Bone development and bone homeostasis (BH) are highly integrated processes that support skeletal formation, maintenance and repair during the whole lifespan. BH depends on the concerted action of specialized cell populations responsible for either bone apposition (osteoblasts) or resorption (osteoclasts). Osteoblasts and osteoclasts coordinate their action through endocrine and paracrine soluble molecular signals. Similar molecular cues also affect the differentiation of mesenchymal and hematopoietic progenitors influencing the expression and activity of key osteogenic transcription factors.1
BH requires coordination of three major processes: (1) osteoblast differentiation and bone matrix production, (2) osteoblast-dependent regulation of osteoclastogenesis through the secretion of Receptor Activator of NF-κB Ligand (RANKL) and osteoprotegerin (OPG2), and (3) cell-autonomous regulation of osteoclast activity.3
Defects in BH are at the origin of common and severe pathologies such as, for example, osteoporosis, a complex disease characterized by a reduction in bone mass with associated bone microarchitectural deterioration and a high risk of fractures. Although environmental factors such as nutrition and mechanical load, as well as lifestyle, may influence bone development and BH, family and twin pair studies have suggested a strong genetic component for these processes.
Bone-related disorders are a major social and economic burden for any health-care system as they affect particularly the aging population, which is increasing in most societies. New insight into the transcriptional programs supporting BH and bone remodelling is, therefore, essential to tackle the challenge posed by the increasing incidence of bone disease.
Mammalian Dlx homeobox transcription factors constitute a gene family formed by three bigenic clusters: Dlx1 and Dlx2; Dlx5 and Dlx6; Dlx3 and Dlx4.4, 5, 6 These six Dlx genes are differentially expressed by the cellular components of bone: chondrocytes, osteoblasts and osteoclasts.7 Dlx transcription factors my well play different and synergic roles in the control of chondrogenesis and/or osteogenesis.8, 9
In particular, mutations in DLX3 result in Tricho-Dento-Osseous syndrome, an autosomal dominant ectodermal dysplasia characterized by curly kinky hair, enamel hypoplasia, taurodontism and increased bone mineral density (BMD) in intramembranous and endochondral bones.10 The importance of DLX3 in bone is also supported by its capacity to regulate directly in vitro critical determinants of bone differentiation such as osteocalcin (Ocn), Runx2 and osteoactivin,11, 12, 13 and by the observation that overexpression of DLX3 in osteoprogenitors stimulates transcription of osteogenic markers.
Most in vivo functional studies on the skeletal regulatory function of Dlx genes have been performed either during embryonic development (e.g.: Samee et al.9) or in organ culture (e.g.: Hsu et al.14) as early lethality of null mutant mice has often hampered the elucidation of their postnatal role.
In this issue of Cell Death Differentiation, Isaac et al.15 perform the first in vivo study of the postnatal role of DLX3 in the appendicular skeleton. They generate and analyse mice carrying conditional loss-of-function mutations of Dlx3 in mesenchymal cells (Dlx3Prx1-cKO) or in osteogenic lineage cells (Dlx3OCN-cKO). Both models present a significant increase in bone mass throughout their lifespan. In the absence of Dlx3 endochondral bone formation still takes place at the growth plate, but bone deposition is significantly higher, with trabecular bone extending into the medullary cavity. Cortical bone is thicker, but more porous and with decreased BMD.
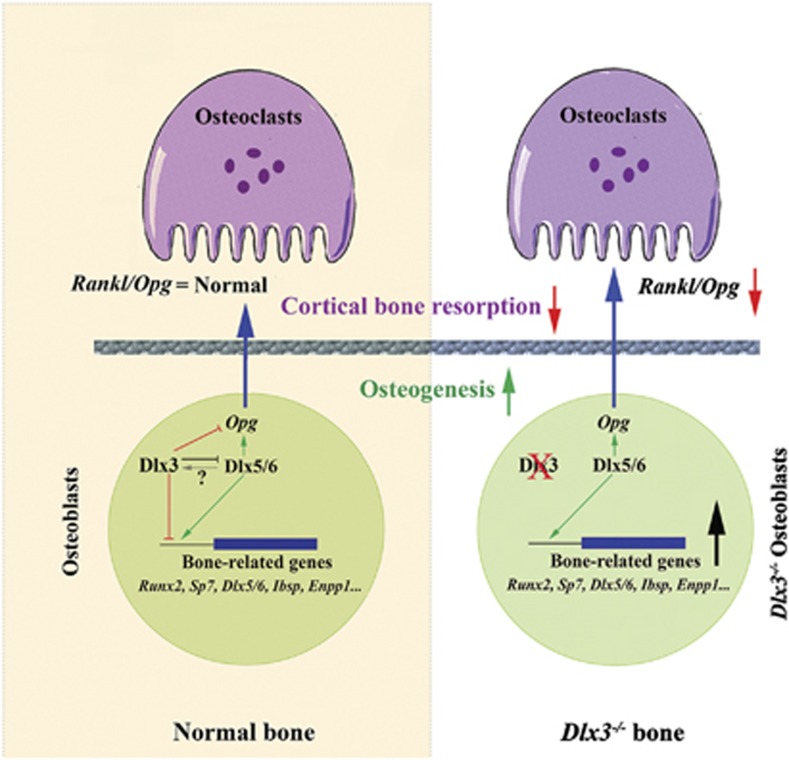
By combining in vivo gene profiling and ex vivo cellular analyses, the authors conclude that the increase in trabecular bone mass observed in Dlx3OCN-cKO mice does not arise from impaired osteoclastic activity but from direct enhancement of the bone-forming osteoblast activity with an imbalance in BH in favour of bone apposition. RNA-Seq analysis of Dlx3OCN-cKO metaphysis shows upregulation of transcription factors essential for osteoblastogenesis, including Runx2,16 its downstream osteoblast-specific target Sp717 and Dlx5/Dlx6, two positive regulators of chondrocyte and osteoblast differentiation (see Figure 1).8, 9, 18, 19
Figure 1.
Summary diagram of the effects of Dlx3 inactivation in osteogenic cells. In osteoblast (lower part of the figure) Dlx3 inhibits the expression of positive regulators of osteogenesis. Dlx3 inactivation results in higher levels of expression of Runx2, Sp7, Dlx5/6, and so on, and increases osteogenesis. Cortical bone osteobalsts of Dlx3−/− mice present also a higher level of expression of Opg, suggesting that, in parallel with increased bone deposition, this bone compartment is also characterized by reduced activity of osteoclasts (upper part of Figure 1). Dlx3 is, therefore, a negative modulator of osteogenesis at multiple levels; its coordinated action with other activators of osteogenesis, such as Dlx5, is required for the maintenance of bone homeostasis
The fact that Dlx3 plays an important role in regulating osteoblast activity is further supported by the analysis of Dlx3-deleted bone marrow stroma cells (BMSCs), which also shows increased osteoblast differentiation and bone-forming activity associated with increased gene expression of Runx2 and Dlx5. The notion that DLX3 acts as a negative regulator of osteoblastogenesis by reducing Runx2, Sp7, Dlx5 and Dlx6 gene expression is further supported by ChIP analysis on BMSCs, which shows that DLX3 binds to the promoters of Sp7, Dlx5 and Dlx6 and Runx2, directly modulating their activity (see also Hassan et al.12).
An interesting concept that emerges from this study is that the combined action of Dlx3 and Dlx5/6 generates a regulatory network essential for the maintenance of BH. First of all, the data (both in vivo and in vitro) indicate that Dlx3 negatively regulates Dlx5/6 in bone, whereas previous studies showed that the targeted simultaneous inactivation of Dlx5 and Dlx620 results in the loss of Dlx3 expression. Taken together, these results suggest that bone formation and homeostasis implicate reciprocal regulatory loops between Dlx5/Dlx6 and Dlx3. In vitro studies have shown that Dlx3 and Dlx5 associate with the Ocn promoter at the onset of transcriptional activation, concomitant with Runx2 occupancy, and, while Dlx3 occupancy on the Ocn promoter decreases from osteoblast maturation to mineralization, Dlx5 occupancy is maximal during the mineralization stage.12
Isaac et al.15 demonstrate that Dlx3 deletion in calvaria osteoblasts results in increased occupancy of Dlx5 and increased and premature occupancy of Runx2 on regulatory elements on the Ocn promoter. Thus, Dlx3 deletion could directly affect the network of molecular switches and promote osteoblastic differentiation and bone-forming activity via an enhancement of the occupancy of bone-activator TFs such as Dlx5 and Runx2. Therefore, Dlx3 and Dlx5 seem to have coordinated and opposite roles in the transcription of bone-related genes via molecular switches at their promoter regions.
Inactivation of Dlx3 results in an increased thickness of cortical bone associated with increased mineral apposition rate in the periosteum, decreased BMD, increased porosity and vascularization. RNA-Seq analysis of cortical bone of mutant mice revealed upregulation of Opg and Mepe, two major inhibitors of osteoclastogenesis strongly expressed by late-differentiated osteoblasts and osteocytes. ChIP analysis demonstrates that Dlx3 binds to several sites on the Opg promoter. These findings bolster the concept that Dlx3 inactivation in osteoblasts induces increased levels of Opg and Mepe in cortical bone, thereby deregulating homeostasis in favour of enhanced bone deposition (see Figure 1). Indeed, Dlx5−/− mice have an increased number of osteoclasts while Dlx5−/− osteoblasts exhibit decreased Opg expression, resulting in a higher Rankl/Opg ratio.9 It seems plausible that, besides Dlx5,9 Dlx3 is also a central regulator of osteoblast/osteoclast coupling. However, conversely to Dlx5, Dlx3 would inhibit osteoblast bone-forming activity via a negative transcriptional control of genes responsible for bone formation, while simultaneously activating bone resorption through osteoblast-activated regulation of osteoclastogenesis. It seems therefore that, throughout life, Dlx3 is a negative modulator of osteogenesis and that its coordinated action with other TFs, such as Dlx5, is required for the maintenance of BH. DLX3 is therefore an interesting target for future translational research in human skeletal disorders such as osteopososis.
The authors declare no conflict of interest.
References
- Karsenty G, et al. Annu Rev Cell Dev Biol. 2009. pp. 629–648. [DOI] [PubMed]
- Boyle WJ, et al. Nature. 2003. pp. 337–342. [DOI] [PubMed]
- Negishi-Koga T, Takayanagi H. Immunol Rev. 2009. pp. 241–256. [DOI] [PubMed]
- Cohen SM, et al. Nature. 1989. pp. 432–434. [DOI] [PubMed]
- O'Hara E, et al. Development. 1993. pp. 847–856. [DOI] [PubMed]
- Panganiban G, Rubenstein JL. Development. 2002. pp. 4371–4386. [DOI] [PubMed]
- Li H, et al. Dev Biol. 2008. pp. 458–470. [DOI] [PMC free article] [PubMed]
- Samee N, et al. Crit Rev Eukaryot Gene Expr. 2007. pp. 173–186. [DOI] [PubMed]
- Samee N, et al. Am J Pathol. 2008. pp. 773–780. [DOI] [PMC free article] [PubMed]
- Haldeman RJ, et al. Bone. 2004. pp. 988–997. [DOI] [PubMed]
- Hassan MQ, et al. Mol Cell Biol. 2004. pp. 9248–9261. [DOI] [PMC free article] [PubMed]
- Hassan MQ, et al. J Biol Chem. 2006. pp. 40515–40526. [DOI] [PubMed]
- Singh M, et al. J Cell Physiol. 2012. pp. 390–399. [DOI] [PubMed]
- Hsu SH, et al. Mech Dev. 2006. pp. 819–830. [DOI] [PubMed]
- Isaac J, et al. Cell Death Differ 2014. e-pub ahead of print 20 June 2014;doi: 10.1038/cdd.2014.82 [DOI]
- Otto F, et al. Cell. 1997. pp. 765–771. [DOI] [PubMed]
- Nakashima K, et al. Cell. 2002. pp. 17–29. [DOI] [PubMed]
- Lee MH, et al. J Biol Chem. 2005. pp. 35579–35587. [DOI] [PubMed]
- Tadic T, et al. J Bone Miner Res. 2002. pp. 1008–1014. [DOI] [PubMed]
- Depew MJ, et al. Science. 2002. pp. 381–385. [DOI] [PubMed]