The number of coding genes is specific and limited in each individual species, and thus to increase the repertoire of proteins required for efficient and effective cellular activity, cells developed several molecular mechanisms to address this issue.1 Transcriptional control is one such common mechanism where alternative splicing takes advantage of the fact that within a single given gene, intronic sequences can be regarded as exonic coding sequences.2 Another mechanism involves posttranslational modifications that permit the generation of protein families encoded by a single gene.3
The p53 tumor suppressor that was regarded for a long time as single protein, is encoded by one functional gene and was shown to be involved in a broad range of cellular activities.4 As p53 is a transcription factor,5 it is well possible that it can transactivate a family of individual specific target genes that are associated with a variety of functional pathways in the cell. Increasing the repertoire of the p53 protein types by alternative splicing is another anticipated mechanism.6 Indeed, in the mouse model, an alternative spliced p53 mRNA form that is shorter in size and represents an altered C′-terminus was described.7, 8 The latter exhibited an augmented DNA binding activity.9 However, the lack of indications of such an analogous p53 molecule in humans made researches neglecting this mouse-specific issue.
Interestingly, however, searching for p53 functionally and structurally related proteins led to the identification of families of protein members such as p63 and p73 as well as a variety of isoforms that are generated by diverse mechanisms in individual cell types under defined conditions.10
As wild-type p53 has diverse functionalities in regulating cell growth, cell death, DNA repair, differentiation, development and more, and it is a key player in cancer development,4 it is reasonable to speculate that such a nature of activities may be mediated by multiple p53 protein molecules. Indeed, p53 isoforms were found to regulate its tumor-suppressive functions, and their dysregulation was found in many cancers.11
Using sensitive GeneRacer PCR technique it was possible to revisit the analysis of the human p53 gene structure. This further indicated that the human p53 gene contains 11 exons with alternative splicing site in intron 9 and intron 2, internal promoter in intron 4 and several ATG codons encoding for initiation of translation.11 Hitherto, 12 p53 isoforms were described as follows: the full-length p53 (FLp53), also named p53α, additional p53 variants lacking either the N′-terminus (Δ40p53, Δ133p53 and Δ160p53), the C′terminus (p53β and p53γ), or the combination of both (Δ40p53β, Δ40p53γ, Δ133p53β, Δ133p53γ, Δ160p53β, and Δ160p53γ). Three conserved mechanisms were suggested to underlie the formation of p53 isoforms; alternative splicing of intron 9 or intron 2, alternative promoter usage, and alternative initiation of translation.11 These mechanisms yield shorter p53 proteins, lacking different domains, that are found to modulate the normal function of the abundantly expressed, full-length p53α.10 This was manifested by the expression of p53 target genes, and by the execution of normal p53 functions. For example, p53β and p53γ were found to induce the FLp53-dependent transactivation of Bax and p21, thus promoting apoptosis and senescence, and retaining tumor-suppressive functions. Alternatively, Δ40p53 and Δ133p53 exert dominant negative functions toward FLp53, by inhibiting transactivation of its target genes and by interfering with p53-dependent growth suppression. The molecular mechanism that underlies these functions was suggested to involve physical interaction with FLp53 in modulating its biochemical function.10, 12, 13
In addition to the well-accepted notion that mutant p53 has a central role in the initiation and development of 50% of human cancers,14 there are indications that the balance between the expression of the various wild-type p53 isoforms may have a role in carcinogenesis.11 Indeed, abnormal expression of p53 isoforms was detected in various human cancer types. For example, higher levels of Δ133p53 were found in colon carcinoma and renal cell carcinoma compared with healthy cells, and higher levels of Δ40p53 were found in melanoma. p53β was found to be highly expressed in tumors such as renal cell carcinoma and melanoma and to be associated with poor ovarian cancer prognosis, whereas in colon carcinoma its levels were found reduced.11 These data indicate p53 isoforms as additional mechanism of deregulating wild-type p53 function during tumorigenesis and may explain tumorigenesis in wild-type p53-expressing tumors. Moreover, in some tumors p53 isoforms can overcome mutant p53 oncogenic gain-of-function activities. In breast tumors, the expression of p53γ conferred patients with low cancer recurrence and an overall survival as good as that of patients with wild-type p53 breast cancer.15 Recently, p53β expression was found to be associated with breast cancer disease-free survival and was further found as protective in patients expressing mutant p53.16 In addition, it was shown that Δ133p53 is associated with improved prognosis and survival of mutant p53 expressing advanced serous ovarian cancers.17 Altogether, these examples point p53 isoforms as biomarkers for cancer prognosis and suggest that uncovering the molecular mechanism controlling the expression of the various p53 variants could contribute to the development of personalized medicine.
Splicing of pre-mRNA into a mature mRNA involves large and complex molecular machinery, consisting of small nuclear ribonucleic particles and proteins.18 As splicing determines the expression of variants of many tumor suppressors and oncogenes, and a strong correlation between aberrant alternative splicing and cancer development was suggested, understanding of its regulation is important for revealing mechanisms underlying cancer development and therapy. It is accepted that splicing alteration in cancer cells could be due to genomic instability or by defects in spliceosome regulation.19 Indeed, several components of the spliceosome were defined as oncogenes or tumor suppressors. One of them is SRSF1 in which its expression is upregulated in various tumors, leading to activation of pro-malignant pathways.20
In the current issue of Cell Death & Differentiation, Marcel and co-authors highlighted the role of p53β and p53γ in regulating cell fate. They have shown that modulation of alternative splicing components using small drug TG003 that inhibits the alternative splicing pre-mRNA pathway regulator Cdc2-like kinases (Clk) or silencing the splicing factor SFRS1, lead to increased expression of p53β and p53γ isoforms. Using advanced PCR and cloning technologies Marcel and co-authors found that alternative splicing of intron 9 of p53 resulted in higher expression of p53β and p53γ. As p53β and p53γ were already suggested to exert tumor-suppressive functions, this study suggests that the basal expression of these isoforms was accompanied with inhibition of cell cycle progression and induction of apoptosis. This study adds additional layer of understanding into the mechanism by which p53β and p53γ function. It suggests that p53β and p53γ can bind to p53α in a DNA-mediated manner, in which the p53-responsive DNA elements enabled the creation of p53β/p53α and p53γ/p53α complexes. This is suggested to enhance p53α transcriptional activity on p53 target genes p21 and Bax that regulate cell cycle progression and apoptosis, respectively. As mentioned above, p53β and p53γ are differentially expressed in cancer cells. Specifically, breast cancer studies show that their levels are associated with better disease outcome.15, 16 This study (Marcel et al.) examined a cohort of 85 primary breast cancer tumors and found positive correlation of SFRS1 with p53α and with lack of detection of p53β. This supports the suggested mechanism underlying p53 splicing of intron 9, and show that it is relevant to human pathological conditions. Finally, this study suggests that modulation of p53 splicing can change the endogenous cellular levels of p53 isoforms, and therefore to affect cellular response.
Most p53-dependent therapy strategies are based on the abolishment of mutant p53. In the era of precision medicine, the existence of such knowledge, pertaining to the expression of p53 isoforms within individual tumor, is of great relevance for designing the most suitable p53-based therapy (Figure 1). Therefore, when searching for new cancer therapy, understanding p53 status is essential, and modulation of p53 splicing can contribute to the development of efficient drugs.
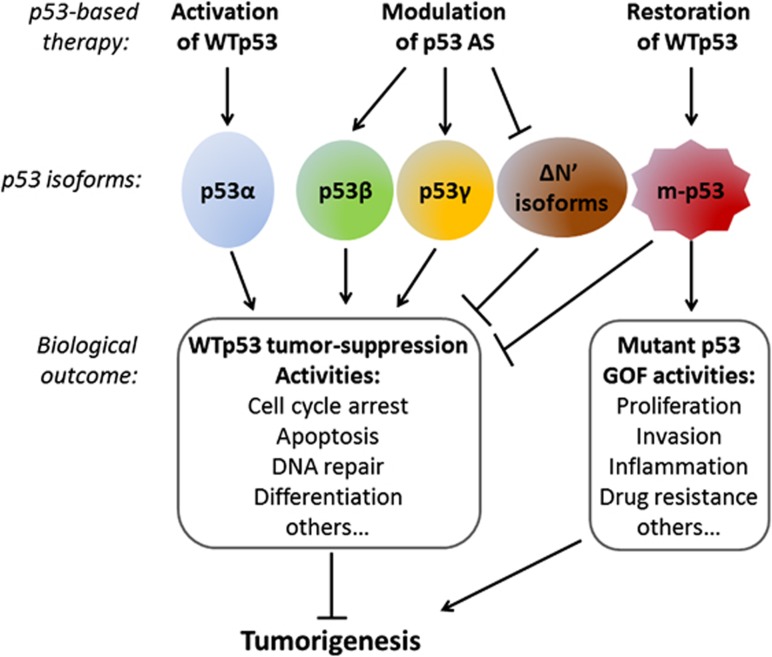
Figure 1.
The roles of p53 isoforms in carcinogenesis. Whereas the full-length wild-type p53 (WTp53) tumor suppressor (also named p53α) has a key role in preventing tumorigenesis, mutant p53 (m-p53) can inhibit these activities in a dominant negative manner and can exert oncogenic gain-of-function (GOF) activities. The expression of p53 isoforms is aberrant in tumor cells and could either enhance or inhibit the tumor-suppressive activities of WTp53, dependent on the cellular context. In principle, p53β and p53γ isoforms were found to enhance WTp53 tumor-suppressive functions, whereas isoforms deleted in p53 N′-terminal (such as Δ40p53 and Δ133p53) were found to exert dominant negative effects on WTp53. Apparently, the balance between the expression of the various isoforms has a role in carcinogenesis. Accordingly, the developing p53-based cancer therapies are focused on the following: over-activating WTp53 (e.g., Nutlin-3), restoring WTp53 in mutant p53-expressing tumors by re-activating compounds (e.g., PRIMA-1) and modulating p53 alternative splicing (AS) to reach the p53 isoform ration that allows beneficial therapy11
Acknowledgments
Research in the laboratory of VR is supported by a center of excellence grant from the Israel Science Foundation (ISF) center from the Israeli Academy of Science and a center of Excellence grant from the Flight Attendant Medical Research Institute (FAMRI). VR is the incumbent of the Norman and Helen Asher Professorial Chair Cancer Research at The Weizmann Institute.
The authors declare no conflict of interest.
References
- Leff SE, Rosenfeld MG, Evans RM. Annu Rev Biochem. 1986. pp. 1091–1117. [DOI] [PubMed]
- Blencowe BJ. Cell. 2006. pp. 37–47. [DOI] [PubMed]
- Olsson A, Manzl C, Strasser A, Villunger A. Cell Death Differ. 2007. pp. 1561–1575. [DOI] [PubMed]
- Goldstein I, et al. Cancer Gene Ther. 2011. pp. 2–11. [DOI] [PubMed]
- Beckerman R, Prives C. Cold Spring Harb Perspect Biol. 2010. p. a000935. [DOI] [PMC free article] [PubMed]
- Marcel V, et al. Cell Death Differ. 2011. pp. 1815–1824. [DOI] [PMC free article] [PubMed]
- Wolf D, Harris N, Goldfinger N, Rotter V. Molecular Cell Biol. 1985. pp. 127–132. [DOI] [PMC free article] [PubMed]
- Arai N, et al. Molecular Cell Biol. 1986. pp. 3232–3239. [DOI] [PMC free article] [PubMed]
- Wolkowicz R, Peled A, Elkind NB, Rotter V. Proc Natl Acad Sci USA. 1995. pp. 6842–6846. [DOI] [PMC free article] [PubMed]
- Bourdon JC, et al. Genes Dev. 2005. pp. 2122–2137. [DOI] [PMC free article] [PubMed]
- Surget S, Khoury MP, Bourdon JC. Onco Targets Ther. 2013. pp. 57–68. [DOI] [PMC free article] [PubMed]
- Courtois S, et al. Oncogene. 2002. pp. 6722–6728. [DOI] [PubMed]
- Fujita K, et al. Nat Cell Biol. 2009. pp. 1135–1142. [DOI] [PMC free article] [PubMed]
- Brosh R, Rotter V. Nat Rev Cancer. 2009. pp. 701–713. [DOI] [PubMed]
- Bourdon JC, et al. Breast Cancer Res. 2011. p. R7. [DOI] [PMC free article] [PubMed]
- Avery-Kiejda KA, et al. Carcinogenesis. 2014. pp. 586–596. [DOI] [PubMed]
- Hofstetter G, et al. Br J Cancer. 2011. pp. 1593–1599. [DOI] [PMC free article] [PubMed]
- Matera AG, Wang Z. Nat Rev Mol Cell Biol. 2014. pp. 108–121. [DOI] [PMC free article] [PubMed]
- Dehm SM. Cancer Res. 2013. pp. 5309–5314. [DOI] [PMC free article] [PubMed]
- Ladomery M. Int J Cell Biol. 2013. p. 463786. [DOI] [PMC free article] [PubMed]