Abstract
Accumulating evidence demonstrated that while a small minority of the human genome encodes proteins a large portion of the genome encodes non-protein coding transcripts. Thus it has been suggested that such RNAs, including the recently described family of long non-coding RNAs (lncRNA) are key regulators of cellular homeostasis and transformation leading to tumorigenesis. Here we tested this hypothesis focusing on the role of lncRNAs in T cell leukemia (T-ALL), a pediatric blood cancer, characterized by Notch pathway activation. Using RNA-Sequencing we identified and catalogued a large number of T-ALL-specific lncRNAs targeted by the NOTCH pathway. We further focused on a single such transcript, LUNAR1 (LeUkemia-induced Non-coding Activator RNA-1), and demonstrated that through chromosomal looping it is able to control the expression of the IGF1R gene, IGF1 signaling and growth of T-ALL, providing evidence that suggests that lncRNAs could be used as biomarkers as well as therapy targets in human cancer.
INTRODUCTION
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematological neoplasm that results from the malignant transformation of T-lymphocyte progenitors. T-ALL accounts for only 15–20% of all acute lymphoblastic leukemia (ALL) cases and is associated with a disproportionate amount of treatment failures and increased mortality. Although the genetic events leading to transformation in T-ALL are complex, aberrant NOTCH1 signaling is a unifying feature, with activating mutations found in more than 50% of cases(Ferrando, 2009; Weng et al., 2004). Previous efforts by our group and others have aimed at dissecting additional genetic(De Keersmaecker et al., 2013; Ntziachristos et al., 2012; Zhang et al., 2012) and molecular(Ferrando et al., 2002; Look, 2004) changes that contribute to induction of T-ALL. Such efforts have yielded important prognostic tools and a more complete understanding of the molecular basis of ALL, including the identification of T-ALL oncogenes (NOTCH1, MYC), tumor suppressors (FBXW7, CYLD, EZH2, SUZ12) and the identification and characterization of the leukemia initiating cell(King et al., 2013). To date, most efforts towards understanding T-ALL have focused on genetic and epigenetic alterations, which ultimately impact function of protein-coding genes. Additionally, several groups have investigated the involvement of non-coding micro-RNAs (miRs) in T-ALL(Fragoso et al., 2012; Li et al., 2011; Yu et al., 2011), however, the requirement of other classes of non-coding RNA for T cell transformation and maintenance has not been investigated thus far.
Recent evidence has revealed that a large portion of the human genome is transcriptionally active despite that fact that only a small portion contains protein-coding genes (Carninci et al., 2005; Clark et al., 2011; Djebali et al., 2012; Katayama et al., 2005). This observation has led many to hypothesize that both human and mouse genomes contain thousands of long non-coding RNA (lncRNA) genes (Cabili et al., 2011; Guttman et al., 2009). Although some have suggested that a subset of lncRNAs are the result of divergent transcription at protein-coding gene promoters(Almada et al., 2013; Seila et al., 2008), this does not explain the existence of many intergenic lncRNAs which do not originate from bidirectional promoters (Cabili et al., 2011; Sigova et al., 2013). In general, lncRNAs are a heterogeneous class of transcripts without any single unifying feature except for an arbitrary minimum length of 200 nucleotides and an apparent lack of protein-coding potential (Guttman et al., 2013; Rinn and Chang, 2012; Ulitsky and Bartel, 2013). Despite this, lncRNAs have been shown to be important in development (Boumil and Lee, 2001; Grote et al., 2013; Guttman et al., 2011; Klattenhoff et al., 2013; Kretz et al., 2013; Loewer et al., 2010; Rinn et al., 2007) and disease (Gomez et al., 2013; Gupta et al., 2010; Huarte et al., 2010; Lee et al., 2012; Yildirim et al., 2013) in many different cell types, suggesting a ubiquitous role in regulation of cellular state. Although the molecular mechanisms by which lncRNAs act are poorly understood, they have been implicated as regulators of diverse cellular processes including regulation of cell cycle (Hung et al., 2011), RNA stability (Kretz et al., 2013), and chromatin structure (Gupta et al., 2010; Lee, 2012; Tsai et al., 2010; Wang et al., 2011b; Yang et al., 2011). Thus, further efforts towards accurate annotation and functional significance of lncRNAs will be critical for our understanding of these processes.
While several groups have investigated possible roles for lncRNAs as players in the TP53 tumor-suppressor transcriptional program (Huarte et al., 2010; Hung et al., 2011) and solid tumors (Du et al., 2013; Prensner et al., 2011; Prensner et al., 2013; Yang et al., 2013), our overall knowledge of lncRNAs in cancer, including leukemia, remains extremely limited (Garding et al., 2013; Lee et al., 2012). Here we have used multiple genome-wide data sets in order to create the first all-inclusive lncRNA annotation and mapping in human T-ALL. We have used this annotation to examine lncRNA expression profiles in the context of oncogenic NOTCH1 signaling and have identified Notch-dependent lncRNA expression programs that are deregulated in T-ALL. Finally using functional studies we have identified a novel T-ALL specific activator lncRNA, which has a key role in promoting tumor maintenance. Altogether these studies provide evidence to support lncRNAs as key regulators of the pathogenic state in acute leukemia and may carry important clinical relevance as biomarkers or future therapeutic targets.
RESULTS
Comprehensive mapping of lncRNAs in T-ALL
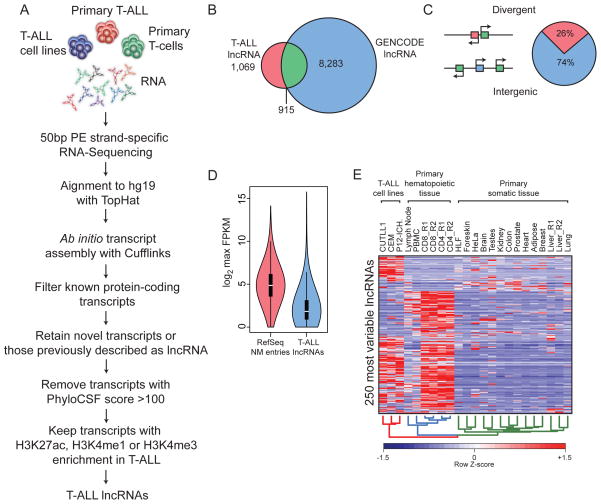
To gain insights into possible functional roles for lncRNAs in T-ALL, we first sought to create a high quality map of such transcripts for use in subsequent analyses and functional studies. We have achieved this goal by utilizing the workflow outlined in Figure 1A. Briefly, ultra-high depth RNA-Seq datasets were generated from multiple human T-ALL cell lines and primary leukemia samples. These data were then used to generate the most comprehensive T-ALL transcriptome assembly to date using an “align then assemble” (Trapnell et al., 2010) approach. To isolate only putative lncRNA genes, we removed all known protein-coding genes and retained all transcripts previously reported as lncRNAs or that have not been identified thus far in other annotation efforts. Single-exon transcripts were eliminated to focus on products of splicing events. Finally, since lncRNA transcription has been suggested to occur by a mechanism very similar to that of coding genes (Guttman et al., 2009), we removed any transcript that did not display enrichment of promoter-associated histone modifications (H3K4me3, H3K4me1, H3K27ac). These efforts have yielded a high confidence T-ALL lncRNA annotation comprised of 6,023 isoforms derived from 1,984 unique gene loci. Out of all of these loci, 46% were identified in the GENCODE v18 lncRNA annotation(Harrow et al., 2012) (Figure 1B), suggesting the presence of many novel lncRNAs in T-ALL. Additionally, we have observed that approximately 26% of lncRNAs detected here were expressed in a divergent orientation with respect to protein-coding genes, while the remaining 74% were true intergenic gene loci with their own regulatory elements (Figure 1C) and this ratio of divergent to intergenic lncRNAs is approximately the same in T-ALL and normal T-cell progenitors (Supplementary Figure 1D). Additionally, 40% of intergenic lncRNAs were identified by GENCODE while the remaining 60% represent novel lncRNA loci (Supplementary Figure 1C).
Figure 1. The long non-coding transcriptome in T-ALL.
(A) A schematic illustration of the procedure used to discover and define lncRNAs in TALL. (B) Venn diagram depicting the overlap between our catalogue of T-ALL associated lncRNAs and those in the Gencode v18 lncRNA collection. (C) Pie chart representation showing the proportion of T-ALL associate lncRNAs that are transcribed in a divergent orientation (red) or intergenic (blue) with respect to protein-coding genes (green, diagram left). (D) Violin plot of log2 maximum expression values (FPKM) for protein coding (red) and T-ALL lncRNA (blue) genes. Boxes represent first and third quartile. Whiskers are 1.5 times the interquartile range. (E) A heat map representation of the 250 lncRNAs with the most variable (IQR/median) expression across T-ALL, normal T-cells and various other somatic tissues.
We next examined the expression patterns of our T-ALL lncRNA catalogue over a diverse panel of cell types including T-ALL, primary T-cells and many other somatic tissues (Human Body Map data). In agreement with previous reports (Cabili et al., 2011), we have observed that average lncRNA expression was lower than protein-coding gene expression (Figure 1D). Additionally, putative lncRNAs showed very little protein-coding potential as measured by the PhyloCSF algorithm (Lin et al., 2011) (Supplementary Figure 1A). In agreement with possible roles in regulating gene expression, many lncRNAs were enriched in RNA from nuclear extracts compared to total RNA (Supplementary Figure 1B). Finally, upon examination of the 250 most variable lncRNAs, as measured by quartile coefficient of dispersion, we observed striking specificity for T-ALL and normal T-cells (Figure 1E), which is consistent with the notion (Guttman et al., 2009) that lncRNA expression is highly tissue specific. However, many lncRNAs were specifically expressed in human T-ALL when compared to untransformed peripheral T cells (Figure 1E) suggesting that T cell transformation is dynamically changing the lncRNA landscape in this cell type. Overall, this effort catalogued T-ALL-specific non-coding RNAs and represented the first comprehensive mapping of lncRNA expression in this aggressive subtype of ALL.
T-ALL-specific lncRNAs are part of the NOTCH1 oncogenic network
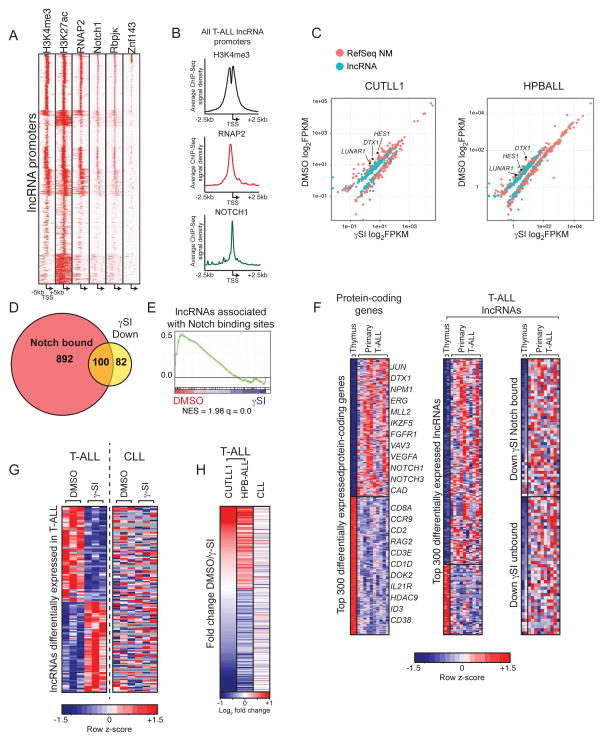
Since oncogenic NOTCH1 activity is one of the key features in T-ALL and NOTCH pathway activity characterizes the vast majority (>90%) of human T-ALL cases, we reasoned that the lncRNA expression program in this disease may be influenced by the NOTCH1/RBPJκ transcriptional activator complex. Upon examination of lncRNA promoters we noticed a high density of NOTCH1/RBPJκ ChIP-Seq signal at many of these regions (Figure 2A, 2B). Additionally, most lncRNA promoters displayed high enrichment of H3K4me3, H3K27ac and RNA polymerase II (RNAP2) signal proximal to their TSS. We also observed a subset of lncRNA, which showed co-occupancy of NOTCH1 and ZNF143 (Figure 2A, Supplementary Figure 2C), a protein that has been previously reported to co-occupy the genome with Notch, although its role in gene regulation is unclear(Wang et al., 2011a). We therefore hypothesized that expression of a subset of lncRNA genes may be dependent on NOTCH1-mediated signaling. To test this hypothesis we used chemical inhibition of the γ–Secretase complex (Kopan and Ilagan, 2009) to perturb NOTCH1 cleavage and nuclear translocation in two prototypical human T-ALL cell lines (CUTLL1 and HPB-ALL) and carried out RNA-Seq to measure lncRNA expression following treatment and NOTCH inhibition. Such approaches have been extensively used in T-ALL and are proven to specifically target NOTCH1 signaling (Palomero et al., 2006; Weng et al., 2004). Indeed, we observed that in addition to protein-coding genes, many lncRNA genes were differentially expressed upon γ–Secretase inhibitor (γSI) treatment compared to vehicle controls (Figure 2C, Supplementary Figure 2A), suggesting direct regulation and possible roles downstream of NOTCH1 activation in this disease. In agreement with this notion, we observed that approximately 55% of lncRNAs whose expression was Notch-dependent were also directly occupied by NOTCH1 (Figure 2D). In support of our hypothesis that NOTCH1 controls the lncRNA transcriptional program in T-ALL, we observed that lncRNAs associated with the top 1000 most enriched NOTCH1 binding sites were significantly down regulated upon administration of γSI according to gene set enrichment analysis (GSEA) (Figure 2E). Upon closer examination of many of these lncRNA loci, we observed strong NOTCH1/RBPJκ binding sites (Wang et al., 2011a) both at promoters and intragenic enhancer elements (Supplementary Figure 2B highlighted yellow), suggesting direct transcriptional control by Notch signaling. Together these data suggest the presence of a Notch-dependent T-ALL lncRNA expression program, members of which, we believe may carry important biological functions.
Figure 2. LncRNAs are a component of the Notch1 transcriptional network.
(A) Heat map representation of ChIP-Seq signal density for H3K27ac, H3K4me3, RNAP2, NOTCH1, RBPJκ and ZNF143 centered on lncRNA TSSs +/− 5kb. All ChIP experiments are from CUTLL1 cells. (B) Histogram depicting ChIP-Seq signal density on all T-ALL lncRNA promoters for H3K4me3, RNAP2 and NOTCH1 (C) Scatterplots showing both lncRNA (green) and protein coding (orange) genes whose expression is significantly altered following addition of γSI in CUTLL1 (left) and HPBALL (right) cells. (D) Venn diagram showing the proportion of Notch-occupied lncRNAs whose expression is significantly down regulated in response to γSI treatment. (E) GSEA enrichment plot showing significant down regulation of lncRNAs associated with the top 1000 NOTCH1 peaks upon γSI treatment. (F) Heat maps showing top differentially expressed protein coding (left) and lncRNA (right) genes in primary T-ALL compared to thymic progenitors. (G) Expression profiles of lncRNAs that are differentially expressed in response to γ-SI treatment in T-ALL are shown in heat map either in T-ALL (left) or CLL (right). Gray indicates no detectable expression. (H) Expression fold change (DMSO/γ-SI) in T-ALL (CUTLL1 And HPBALL) and CLL of all lncRNAs consistently regulated upon Notch inhibition in T-ALL.
To address whether these Notch-regulated lncRNA genes might be expressed in primary NOTCH1-induced T-ALL, we measured their expression across ten patient samples which harbored activating NOTCH1 mutations as well as 2 primary human thymus samples, as a matched physiological tissue with low Notch activation (Ntziachristos et al., 2012). These analyses yielded a subset of lncRNAs, which, displayed differential expression in primary T-ALL compared to normal thymic T cells (Figure 2F). We also observed that a subset of lncRNAs are differentially expressed among distinct subtypes of T-ALL as defined by aberrant expression of TLX1 or TLX3 (Supplementary Figure 2D), suggesting a possible role in specifying discreet subclasses within this disease.
Since activating mutations on NOTCH1 have been recently discovered in chronic lymphocytic leukemia (CLL)(Fabbri et al., 2011), a tumor of the B-lymphocyte compartment, we hypothesized that Notch-dependent lncRNAs we have described in TALL might also show similar expression patterns in CLL. To test this hypothesis we cultured Notch-mutant CLL cells on OP9-DL1 stromal cells, which express the Notch ligand delta-like 1 (DL1), and added vehicle control (DMSO) or γ–SI followed by RNA-Seq (Figure 2G). By comparing our T-ALL and CLL γ-SI treatment experiments, we observed that Notch-dependent changes in lncRNA expression in T-ALL are different from those in CLL (Figure 2G, 2H). To our surprise we observed very little conservation of the Notch-dependent T-ALL lncRNA expression program in Notch mutant CLL cells, suggesting that although these two tumor harbor similar oncogenic lesions, the downstream consequences of Notch activation are distinct.
LUNAR1 is a novel, NOTCH-regulated lncRNA transcript in human T-ALL
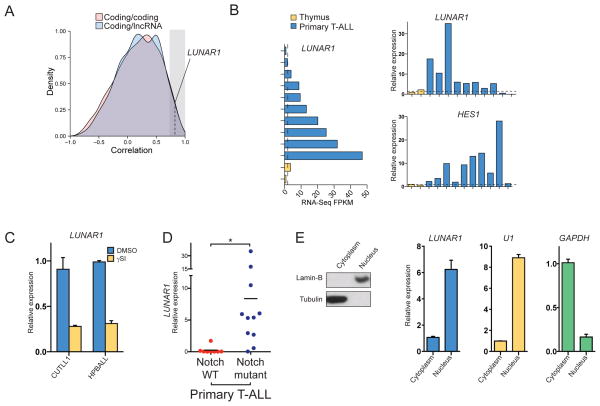
Since lncRNAs have been previously shown to enhance expression of nearby genes through cis regulation(Gomez et al., 2013; Lai et al., 2013; Orom et al., 2010), we next asked whether any of the lncRNAs identified here display a high Pearson correlation with neighboring protein-coding genes. By comparing correlation density plots for all coding/lncRNA and coding/coding gene pairs, we found that in general lncRNAs are no more correlated with neighboring genes than their coding counterparts (Figure 3A). However, since enhancer-like activity for lncRNAs has been proposed before, we considered all lncRNAs, with at least 0.75 correlation (Figure 3A, shaded area) with a coding neighbor as candidates for further study as cis-regulators. From these candidates we noticed one lncRNA gene, which we have termed LUNAR1 (LeUkemia-induced Non-coding Activator RNA) that showed high correlation with its coding neighbor gene the insulin-like growth factor receptor 1 (IGF1R) (r = 0.77), a gene which as been previously suggested to play a role in T-ALL(Medyouf et al., 2011). We noted several other features which made LUNAR1 appear attractive as a candidate. It was down regulated upon Notch inhibition (Figure 3B), over-expressed in primary T-ALL (Figure 3C) and expressed significantly higher in T-ALL samples that harbored a Notch mutation compared to those without mutations (Figure 3D). Additionally, we also found an enrichment of LUNAR1 in the nucleus (Figure 3E), to a degree similar to that of U1, a component of a snRNP splicing complex which supported our hypothesis that this transcript may be involved in gene regulation. By examining the chromatin state at the LUNAR1 locus, we observed typical features of RNAP2-dependent genes in T-ALL (Supplementary Figure 3A), as well as, a transcriptionally active state in several related hematopoietic cell types, according to the chromHMM algorithm (Supplementary Figure 3B). While a transcriptionally active chromatin state was restricted to very few samples, nearly all of the cell types for which chromHMM data was available showed signs of active promoter-associate chromatin structure proximal to the LUNAR1 TSS, indicating a true transcriptional unit (Supplementary Figure 3B). Although the LUNAR1 locus shows signs of promoter activity in diverse tissues, its expression is highly restricted, suggesting that specific factors are required for its activation. Using standard 5′ and 3′ RACE approaches we were able to clone a 491-nucleotide transcript containing 4 exons and a polyA tail (FASTA sequence in supplement). In order to address whether LUNAR1 is indeed non-coding, we utilized the PhyloCSF algorithm(Lin et al., 2011), which yielded a score of −48.12 indicating lack of an ORF with selective pressure for codon preservation. Additionally we have translated the RNA sequence in three frames and searched for known protein domains using PFAM, of which we found none (not shown). These data strongly suggest that the LUNAR1 transcript is unlikely to encode any protein product.
Figure 3. LUNAR1 is a novel lncRNA gene controlled by NOTCH1 in T-ALL.
(A) Correlation density plot showing expression correlation of protein coding (red) or lncRNA (blue) genes with the nearest coding neighbor. (B) RNA-Seq expression values for LUNAR1 in primary T-ALL and thymic progenitors (left). Over expression was validated by qPCR (right, top) and over activation of Notch signaling was verified by measuring HES1 expression (bottom, right). (C) qPCR for LUNAR1 following treatment with vehicle (blue) or γSI (red) in CUTLL1 (left) and HPBALL (right) cells. (D) qPCR for LUNAR1 in Notch WT vs. Notch mutant tumors. (E) Immunoblot (left) for Lamin-B and Tubulin on cytoplasmic and nuclear fractions from CUTLL1 cells. qPCR (right) for LUNAR1, U1 and GAPDH from RNA extracted from cytoplasmic and nuclear fractions. * indicates p-value < 0.05.
LUNAR1 expression is controlled by an intronic enhancer in the IGF1R locus
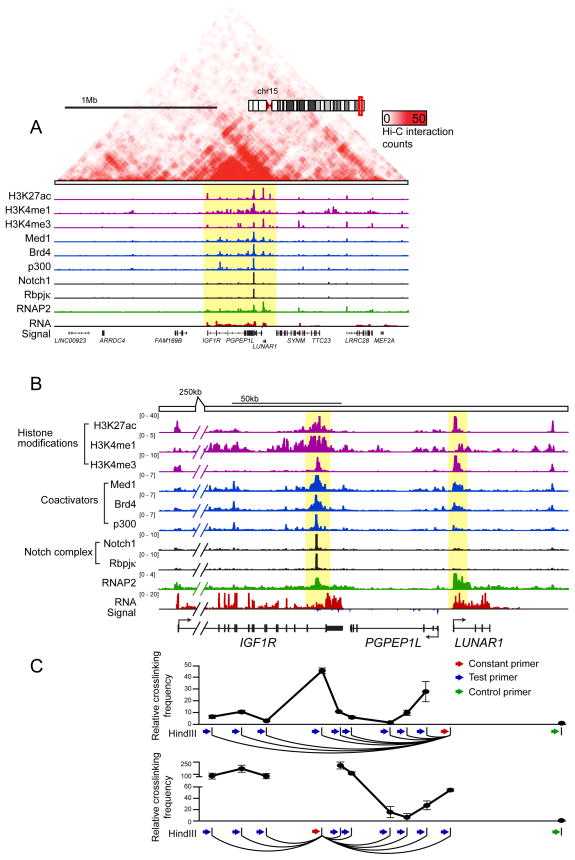
To further study possible functional significance of LUNAR1 we utilized genome-wide chromosome conformation capture (Hi-C) in T-ALL cells (CUTLL1) to examine the higher order chromatin context in T-ALL (I.A., P.N., A.T. manuscript in preparation). Similar to other chromosome capture methods, Hi-C utilizes restriction enzyme digestion of cross-linked chromatin followed by intramolecular ligation to physically link genomic regions that were in close spatial proximity to one another(Lieberman-Aiden et al., 2009). By coupling this method to high throughput sequencing, we created a map of 3D chromatin structure in human T-ALL cells. In doing so we discovered that LUNAR1 resides in a 500kb topologically associating domain(Dixon et al., 2012) on chromosome 15, which also includes neighboring genes IGF1R and PGPEP1L (Figure 4A). Of these two neighboring genes, only IGF1R is transcriptionally active, therefore we reasoned that LUNAR1 might play a role in enhancing expression of this gene. Closer examination of the LUNAR1/IGF1R locus revealed the presence of a highly active enhancer in the last intron of IGF1R, which showed a high degree of occupancy by NOTCH1 (previously reported (Medyouf et al., 2011)), Mediator subunit MED1, histone acetyltransferase P300, RNA Pol2 and the histone reader BRD4 (Figure 4B, Supplementary Figure 4A), all of which have been suggested to be hallmarks of active enhancer elements when located outside of promoter regions(Heintzman et al., 2009; Rada-Iglesias et al., 2011; Whyte et al., 2013). In addition to showing enrichment for enhancer-associated chromatin factors and a chromatin signature characteristic of active enhancer elements (high H3K4me1, low H3K4me3, high H3K27ac), we showed that this element is able to drive expression in a reporter assay in a Notch-dependent manner (Supplementary Figure 4B, C).
Figure 4. LUNAR1 is physically associated with a nearby Notch-occupied enhancer.
(A) Gene track of a 2Mb region surrounding the LUNAR1 locus including Hi-C interaction density heat map (red upper panel), ChIP-Seq tracks for H3K27ac, H3K4me1, H3K4me3, MED1, BRD4, P300, NOTCH1, RBPJκ, RNAP2 and RNA-Seq track. (B) Gene track view of an approximately 150kb region which contains LUNAR1 (highlighted yellow, right and a Notch-occupied enhancer in the last intron of IGF1R (highlighted yellow, left). (C) Relative cross-linking frequency as measured by 3C-qPCR using a constant primer in a HindIII fragment at the LUNAR1 TSS (top) or at the Notch-occupied enhancer (bottom). Crosslinking frequency is relative to a negative region (green). Error bars indicate the +/− SEM of three experiments.
Using chromosome conformation capture (3C) followed by qPCR we validated our Hi-C findings and identified a peak of high cross-linking frequency at the IGF1R enhancer when using a constant HindIII fragment located close to the LUNAR1 promoter, indicating the presence of a chromatin loop that places these regions in close physical proximity within the 3-D organization of the nucleus (Figure 4C). This finding was verified using a constant HindIII fragment in the IGF1R enhancer, suggesting a specific interaction (Figure 4C, lower panel). These results suggest that this Notch-occupied enhancer element in the IGF1R locus is able to control LUNAR1 expression through promoter/enhancer contacts.
LUNAR1 controls IGF1R expression and is essential for T-ALL maintenance
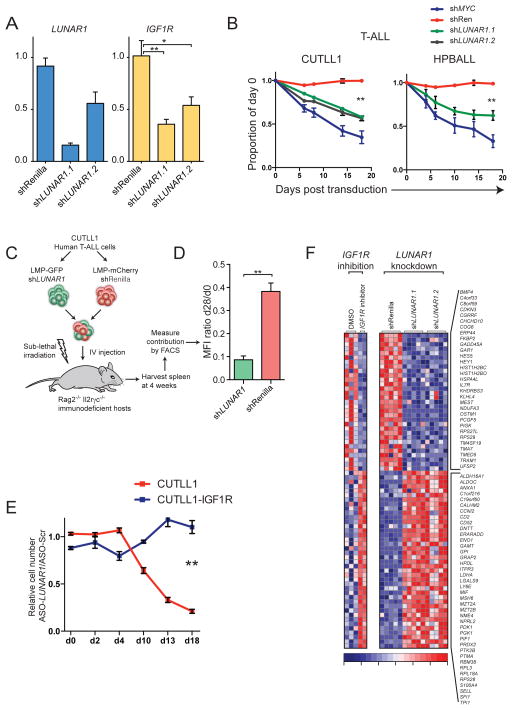
To provide evidence further supporting a more causal relationship between LUNAR1 and IGF1R we used RNAi to attenuate LUNAR1 expression. In doing so we noticed that abrogation of LUNAR1 led to repression of IGF1R, an effect that was observed using two independent shRNAs targeting distinct regions of the transcript (Figure 5A). Ectopic expression of LUNAR1 using retroviral vectors, which integrate randomly into the genome, did not yield significantly different expression of IGF1R mRNA (Supplementary Figure 5A), supporting our hypothesis of a cis-activation mechanism. Additionally, LUNAR1 depletion resulted in a competitive growth disadvantage phenotype in T-ALL cells (CUTLL1, HPB-ALL) in which the transcript is highly expressed (Figure 5B), but did not lead to a similar phenotype in myeloid leukemia cells (HL-60) (Supplementary Figure 5B), in which LUNAR1 expression is limited, suggesting on-target and T-ALL-specific effects (Figure 5B). Depletion of LUNAR1 caused a significant decrease in the number of actively cycling cells as shown by 7AAD staining (Supplementary Figure 5E). These T-ALL growth effects were similar to the ones noted when the IGF1R gene was silenced ((Medyouf et al., 2011)and data not shown). In order to further rule out off target effects of RNAi, we utilized antisense DNA/RNA hybrid oligonucleotide (ASO) gene silencing technology, which triggers RNase-H-mediated degradation of the target transcript upon ASO binding. ASO-mediated depletion of LUNAR1 led to both repression of IGF1R mRNA (Supplementary Figure 5C) and a growth retardation phenotype similar to what we observed using RNAi (Supplementary Figure 5D).
Figure 5. LUNAR1 regulates T-ALL proliferation by enhancing IGF1R expression.
(A) qPCR showing expression of LUNAR1 (blue) and IGF1R (yellow) in the presence of shRNAs targeting Renilla or LUNAR1. (B) Line graphs showing relative contribution from T-ALL cells expressing shRNAs against LUNAR1 grown in competition with cells expressing a non-targeting shRNA. (C) Illustration describing in vivo xenograft competition assay. (D) Ratio of MFI on day 28:day 0. (E) Line graphs showing relative cell number (targeting/Scr) for cells expressing exogenous IGF1R (blue) or empty vector (red) treated with ASO. (F) Genome-wide measurement of differentially expressed genes following pharmacological inhibition of IGF1R or LUNAR1 knockdown. Error bars represent +/− SEM of at least 3 experiments. * indicates p-value < 0.05. ** indicates p-value < 0.01.
In order to test the effects of LUNAR1 depletion on tumor growth in vivo we performed xenograft assays in which human T-ALL cells expressing an shRNA targeting LUNAR1 (GFP) or Renilla (mCherry) control were mixed at a 1:1 ratio and transferred intravenously into sublethally irradiated immunodeficient hosts (Figure 5C). Four weeks after the transplantation we harvested peripheral tumors and measured the relative contribution of cells harboring each shRNA by FACS analysis. Using this assay we observed a significant loss of representation of cells in which LUNAR1 was depleted (Figure 5D), which suggested that this lncRNA is required for efficient tumor growth both in vitro and in vivo.
To test whether IGF1R is the relevant target of LUNAR1 as we have hypothesized, we performed ASO-mediated lncRNA depletion in T-ALL cells ectopically expressing IGF1R using retroviral constructs or empty vector control. Following ASO delivery, we observed that ectopic expression of IGF1R efficiently rescued the growth defects observed following LUNAR1 depletion (Figure 5E), suggesting that IGF1R is indeed a key target of this lncRNA.
To characterize the overall cellular response to LUNAR1 silencing we performed global gene expression analysis (RNA-Seq) in T-ALL cells following LUNAR1 depletion with two independent shRNAs. For comparison, we also performed experiments in which we blocked IGF1 signaling using pharmacological inhibition (BMS-536924) followed by RNA-Seq. The outcome of these studies was striking as we noted significant repression of IGF1R mRNA, in agreement to our qPCR studies (Figure 5A). We also identified a subset of genes, which were similarly regulated following either IGF1R inhibition or LUNAR1 depletion (Figure 5F), suggesting that this lncRNA promotes IGF1 signaling by transcriptional regulation of IGF1R. Finally, using gene set enrichment analysis (GSEA) we found significant enrichment for gene-sets containing genes significantly down-regulated upon IGF1 inhibition in T-ALL (Supplementary Figure 5F), and other publicly available gene-sets containing targets of IGF1/2 (Supplementary Figure 5F).
LUNAR1 is an activator RNA capable of stimulating gene activity
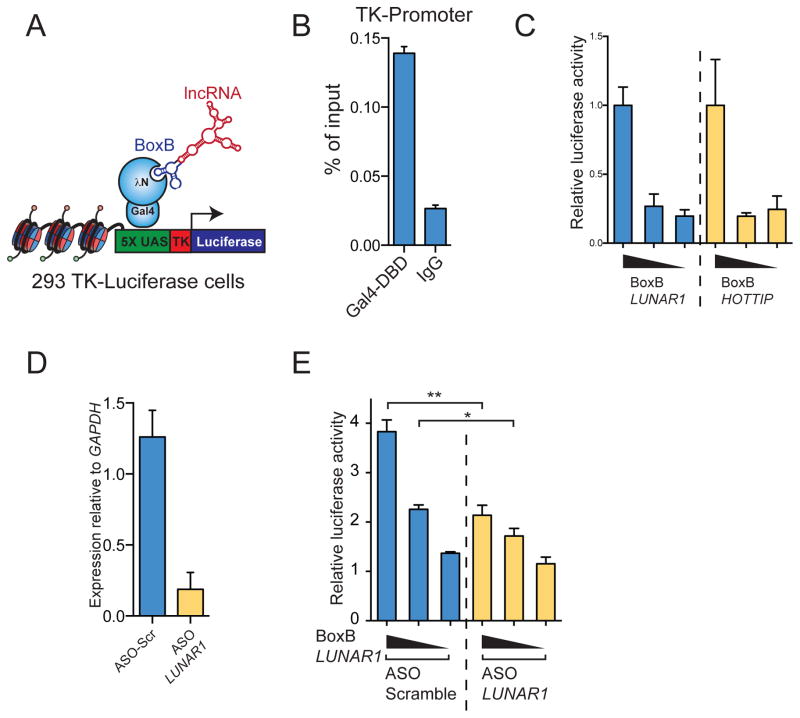
In order to test whether LUNAR1 displayed intrinsic ability to promote gene activity, we used the Gal4-λN/BoxB system to tether this lncRNA to a heterologous reporter promoter(Li et al., 2013; Wang et al., 2011b). In this system the BoxB RNA stem loop is fused to a lncRNA, which allows it to bind specifically and with high affinity to the phage λ N-peptide (λN). By fusing λN to the DNA binding domain of the yeast Gal4 protein, a BoxB tagged lncRNA can be tethered to UAS DNA sequences (Figure 6A). We co-transfected vector containing the Gal4-λN fusion with either BoxB tagged LUNAR1 or known activator lncRNA HOTTIP into HEK 293 cells stably expressing 5 UAS sites upstream of a TK-Luciferase reporter gene(Vaquero et al., 2004). Upon transfection we could detect binding of the Gal4-λN fusion at the reporter promoter as expected (Figure 6B). Tethering LUNAR1 to this reporter gene stimulated transcription of the reporter to a similar degree as HOTTIP (Figure 6C). Additionally ASO-mediated depletion of LUNAR1 (Figure 6D) significantly reduced this stimulatory effect compared to a non-targeting ASO (Figure 6E), suggesting that molecules of LUNAR1 are functionally important for the stimulation of transcription seen here.
Figure 6. LUNAR1 is able to stimulate transcription of a reporter gene.
(A) Illustration describing the BoxB Gal4-λN RNA tethering system used. (B) ChIP assay for Gal4-DBD at the reporter gene promoter. (C) Luciferase reporter activity in experiments where BoxB tagged LUNAR1 (blue) or HOTTIP (yellow) were co-transfected with Gal4-λN. (D) qPCR following ASO knockdown of LUNAR1 in luciferase assay. (E) Reporter assay showing relative reporter gene activity when BoxB-LUNAR1 was co-transfected with either non-targeting (blue) or LUNAR1-specific (yellow) ASOs. Error bars represent SEM of at least 3 experiments. * indicates p-value < 0.05. ** indicates p-value < 0.01.
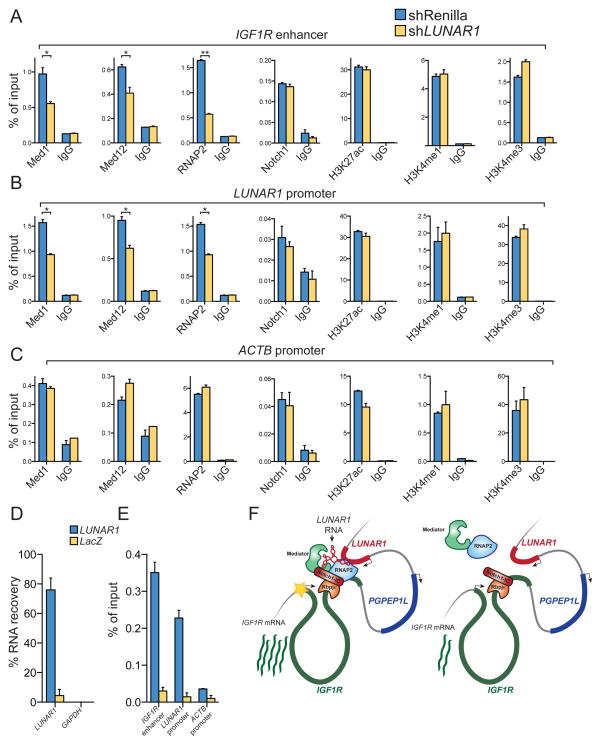
In order to investigate the mechanism by which endogenous LUNAR1 promotes transcriptional activity of the IGF1R gene, we performed chromatin studies following depletion of the lncRNA in T-ALL cells. Since we hypothesized that LUNAR1 might be an important component of the intronic IGF1R enhancer we reasoned that depletion of this transcript might influence chromatin state or occupancy of one of the activators at that locus. Following depletion of LUNAR1 we observed a significant reduction in Mediator complex (MED1 and MED12) and RNA Pol2 occupancy at both the IGF1R enhancer and the LUNAR1 promoter (Figure 7A, 7B). Additionally we observed significant loss of RNA Pol2 binding at the IGF1R promoter (not shown). We did not observe any changes in NOTCH1 binding or levels of histone modifications H3K27ac, H3K4me1 or H3K4me3 at these loci (Figure 7A, 7B), suggesting specific destabilization of Mediator and RNAP2 following LUNAR1 depletion. We observed no changes in the status of any of these factors at the ACTB promoter, again suggesting a locus specific effect. Since depletion of LUNAR1 led to locus specific loss of Mediator and RNA Pol2 binding, we hypothesized that LUNAR1 RNA might co-occupy the chromatin in those loci. Using chromatin isolation by RNA purification (ChIRP), we were able to efficiently retrieve LUNAR1 RNA (Figure 7D) and observed specific enrichment of LUNAR1 at the IGF1R enhancer and LUNAR1 promoter (Figure 7E) which supports a mechanism by which LUNAR1 exploits chromatin configuration to reach its targets (Figure 7F)(Engreitz et al., 2013). Together these data suggest that the intronic IGF1R enhancer activates LUNAR1, which then co-occupies the element and further recruits Mediator in order to sustain full activation of the IGF1R promoter.
Figure 7. LUNAR1 modulates Mediator and RNA PolII binding at the IGF1R enhancer.
ChIP assays for MED1, MED12, RNAP2, NOTCH1, H3K27ac, H3K4me1 and H3K4me3 in T-ALL cells harboring non-targeting (blue) or LUNAR1-specific shRNAs followed by locus-specific qPCR at (A) IGF1R enhancer, (B) LUNAR1 promoter or (C) ACTB promoter. (D) Percent recovery of LUNAR1 following ChIRP. (E)ChIRP assay using probes targeting LUNAR1 (blue) or LacZ (yellow) followed by qPCR at the IGF1R enhancer, LUNAR1 promoter and ACTB promoter. (F) A model for cis-regulation of gene expression by LUNAR1. Error bars represent SEM of at least 3 experiments. * indicates p-value < 0.05. ** indicates p-value < 0.01.
These studies revealed LUNAR1 as a novel regulator of IGF1 signaling and T-ALL cell growth, identifying the first putative lncRNA that could be therapeutically targeted in acute leukemia. Additionally we provide further evidence for the ubiquitous presence and functional importance of lncRNAs in human disease and provide evidence that lncRNAs, in addition to protein-coding genes, are key downstream targets of Notch signaling.
DISCUSSION
The last decades were characterized by an extensive characterization of oncogenic pathways in different cancer types, focusing on protein-coding transcripts and more recently on non-coding micro-RNA networks. However, very little was known on expression patterns and biological significance of lncRNAs in human tumorigenesis. Also, very little was reported on the regulation of such lncRNAs by well-described oncogenic signaling pathways. Here we map the expression of lncRNAs in acute leukemia, using T-ALL, a disease characterized by activation of the NOTCH pathway. Using integration of whole transcriptome analysis and genome-wide chromatin state maps we systematically identified T-ALL-specific lncRNA genes and characterized non-coding transcripts regulated directly by the binding and the activation of NOTCH1. To further suggest biological significance we used RNA-Seq of primary human T-ALL, characterize by NOTCH1 and FBXW7 mutations (leading to NOTCH activation) and were able to prove the existence of T-ALL-specific lncRNAs also regulated by NOTCH1 activity, suggesting that NOTCH signaling is able to shape not only the protein-coding but also the lncRNA landscape in this disease. These results suggest that – at least a fraction- of the NOTCH1-triggered oncogenic activity could be due to its ability to regulate such non-coding transcripts.
To test this assumption we have selected LUNAR1, a lncRNA that shows T-ALL-specific Notch-dependent expression patterns, is localized in the nucleus and displayed high correlation with IGF1R, a receptor previously suggested to play a role in T-ALL (Medyouf et al., 2011). Using assays that can map chromosome conformation and looping we were able to show an interaction between the TSS of LUNAR1 and an enhancer element located within the IGF1R locus, characterized by NOTCH1 binding (Figure 4, Supplementary Figure 4). To prove the connection between LUNAR1 expression and IGF1R, we were able to demonstrate that the silencing of this lncRNA led to a significant down-regulation of IGF1R expression in T-ALL cells and diminished IGF1 pathway activity. Although it is possible that IGF1R is not the sole target of LUNAR1, we propose that one of its key functions is to modulate IGF1 signaling in TALL by regulation transcription of the receptor gene (Figure 5).
Using various assays (Figures 6 and 7) we have provided evidence that LUNAR1 belongs to a sub-class of enhancer-like lncRNAs, which have been reported previously(Lai et al., 2013; Orom et al., 2010; Wang et al., 2011b; Yang et al., 2014). Among enhancer-like lncRNAs there appear to be two distinct functional mechanisms at play. One mechanism involves lncRNA-dependent recruitment of WDR5-containing methyltrasferase complexes, which catalyzed methylation of the tails of histone 3 on lysine 4(Gomez et al., 2013; Wang et al., 2011b; Yang et al., 2014). A second mechanism involves stabilization of Mediator complexes and RNA Pol2 at enhancer elements(Lai et al., 2013). Based on chromatin experiments following LUNAR1 depletion this lncRNA is most likely functionally similar to the non-coding RNA-activators (nc-RNA-a) which have been described previously(Lai et al., 2013; Orom et al., 2010). Since there are currently no methods for predicting lncRNA function based on sequence alone we believe our results represent an incremental, yet important contribution to the overall understanding of lncRNA biology.
Our mapping of lncRNAs in human T-ALL opens the possibility that such previously uncharacterized transcripts are key modulators of cellular transformation, through their interaction with oncogenic and tumor suppressor programs in leukemia. Additionally, the suggested (and reported also here) tissue and cell-type specificity of lncRNA expression would suggest that such transcripts could be powerful and specific biomarkers used to categorize cancer subtypes and stratify patients for clinical trials and therapeutic protocols.
MATERIALS AND METHODS
RNA extraction preparation for next generation sequencing
Total RNA was extracted from samples using the RNeasy Plus mini kit (Life Technologies, Carlsbad, CA). Samples were then subject to PolyA selection (Figures 1E, 5F and 5G only) using oligo-dT beads (Life Technologies, Carlsbad, CA) or rRNA removal (all other samples) using the Ribo-Zero kit (Epicentre, Madison, WI) according to the manufacturers instructions. The resulting RNA samples were then used as input for library construction using the dUTP method as described(Parkhomchuk et al., 2009). RNA libraries were then sequenced on the Illumina HiSeq 2000 or 2500 using 50bp paired-end reads.
RNA-Sequencing data analysis
All RNA-Seq data was aligned to hg19 using TopHat (Trapnell et al., 2009) v1.4 with default parameters. We used Cuffdiff (Parkhomchuk et al., 2009) v1.3 for all differential expression (DE) analyses with our custom annotation consisting of RefSeq entries plus T-ALL lncRNAs as described below. In all DE tests, a gene was considered significant if the q-value was less than 0.05 (Cuffdiff default). Data have been deposited at NCBI Gene Exprerssion Omnibus accession number XXXXXX.
LncRNA discovery
We sequenced two samples from T-ALL cell lines (CUTLL1 and HPBALL), two primary human thymus samples to ultra high depth (>200 million million mate pairs each), ten primary pediatric T-ALL samples (60–80 million mate pairs each) and data generated by the Roadmap Epigenomics project for Naïve CD4+ and CD8+ T-cells to be used for ab initio transcirptome assembly with Cufflinks v1.3. Briefly, Cufflinks was run with the following options: -u, -N, -g (RefSeq GTF file provided as guide), -M (rRNA and 7SK RNA mask file provided). We generated transcriptome assemblies for each of these samples separately and then used Cuffmerge to combine all annotations. For all subsequent filtering and processing steps please se Supplementary Methods.
Chromatin immunoprecipitation (ChIP)
The following antibodies were used for chromatin immunoprecipitation experiments: Notch1 C-20 (Santa Cruz sc-6014), Med1 (Bethyl, A300-793A), Med12 (Bethyl, A300-774A), RNA PolII N-20 (Santa Cruz, sc-899), H3K4me1 (Abcam, ab8895), H3K4me3 (Active Motif, 39159), H3K27ac (Abcam, ab4729). ChIP assays were performed essentially as described previously(Whyte et al., 2013). See supplementary methods for detailed description of ChIP assay.
Chromatin isolation by RNA purification (ChIRP)
ChIRP assays were performed as described (Chu et al., 2011) with the following modifications. Cells were double crosslinked first with 2mM EGS for 45 minutes at room temperature and washed twice with ice cold PBS. Cells were then further cross liked with 3% formaldehyde for 30 minutes at room temperature and reaction was stopped by the addition of 0.125M glycine followed by two more washes in PBS. Crosslinked cells were then lysed in sonication buffer (see ChIP method) supplemented with SUPERaseIn (Life technologies) and chromatin was sheared exactly as for ChIP assays. Chromatin was cleared by centrifugation and supernatant was used for ChIRP reactions. All following steps were performed exactly as described by Chu et al. with the following exceptions: in the hybridization buffer 1% Triton with 0.1% SDS was used instead of 1% SDS, in the wash buffer SDS concentration was lowered from 0.5% to 0.1%. Probes used for ChIRP assays are listed in supplemental table.
Chromosome conformation capture (3C)
3C experiments were performed essentially as described previously(Hagege et al., 2007). Please see Supplementary methods for detailed description of this method.
Supplementary Material
SUMMARY.
Notch signaling is a key developmental pathway, which is subject to frequent genetic and epigenetic perturbations in many different human tumors. Here we investigate whether lncRNA genes, in addition to mRNAs are key downstream targets of oncogenic Notch1 in human T-ALL. By integrating transcriptome profiles with chromatin state maps we have uncovered many previously unreported T-ALL-specific lncRNA genes, a fraction of which are directly controlled by the Notch1/Rpbjκ activator complex. Finally we have shown that one specific Notch-regulated lncRNA, LUNAR1, is required for efficient T-ALL growth in vitro and in vivo due to its ability to enhance IGF1R mRNA expression and sustain IGF1 signaling. These results confirm that lncRNAs are important downstream targets of the Notch signaling pathway and additionally they are key regulators of the oncogenic state in T-ALL.
HIGHLIGHTS.
Many previously unreported lncRNA genes are expressed in human TALL
Notch1 oncogene directly controls lncRNA expression in T-ALL
LUNAR1 is a Notch-regulated pro-oncogenic lncRNA which is essential for T-ALL growth
LUNAR1 maintains high expression of IGF1R mRNA through a cis-activation mechanism
Acknowledgments
We would like to thank the members of the Aifantis laboratory for helpful discussions throughout the duration of the project. We thank Drs. Danny Reinberg, Roberto Bonasio, Howard Chang and Adam Schmidtt for valuable discussions, technical advice and reagents. We thank Drs. Sergei Koralov and Jane Skok for their continued support and helpful insight. We are grateful to Dr. Mignon Loh and the Children’s Oncology Group for providing patient samples. Dr. A. Heguy and the NYU Genome Technology Center (supported in part by NIH/NCI P30 CA016087-30 grant) for assistance with sequencing experiments. I.A. was supported by the National Institutes of Health (1RO1CA133379, 1RO1CA105129, 1RO1CA149655, 5RO1CA173636, 5RO1CA169784, and 1RO1GM088847). I.A. was also supported by the William Lawrence and Blanche Hughes Foundation, The Leukemia & Lymphoma Society (TRP#6340-11, LLS#6373-13), The Chemotherapy Foundation, The Irma T. Hirschl Trust, The V Foundation for Cancer Research and the St. Baldrick’s Foundation. TT is supported by the NIH training grant 5T32CA009161-37. I.A. is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013 doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumil RM, Lee JT. Forty years of decoding the silence in X-chromosome inactivation. Hum Mol Genet. 2001;10:2225–2232. doi: 10.1093/hmg/10.20.2225. [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Molecular cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, et al. The reality of pervasive transcription. PLoS Biol. 2011;9:e1000625. doi: 10.1371/journal.pbio.1000625. discussion e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, Girardi T, Gianfelici V, Geerdens E, Clappier E, Porcu M, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nature genetics. 2013;45:186–190. doi: 10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, Chen Y, Liu XS. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20:908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J, Grunn A, Fangazio M, Capello D, Monti S, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. The Journal of experimental medicine. 2011;208:1389–1401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando AA. The role of NOTCH1 signaling in T-ALL. Hematology / the Education Program of the American Society of Hematology. American Society of Hematology Education Program; 2009. pp. 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, Behm FG, Pui CH, Downing JR, Gilliland DG, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- Fragoso R, Mao T, Wang S, Schaffert S, Gong X, Yue S, Luong R, Min H, Yashiro-Ohtani Y, Davis M, et al. Modulating the strength and threshold of NOTCH oncogenic signals by mir-181a-1/b-1. PLoS genetics. 2012;8:e1002855. doi: 10.1371/journal.pgen.1002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garding A, Bhattacharya N, Claus R, Ruppel M, Tschuch C, Filarsky K, Idler I, Zucknick M, Caudron-Herger M, Oakes C, et al. Epigenetic upregulation of lncRNAs at 13q14.3 in leukemia is linked to the In Cis downregulation of a gene cluster that targets NF-kB. PLoS genetics. 2013;9:e1003373. doi: 10.1371/journal.pgen.1003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152:743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome Profiling Provides Evidence that Large Noncoding RNAs Do Not Encode Proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nature protocols. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome research. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature genetics. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- King B, Trimarchi T, Reavie L, Xu L, Mullenders J, Ntziachristos P, Aranda-Orgilles B, Perez-Garcia A, Shi J, Vakoc C, et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell. 2013;153:1552–1566. doi: 10.1016/j.cell.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey L, Haas S, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Ungewickell A, Bhaduri A, Qu K, Webster DE, Armstrong R, Weng WK, Aros CJ, Mah A, Chen RO, et al. Transcriptome sequencing in Sezary syndrome identifies Sezary cell and mycosis fungoides-associated lncRNAs and novel transcripts. Blood. 2012;120:3288–3297. doi: 10.1182/blood-2012-04-423061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Sanda T, Look AT, Novina CD, von Boehmer H. Repression of tumor suppressor miR-451 is essential for NOTCH1-induced oncogenesis in T-ALL. The Journal of experimental medicine. 2011;208:663–675. doi: 10.1084/jem.20102384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nature genetics. 2010;42:1113–1117. doi: 10.1038/ng.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Look AT. Molecular pathways in T-cell acute lympho-blastic leukemia: ramifications for therapy. Clinical advances in hematology & oncology: H&O. 2004;2:779–780. [PubMed] [Google Scholar]
- Medyouf H, Gusscott S, Wang H, Tseng JC, Wai C, Nemirovsky O, Trumpp A, Pflumio F, Carboni J, Gottardis M, et al. High-level IGF1R expression is required for leukemia-initiating cell activity in T-ALL and is supported by Notch signaling. The Journal of experimental medicine. 2011;208:1809–1822. doi: 10.1084/jem.20110121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D, da Ros V, Tang Z, Siegle J, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nature medicine. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T, Barnes KC, Real PJ, Glade Bender JL, Sulis ML, Murty VV, Colovai AI, Balbin M, Ferrando AA. CUTLL1, a novel human T-cell lymphoma cell line with t(7;9) rearrangement, aberrant NOTCH1 activation and high sensitivity to gamma-secretase inhibitors. Leukemia. 2006;20:1279–1287. doi: 10.1038/sj.leu.2404258. [DOI] [PubMed] [Google Scholar]
- Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic acids research. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nature genetics. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, Guenther MG, Almada AE, Lin C, Sharp PA, Giallourakis CC, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP. lincRNAs: Genomics, Evolution, and Mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Molecular cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Wang H, Zou J, Zhao B, Johannsen E, Ashworth T, Wong H, Pear WS, Schug J, Blacklow SC, Arnett KL, et al. Genome-wide analysis reveals conserved and divergent features of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia cells. Proceedings of the National Academy of Sciences of the United States of America. 2011a;108:14908–14913. doi: 10.1073/pnas.1109023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011b;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YW, Flynn RA, Chen Y, Qu K, Wan B, Wang KC, Lei M, Chang HY. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. eLife. 2014;3:e02046. doi: 10.7554/eLife.02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Slovak ML, Mannoor K, Chen C, Hunger SP, Carroll AJ, Schultz RA, Shaffer LG, Ballif BC, Ning Y. Microarray detection of multiple recurring submicroscopic chromosomal aberrations in pediatric T-cell acute lymphoblastic leukemia. Leukemia. 2011;25:1042–1046. doi: 10.1038/leu.2011.33. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.