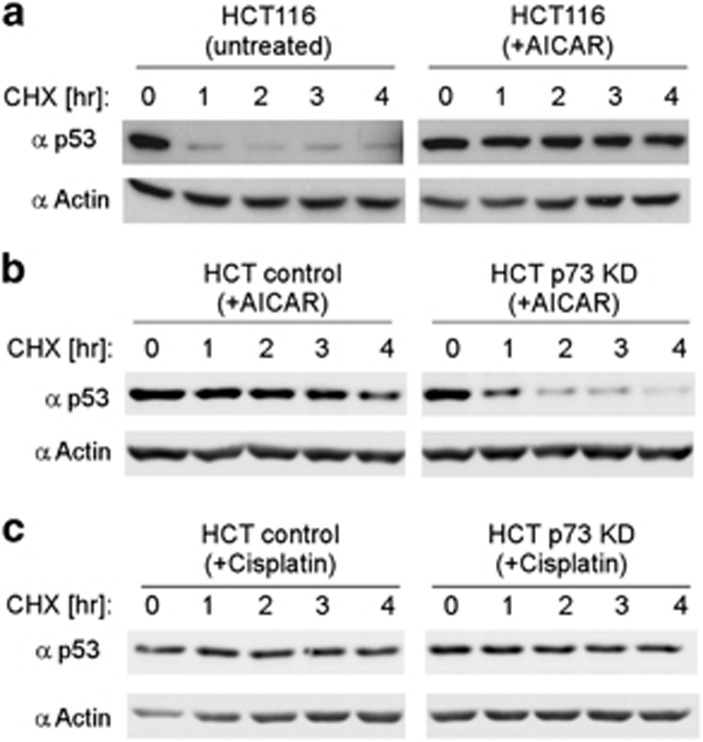
AICAR-induced p53 accumulation is p73 dependent
As shown above (Figure 4a), in p73 knockdown cells, p53 accumulation in response to AICAR treatment was attenuated. Similar results were also obtained with a cell line constitutively expressing a different p73 shRNA cassette (Supplementary Figure S7). Interestingly, in the AICAR-treated p73-depleted cells, phosphorylation of p53 on S15, which disrupts the p53–MDM2 interaction and enhances its stability,47, 48 was reduced (Supplementary Figure S7). In HCT116-untreated cells, p53 half-life was very short, <1 h (Figure 6a). In sharp contrast, under AICAR treatment, p53 was highly stable up to 4 h, the last measured time point. The finding that p53 escapes degradation and accumulates to high levels is consistent with previous observations.29, 30 Remarkably, p53 accumulation was severely reduced in p73 knockdown cells (Figure 6b), suggesting that under AMPK activation, p53 accumulation is p73 dependent. In sharp contrast, p53 stability was refractory to the p73 knockdown following cisplatin treatment (Figure 6c). These results indicate that AICAR-mediated p53 stabilization depends on p73, and can account for the reduced apoptotic response that we observed in p73 knockdown cells. Taken together, our findings suggest that under metabolic but not DNA damage stress p53 accumulation requires p73.
Figure 6.
p73 is required for p53 stabilization under AICAR but not under cisplatin treatment. (a) HCT116 cells were either untreated or incubated with 0.5 mM AICAR for 24 h. CHX (20 μg/ml) was then added, and cells were collected at different time points and analyzed by immunoblotting to measure p53 protein half-life. Actin serves as a loading control. (b) HCT116 cells stably expressing a non-silencing cassette (control) or an shRNA directed against p73 (p73 KD) were treated with 0.5 mM AICAR for 24 h. CHX (20 μg/ml) was then added, and cells were collected at different time points to measure p53 protein half-life. Actin serves as a loading control. (c) Same as in (b), only here cells were treated with 25 μM cisplatin for 24 h
Discussion
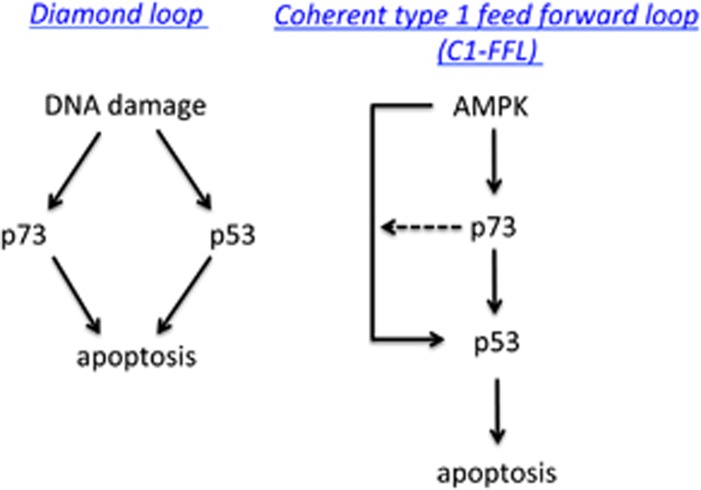
In this study, we provide evidence for p73 being a downstream target of the metabolic LKB1-AMPK signaling pathway. We found that AMPK directly phosphorylates p73 on S426. Furthermore, pharmacologic stimulation of AMPK by AICAR in cells led to p73 accumulation, which was mediated partially by direct phosphorylation of p73 on S426. The underlying mechanism for AMPK-mediated p73 accumulation was escaping the E3 ligase Itch and ubiquitin-dependent proteasomal degradation. By using a p73 knockdown approach, we discovered that p73 supports AICAR-induced apoptosis by positively regulating the stabilization and accumulation of p53. Our findings therefore demonstrate that AMPK couples p73 with p53 in regulating cell fate. Moreover, under these conditions, p53 acted as a critical cell fate decision maker, because apoptosis was not induced in its absence. In addition, accumulation of p73 was not sufficient to elicit apoptosis in the absence of p53. This is different from the cellular response to DNA damage, in which p53 and p73 can act in parallel routes to induce apoptosis. Thus, our study establish a new coherent type 1 feed forward loop (C1-FFL) between p73 and p53, which is different from the diamond pattern that is mostly observed under DNA damage (Figure 7).
Figure 7.
Proposed generalized model for differential induction of apoptosis by p53 and p73 under DNA-damage versus metabolic stress. In response to DNA damage, p73 can activate the apoptotic pathway in the absence of p53, and thus backup p53 function (diamond pattern). In contrast, the apoptotic response under metabolic stress requires the presence of p53, and p73 serves to support its function (C1-FFL)
Accumulation of p53 was previously observed under both pharmacological and physiological activation of AMPK. This accumulation is a result of phosphorylation of serine residues within the trans-activation domain of p53 that negates the p53-MDM2 axis.49 It was shown through in vitro assays that AMPK directly phosphorylates both S15 and S20, however, in cells, phosphorylation of S15 and S46 was detected.30, 31 Here we found that AICAR-mediated AMPK activation in p73 knockdown cells resulted in reduced p53 S15 phosphorylation. Looking at the sequence surrounding S15 reveals that there are no basic residues in neither -4 nor -5 positions (Figure 1a), making this site a less favorable AMPK-phsophorylation substrate motif. Moreover, under conditions in which we detected coimmunoprecipitation between AMPK and p73, we could not detect coimmunoprecipitation between AMPK and p53 (Figure 1e). This is in agreement with a previous report demonstrating that, although AMPK interacts with the C-terminal domain of p73, it does not interact with p53.50 It is possible therefore that p73 improves the substrate preference of AMPK toward p53 (dashed arrow in Figure 7).
In the absence of p53, p73 accumulated to the same level under AMPK activation, suggesting that the AMPK-p73 axis is upstream of p53. Nevertheless, it was surprising that the accumulated p73 was not able to activate transcription of p53-responsive genes and to promote apoptosis in the absence of p53, especially because p73 is known to act independently, at least under DNA damage response and when overexpressed exogenously.43, 51 Moreover, p73 contributes to chemotherapy-induced apoptosis, and dysregulation of p73 transcriptional activity by mutant p53 confers chemoresistance.52, 53 Interestingly, it was previously shown using luciferase assays that AMPK specifically represses p73-induced transcription.50 This may suggest that AMPK suppresses one function of p73, induction of p53-responsive genes, whereas activating another new function of p73, regulating p53 stability. Nevertheless, the fact that p73 was stabilized and accumulated in the nucleus in response to AMPK suggests that further investigation is needed to fully reveal the more direct function of p73 in this pathway. Another possible reason for the inability of p73 to activate transcription of p53-responsive genes might be that c-Abl, which is known to phosphorylate p73 and stimulate its transcription activity under DNA damage, is not induced under these conditions.
It appears there is a difference between the effect of AMPK activation in normal versus cancer cells, and between physiological versus rapid pharmacological activation of AMPK, with the former being antiapoptotic and the later resulting in a proapoptotic effect.24, 46 Our proposed C1-FFL may therefore lie on a decision-making interface between a pro-survival and a proapoptotic response that depends on the severity of the AMPK pathway activation. Owing to the growing interest in therapeutically manipulating the AMPK pathway to treat cancer, it is important to further understand how this new regulatory AMPK-p73-p53 loop is regulated and affects cell fate decisions.
Materials and Methods
Cell culture, plasmids, and transfection
HEK-293, HCT116, HCT p53−/−,54 and H1299 cell lines were grown and maintained as previously described.19 The plasmids employed were as follows: pCDNA3 Flag-p73α, pCDNA3 HA-p73α, pCDNA3 HA-p73α S426A, pCDNA3 Flag-LKB1, pCDNA3 HA-AMPKα2, constitutively active pCDNA3 HA-AMPKα2 (1-313, T172D), pRK5 Myc-Itch, and pCDNA3 HA-Ubiquitin. Transient transfections were carried out by the calcium phosphate method in the case of HEK-293 cells, or with jetPEI (Polyplus-transfection SA, Illkirch, France) in the case of HCT cells. Stably expressing control or p73 knockdown HCT cells were created by using the GIPZ Lentiviral shRNAmir system (Open Biosystems, Huntsville, AL, USA), or by using the pLKO shRNA system: ‘2nd clone' (Sigma, St. Louis, MO, USA).
Compounds
AICAR (Biaffin, Kassel, Germany) was dissolved in water to make a final stock solution of 100 mM. MG-132 (Sigma) was dissolved in DMSO to make 10 mM final stock solution. Cisplatin was purchased from ABIC (Jerusalem, Israel). Resveratrol, metformin, and AMP were purchased from Sigma.
In vitro phosphorylation assay
GST-fused constructs were expressed in E. coli using the IPTG induction system, and purified by affinity resin on glutathione agarose (Sigma). Kinase assay was performed according to the manufacturer's protocol using AMPK partially purified from rat liver (Millipore, Billerica, MA, USA; Upstate). (γ-32P)-ATP was purchased from PerkinElmer (Waltham, MA, USA).
Apoptosis analysis
Cells were collected by trypsinization together with growth medium and floating cells, and washed once with PBS. Cells were fixed in cold 70% ethanol, and then rehydrated in PBS for 1 h on ice. The cells were then resuspended in PBS containing 50 μg/ml RNase (Sigma) A and 25 μg/ml propidium iodide (Sigma). In each experiment, at least 50 000 cells were collected by FACScan and analyzed with the Cellquest program (BD CellQuest Pro Analysis program). In analyzing sub-G1 populations, biological triplicates were prepared for each sample. Data are represented as mean±S.D. (n=3). For analysis of AnnexinV/PI double staining, we used the Annexin V-APC Apoptosis Detection Kit (eBioscience, San Diego, CA, USA) according to the manufacturer' instructions. For each measurement, at least 100 000 cells were acquired on FACScan flow cytometer (Becton Dickinson, Mountain View, CA, USA) and analyzed with BD FACSDiva software (Becton Dickinson).
CHX half-life experiment
Cells were treated with 20 μg/ml CHX (Sigma) 24 h after the addition of AICAR or cisplatin. Cells were then collected at different time points, and analyzed together by immunoblotting.
Immunoprecipitation and protein analysis
Immunoprecipitation was performed using anti-HA or anti-Flag-agarose beads (Sigma), or by incubating the extracts with rabbit polyclonal antibody against p73 (BL906, Bethyl Laboratories, Montgomery, TX, USA) for 3 h and then incubating with protein A/G agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for an additional 2 h. Protein extraction and immunoblotting were done as previously described.19 The antibodies used were as follows: rabbit monoclonal antibody anti-cleaved Caspase-3 (Asp175, Cell Signaling, Beverly, MA, USA), monoclonal mouse anti-actin, anti-HA, anti-Flag (Sigma), mouse anti-p53 1801, mouse anti-Myc (made in the Weizmann Institute).
Fluorescence microscopy
Before cell fixation, nuclei were stained with Hoechst 33342 (Molecular Probes, Carlsbad, CA, USA) by brief incubation. The cells were then washed with PBS and fixed with 4% paraformaldehyde for 30 min at room temperature. Fixed cells were permeabilized with 0.5% Triton-X-100 in PBS for 25 min at room temperature, washed with PBS containing 0.2% Tween 20 (PBS-T), and blocked with fetal calf serum containing 10%(v/v) skim milk and 0.2%(v/v) Tween 20 for 45 min. Cells were then incubated with a polyclonal rabbit anti-p73 antibody (diluted 1 : 200) in PBS-T-containing 10% skim milk for 2 h, washed six times for 5 min with PBS-T, and incubated with a Rhodamine red X-conjugated donkey anti-rabbit antibody (Jackson Immunoresearch Laboratories, West Grove, PA, USA; diluted: 1 : 140) for 1 h. The cells were washed as above, and the coverslips were mounted in Aqua-polymount (Polysciences, Eppelheim, Germany). Microscopic images were obtained using a Nikon Eclipse TE300 microscope equipped with Radiance 2100 confocal laser scanning system (Bio-Rad, Hercules, CA, USA) and managed by Laser Sharp 2000 software (Zeiss, Munich, Germany). Representative images with identical laser intensities were taken from each sample.
Subcellular fractionation
Cell pellets were resuspended in five volumes of hypotonic buffer (10 mM NaCl, 1.5 mM MgCl2, 10 mM Tris-HCl, pH 7.5, 210 mM mannitol, 70 mM Sucrose, 1 mM EDTA) and were allowed to swell on ice for 10 min. The cell membrane was then disrupted by homogenization in a prechilled glass Dounce homogenizer by 15–20 strokes. The homogenate was checked microscopically for cell lysis and centrifuged for 5 min at 1000 × g. The supernatant was collected as a cytoplasmic extract. The pellet was washed three times and resuspended in nuclear extraction buffer containing: 50 mM Tris-HCl, pH 7.5, 1% NP-40, 0.25% deoxycholate, 150 mM NaCl, and 1 mM EDTA. After 10 min incubation on ice, the suspension was centrifuged for 15 min at 16 000 × g, and the supernatant was collected as the nuclear fraction.
Analysis of gene expression
Total RNA was isolated from cells by using TriReagent (Molecular Research Center, Cincinnati, OH, USA). RNA was treated with DNaseI and 1 μg was reverse transcribed using the iScript cDNA kit (Bio-Rad). cDNA was amplified and quantified with Roche-Light Cycler 480 system using Sybr-green (KAPA-biosystems, Woburn, MA, USA). Gene expression levels were normalized to the geometric mean of three housekeeping genes TBP, Hprt, Actin, or GAPDH, and expressed relative to control using the ΔΔCt threshold cycle method. Biological triplicates were prepared for each sample, and data are represented as mean±S.D.
Acknowledgments
We thank Professor David Carling for the LKB and AMPKα-expressing plasmids and Professor Gerry Melino for the Itch-expressing plasmid. This study was supported by grants from The McGill-Weizmann Joint Research Program and the Israel Science Foundation (551/11).
Glossary
- AMPK
AMP-activated protein kinase
- LKB1
liver kinase B1
- PGC-1α
peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- AICAR
5-aminoimidazole-4-carboxamide 1-β-D-ribofuranoside
- CHX
cycloheximide
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
Supplementary Material
References
- Bergamaschi D, Samuels Y, Jin B, Duraisingham S, Crook T, Lu X. ASPP1 and ASPP2: common activators of p53 family members. Mol Cell Biol. 2004;24:1341–1350. doi: 10.1128/MCB.24.3.1341-1350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunghi P, Costanzo A, Mazzera L, Rizzoli V, Levrero M, Bonati A. The p53 family protein p73 provides new insights into cancer chemosensitivity and targeting. Clin Cancer Res. 2009;15:6495–6502. doi: 10.1158/1078-0432.CCR-09-1229. [DOI] [PubMed] [Google Scholar]
- Conforti F, Sayan AE, Sreekumar R, Sayan BS. Regulation of p73 activity by post-translational modifications. Cell Death Dis. 2012;3:e285. doi: 10.1038/cddis.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13:962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- Bisso A, Collavin L, Del Sal G. p73 as a pharmaceutical target for cancer therapy. Curr Pharm Des. 2011;17:578–590. doi: 10.2174/138161211795222667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agami R, Blandino G, Oren M, Shaul Y. Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature. 1999;399:809–813. doi: 10.1038/21697. [DOI] [PubMed] [Google Scholar]
- Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin WG, Jr, Levrero M, et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- Yuan ZM, Shioya H, Ishiko T, Sun X, Gu J, Huang YY, et al. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature. 1999;399:814–817. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- Shafman T, Khanna KK, Kedar P, Spring K, Kozlov S, Yen T, et al. Interaction between ATM protein and c-Abl in response to DNA damage. Nature. 1997;387:520–523. doi: 10.1038/387520a0. [DOI] [PubMed] [Google Scholar]
- Baskaran R, Wood LD, Whitaker LL, Canman CE, Morgan SE, Xu Y, et al. Ataxia telangiectasia mutant protein activates c-Abl tyrosine kinase in response to ionizing radiation. Nature. 1997;387:516–519. doi: 10.1038/387516a0. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Ozaki T, Furuya K, Nakanishi M, Kikuchi H, Yamamoto H, et al. ATM-dependent nuclear accumulation of IKK-alpha plays an important role in the regulation of p73-mediated apoptosis in response to cisplatin. Oncogene. 2008;27:1183–1188. doi: 10.1038/sj.onc.1210722. [DOI] [PubMed] [Google Scholar]
- Gonzalez S, Prives C, Cordon-Cardo C. p73alpha regulation by Chk1 in response to DNA damage. Mol Cell Biol. 2003;23:8161–8171. doi: 10.1128/MCB.23.22.8161-8171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Urist M, Tanaka T, Poyurovsky MV, Prives C. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev. 2004;18:3041–3054. doi: 10.1101/gad.1221004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Prieto R, Sanchez-Arevalo VJ, Servitja JM, Gutkind JS. Regulation of p73 by c-Abl through the p38 MAP kinase pathway. Oncogene. 2002;21:974–979. doi: 10.1038/sj.onc.1205134. [DOI] [PubMed] [Google Scholar]
- Jones EV, Dickman MJ, Whitmarsh AJ. Regulation of p73-mediated apoptosis by c-Jun N-terminal kinase. Biochem J. 2007;405:617–623. doi: 10.1042/BJ20061778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Datta R, Shioya H, Li Y, Oki E, Biedermann V, et al. p73beta is regulated by protein kinase Cdelta catalytic fragment generated in the apoptotic response to DNA damage. J Biol Chem. 2002;277:33758–33765. doi: 10.1074/jbc.M110667200. [DOI] [PubMed] [Google Scholar]
- Rossi M, De Laurenzi V, Munarriz E, Green DR, Liu YC, Vousden KH, et al. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J. 2005;24:836–848. doi: 10.1038/sj.emboj.7600444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Adamovich Y, Reuven N, Shaul Y. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol Cell. 2008;29:350–361. doi: 10.1016/j.molcel.2007.12.022. [DOI] [PubMed] [Google Scholar]
- Levy D, Adamovich Y, Reuven N, Shaul Y. The Yes-associated protein 1 stabilizes p73 by preventing Itch-mediated ubiquitination of p73. Cell Death Differ. 2007;14:743–751. doi: 10.1038/sj.cdd.4402063. [DOI] [PubMed] [Google Scholar]
- Strano S, Monti O, Pediconi N, Baccarini A, Fontemaggi G, Lapi E, et al. The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA Damage. Mol Cell. 2005;18:447–459. doi: 10.1016/j.molcel.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, et al. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem. 2001;276:15164–15173. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG, Hawley SA. AMP-activated protein kinase: the energy charge hypothesis revisited. Bioessays. 2001;23:1112–1119. doi: 10.1002/bies.10009. [DOI] [PubMed] [Google Scholar]
- Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- Jenne DE, Reimann H, Nezu J, Friedel W, Loff S, Jeschke R, et al. Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat Genet. 1998;18:38–43. doi: 10.1038/ng0198-38. [DOI] [PubMed] [Google Scholar]
- Kuhajda FP. AMP-activated protein kinase and human cancer: cancer metabolism revisited. Int J Obes (Lond) 2008;32 (Suppl 4:S36–S41. doi: 10.1038/ijo.2008.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AS, Chapuis N, Lacombe C, Mayeux P, Bouscary D, Tamburini J. LKB1/AMPK/mTOR signaling pathway in hematological malignancies: from metabolism to cancer cell biology. Cell Cycle. 2011;10:2115–2120. doi: 10.4161/cc.10.13.16244. [DOI] [PubMed] [Google Scholar]
- Imamura K, Ogura T, Kishimoto A, Kaminishi M, Esumi H. Cell cycle regulation via p53 phosphorylation by a 5'-AMP activated protein kinase activator, 5-aminoimidazole- 4-carboxamide-1-beta-D-ribofuranoside, in a human hepatocellular carcinoma cell line. Biochem Biophys Res Commun. 2001;287:562–567. doi: 10.1006/bbrc.2001.5627. [DOI] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Okoshi R, Ozaki T, Yamamoto H, Ando K, Koida N, Ono S, et al. Activation of AMP-activated protein kinase induces p53-dependent apoptotic cell death in response to energetic stress. J Biol Chem. 2008;283:3979–3987. doi: 10.1074/jbc.M705232200. [DOI] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, Norman DG, Hawley SA, Kontogiannis L, Hardie DG. Protein kinase substrate recognition studied using the recombinant catalytic domain of AMP-activated protein kinase and a model substrate. J Mol Biol. 2002;317:309–323. doi: 10.1006/jmbi.2001.5316. [DOI] [PubMed] [Google Scholar]
- Jager S, Handschin C, St-Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proc Natl Acad Sci USA. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zha X, Tan Y, Hornbeck PV, Mastrangelo AJ, Alessi DR, et al. Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J Biol Chem. 2002;277:39379–39387. doi: 10.1074/jbc.M206399200. [DOI] [PubMed] [Google Scholar]
- Um JH, Park SJ, Kang H, Yang S, Foretz M, McBurney MW, et al. AMP-activated protein kinase-deficient mice are resistant to the metabolic effects of resveratrol. Diabetes. 2010;59:554–563. doi: 10.2337/db09-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Myers R, Li Y, Chen Y, Shen X, Fenyk-Melody J, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corton JM, Gillespie JG, Hawley SA, Hardie DG. 5-aminoimidazole-4-carboxamide ribonucleoside. A specific method for activating AMP-activated protein kinase in intact cells. Eur J Biochem. 1995;229:558–565. doi: 10.1111/j.1432-1033.1995.tb20498.x. [DOI] [PubMed] [Google Scholar]
- Hawley SA, Boudeau J, Reid JL, Mustard KJ, Udd L, Makela TP, et al. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol. 2003;2:28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Johnstone SR, Dickerson K, Leiper FC, Fryer LG, Neumann D, et al. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol. 2003;13:2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- Inoue T, Stuart J, Leno R, Maki CG. Nuclear import and export signals in control of the p53-related protein p73. J Biol Chem. 2002;277:15053–15060. doi: 10.1074/jbc.M200248200. [DOI] [PubMed] [Google Scholar]
- Ben-Yehoyada M, Ben-Dor I, Shaul Y. c-Abl tyrosine kinase selectively regulates p73 nuclear matrix association. J Biol Chem. 2003;278:34475–34482. doi: 10.1074/jbc.M301051200. [DOI] [PubMed] [Google Scholar]
- Melino G, Bernassola F, Ranalli M, Yee K, Zong WX, Corazzari M, et al. p73 Induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J Biol Chem. 2004;279:8076–8083. doi: 10.1074/jbc.M307469200. [DOI] [PubMed] [Google Scholar]
- Lapi E, Di Agostino S, Donzelli S, Gal H, Domany E, Rechavi G, et al. PML, YAP, and p73 are components of a proapoptotic autoregulatory feedback loop. Mol Cell. 2008;32:803–814. doi: 10.1016/j.molcel.2008.11.019. [DOI] [PubMed] [Google Scholar]
- Bernassola F, Salomoni P, Oberst A, Di Como CJ, Pagano M, Melino G, et al. Ubiquitin-dependent degradation of p73 is inhibited by PML. J Exp Med. 2004;199:1545–1557. doi: 10.1084/jem.20031943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoshima H, Goldstein BJ, Igata M, Araki E. AMPK and cell proliferation–AMPK as a therapeutic target for atherosclerosis and cancer. J Physiol. 2006;574 (Pt 1:63–71. doi: 10.1113/jphysiol.2006.108324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Maclaine NJ, Hupp TR. The regulation of p53 by phosphorylation: a model for how distinct signals integrate into the p53 pathway. Aging (Albany NY) 2009;1:490–502. doi: 10.18632/aging.100047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YG, Lee SW, Sin HS, Kim EJ, Um SJ. Kinase activity-independent suppression of p73alpha by AMP-activated kinase alpha (AMPKalpha) Oncogene. 2009;28:1040–1052. doi: 10.1038/onc.2008.452. [DOI] [PubMed] [Google Scholar]
- Flinterman M, Guelen L, Ezzati-Nik S, Killick R, Melino G, Tominaga K, et al. E1A activates transcription of p73 and Noxa to induce apoptosis. J Biol Chem. 2005;280:5945–5959. doi: 10.1074/jbc.M406661200. [DOI] [PubMed] [Google Scholar]
- Strano S, Munarriz E, Rossi M, Cristofanelli B, Shaul Y, Castagnoli L, et al. Physical and functional interaction between p53 mutants and different isoforms of p73. J Biol Chem. 2000;275:29503–29512. doi: 10.1074/jbc.M003360200. [DOI] [PubMed] [Google Scholar]
- Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21:1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.