Abstract
Here we report iceFISH, a multiplex imaging method for measuring gene expression and chromosome structure simultaneously on single chromosomes. We demonstrate that chromosomal translocations can alter transcription chromosome-wide, finding substantial differences in transcriptional frequency between genes located on a translocated chromosome in comparison to the normal chromosome in the same cell. Examination of correlations between genes on a single chromosome revealed a cis chromosome-level transcriptional interaction spanning 14.3 megabases.
Researchers generally believe that the transcription of a gene's DNA into RNA is controlled by the interaction of regulatory proteins with DNA sequences proximal to the gene itself. At the same time, genes are organized by the thousands into chromosomes, raising the possibility that the structure or organization of chromosomes themselves may influence transcription1,2; however, little is known about how organization at the chromosome length scale affects gene expression. Here, we describe a method based on RNA fluorescence in situ hybridization 3,4 (RNA FISH) called iceFISH (for intron chromosomal expression FISH) that enabled us to generate per-chromosome transcriptional profiles of 20 genes simultaneously along individual copies of human chromosome 19 in single cells.
To capture the transcriptional activity of each gene, we took advantage of the fact that cells transcribe nascent RNAs comprised of exons and introns and that introns typically degrade rapidly after being spliced out of nascent RNA. Labeling the intron with short, fluorescently labeled oligonucleotides4 (Fig. 1a, Supplementary Fig. 1a) enabled us to measure whether or not the gene is actively transcribing5, and, if active, the three-dimensional coordinates of that gene6-8. A series of control experiments confirmed that the intron spot marked the site of transcriptionally active genes (Supplementary figs. 2-7). Note that even genes considered constitutively active do not always actively transcribe RNA, as transcription occurs in short but intense “bursts” thought to arise from random aspects of the transcriptional process9-12. The overall transcription rate is proportional to the probability of finding such a spot for each gene (see supplementary discussion).
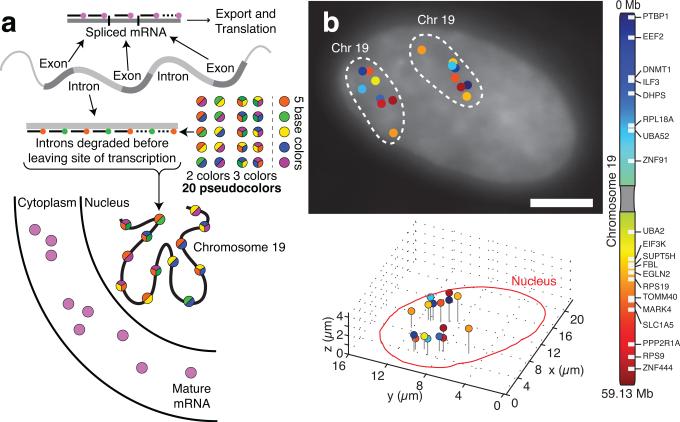
Figure 1. Unique identification of 20 loci on chromosome 19 by RNA FISH targeting introns in human foreskin fibroblasts.
a. Depiction of our pseudocoloring scheme for labeling the site of transcription by targeting gene introns with a series of labeled oligonucleotide probes. b. Using images from each fluorescence channel, we computationally identified the transcriptional activity and location of the 20 genes. Along with probes targeting the introns, we also included probes targeting Cyclin A2 mRNA to determine position in cell cycle and SUZ12 mRNA as a fiducial marker. The scale bar is 5μm long.
In order to concomitantly measure overall chromosome structure, we designed probes targeting the introns of 20 genes along chromosome 19 (Supplementary fig. 1a and Supplementary Table 1). This yielded an average resolution of 3Mb with a minimum of 360Kb, although we also demonstrated the ability to distinguish loci separated by just 30Kb (Supplementary Fig. 6). In order to measure all 20 genes’ transcriptional status simultaneously, we utilized a color-coding approach in which we labeled each gene's introns with a particular “pseudocolor”, which is a distinct code for each gene consisting of either two or three out of a base palette of five spectrally distinguishable fluorophores (Fig. 1A; akin to other schemes13,14). To assign gene identity, we looked for colocalization of two or three spots in the images we acquired for each fluorescence channel (Supplementary Fig. 8). In human foreskin fibroblasts, we could discern two clearly separated chromosomes (Fig. 1b) 78% of the time (introducing no bias; Supplementary fig. 9). On average, we found 6 ± 2 expressing genes (out of the 20 labeled) per chromosome. The positions of these genes appeared more spread out than expected15 (Supplementary Fig. 6, see supplementary discussion). We found that using more probes did not change spot detection efficiency (see Methods), nor does pseudocoloring incur a significant rate of spot misidentification (Supplementary fig. 10). We ensured that the cells we analyzed were in the G0/G1 stage of the cell cycle by co-labeling Cyclin A2 mRNA and examining only cells with low levels of Cyclin A2, which is abundant during the S, G2, and M phases of the cell cycle16 (Supplementary fig. 11).
By grouping actively transcribing genes into territories corresponding to each chromosome, we constructed transcriptional profiles showing which of our 20 genes are on and off per chromosome. Researchers largely believe that gene transcription depends on both chromosome-extrinsic trans factors (such as transcription factors) and local cis factors on the DNA (typically within 1 megabase of the gene itself). Our method enabled us to examine the possibility that non-local mechanisms at the chromosome scale may also regulate transcription.
Translocations provide a means to search for such possibilities: while they disrupt the large-scale structure of a chromosome, the cell's trans environment and local cis DNA regulatory code remains unchanged for most genes on the translocated chromosome. For example, HeLa cells contain two intact copies of chromosome 19 and one copy that is split into two pieces fused to parts of other chromosomes17: one, denoted t(6;19), consists of the first 17-20 megabases of chromosome 19 fused to part of chromosome 6, and the other, denoted t(13;19) consists of the remaining 40-43 megabases of chromosome 19 translocated onto a portion of chromosome 13 (Fig. 2a, Supplementary fig. 12). Our iceFISH data recapitulated these genetic rearrangements (Fig. 2b). We found that most genes on t(13;19) were up to 5 fold more transcriptionally active than those on the normal copies of chromosome 19 (Fig. 2c, replicate in Supplementary fig. 13), consistent with the existence of chromosome-specific transcriptional regulation that the translocation may have disrupted in some way. Intron spot intensities were roughly the same on all the chromosomes we examined (Supplementary fig. 14), suggesting that transcriptional hyperactivation results from an increased probability of a gene being active rather than an increased rate of transcription when the gene is active (see supplementary discussion).
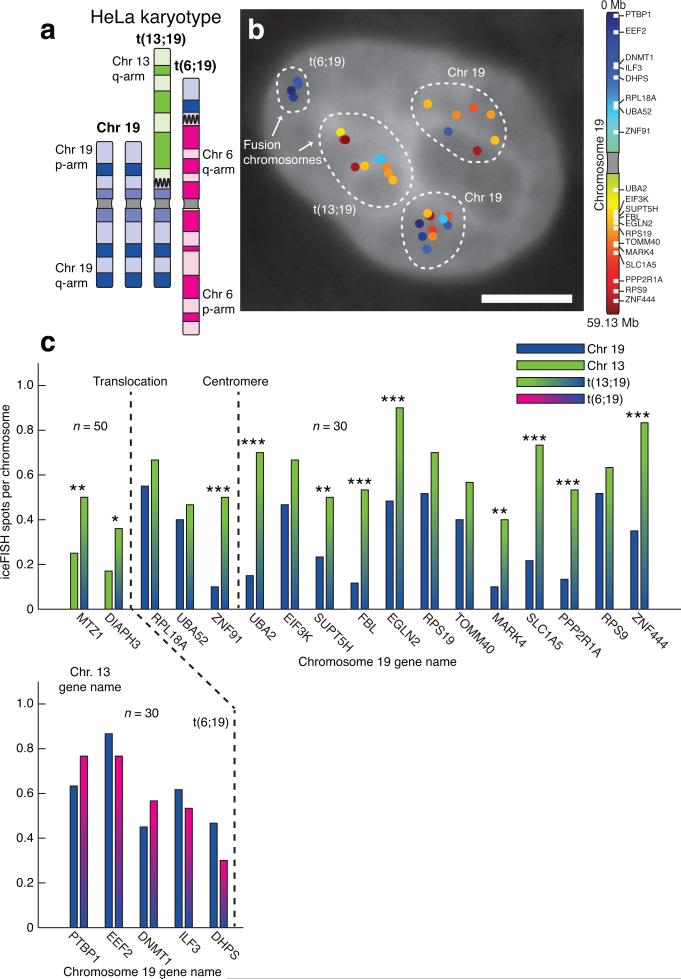
Figure 2. Translocated portions of chromosome 19 can display different expression patterns than intact chromosomes.
a. Schematic showing chromosome 19 and its derivatives in our HeLa cells. b. Computational identification of actively transcribing genes on chromosome 19 revealed the two intact copies and the two translocated pieces of chromosome 19. The scale bar is 5μm long. c. Comparison of the transcriptional activity of the genes on chromosome 19 (as measured by frequency of observing a transcription site per chromosome) on the translocated fragments of chromosome 19 to the intact copies of chromosome 19. For the expression of the two genes on chromosome 13 (MTZ1 and DIAPH3), we measured spot frequency as described in Supplementary fig. 15. We denote p-values for the difference in frequency (using a binomial distribution test) by *** for p<0.001, ** for p<0.01, * for p<0.05.
We then asked whether the portion of chromosome 13 on t(13;19) also displays heightened transcriptional activity. We found that the transcriptional frequency of two genes from chromosome 13 (DIAPH3 and MTZ1) was roughly 2 fold higher on t(13;19) than on the normal copies of chromosome 13 (Fig. 2c, Supplementary fig. 15), suggesting that this translocation resulted in hyperactivation of all genes on t(13;19) irrespective of location. Meanwhile, transcription of the chromosome 19 genes on t(6;19) was similar to the normal copies (Fig. 2c), suggesting that translocations do not necessarily lead to transcriptional changes. We note also that per-chromosome differences in transcription are difficult to observe using bulk assays that average expression from all chromosomes, which may explain why reports of such effects18 are not widespread. We also found that these transcriptional differences did not appear to correlate with any differences in spatial chromosome conformation (Supplementary figs. 6, 16, 17).
We next looked for evidence of interactions governing the transcription of genes within a single chromosome by examining whether the transcriptional status of one gene in our panel affected the transcriptional status of another gene on the same chromosome. Such an interaction would manifest itself as a deviation from independence, with positive correlations signifying that the two genes A and B would be more likely than chance to be actively transcribing at the same time on the same chromosome, and anti-correlations indicating that the transcriptional statuses of genes A and B would be mutually exclusive.
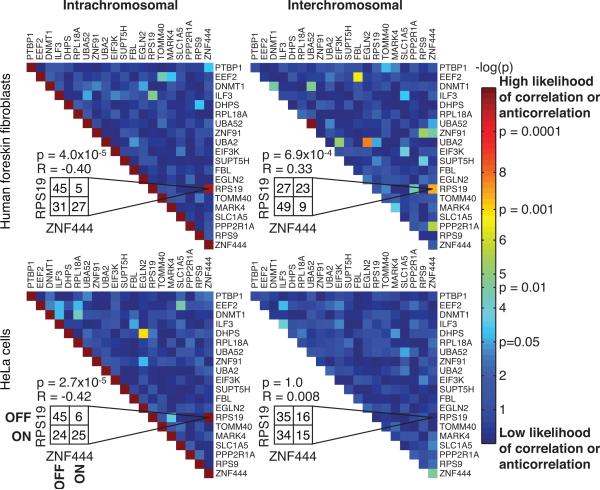
We found that most pairwise interactions on single chromosomes did not show significant deviations from independence (Fig. 3, Supplementary figs. 18 & 19). However, one pair of genes, RPS19 and ZNF444 (separated by 14.3 megabases), showed an anti-correlation (R = -0.40±0.08; p=3.99×10-5, Fisher Exact Test). One explanation for this anti-correlation is fluctuations in a potential trans-acting factor, such as a transcription factor, that activated RPS19 and inactivated ZNF444 in some cells while activating ZNF444 and inactivating RPS19 in others. Such trans factors would, however, also affect the copy of the gene on the other chromosome 1919. Thus, we looked for an anti-correlation between RPS19 on one chromosome and ZNF444 on the other copy of chromosome 19 in the same cell. We found that the inter-chromosomal interaction between the genes was qualitatively different, having a mild and less statistically significant positive correlation (R = 0.33±0.09; p=6.90×10-4, Fisher Exact Test), indicating that the interaction between these genes is indeed a cis effect confined to the chromosome itself. The lack of anti-correlation between the chromosome 19 copies (both between the pair of genes and also each gene with itself; Fig. 3) also precludes the possibility of genetic imprinting. Notably, we also found the exact same pattern of interactions when examining the two intact copies of chromosome 19 in HeLa cells (Fig. 3). The interaction between RPS19 and ZNF444 disappeared, however, on t(13;19) (Supplementary Fig. 20), suggesting that whatever mechanism is responsible is an intrinsic property of a fully intact chromosome 19. The spatial conformation of chromosomes did not correlate with these observations (Supplementary figs. 17, 21).
Figure 3. Identification of a pair of genes showing an intrachromosomal but not interchromosomal expression relationship.
a. Intrachromosomal cis transcriptional interactions between two genes would result in a correlation or anti-correlation in the transcriptional activity of the two genes on the same chromosome, whereas trans effects would yield interchromosomal interactions. b. Heat map showing the deviation from independence of the intra and interchromosomal transcriptional activity of all pairs of genes we measured in human foreskin fibroblasts and the two intact copies of chromosome 19 in HeLa cells (p-value calculated using the Fisher Exact Test; see methods). A smaller p-value indicates a more significant deviation from independence. Here, we have presented data combined from two independent biological replicates (see Supplementary figs. 18 & 19 for replicates).
Our method has enabled us to measure transcriptional activity on individual chromosomes in single cells by spatially segregating intron RNA FISH signals to particular chromosome territories, revealing regulatory mechanisms working at the chromosome-scale. We believe our method provides a nice complement to chromosome conformation assays20 that look for interactions at the DNA (rather than transcriptional) level. We believe that iceFISH and similar tools will allow us to determine the prevalence of these chromosome-level regulatory phenomena and uncover their underlying mechanisms.
Online Methods
Cell culture, fixation, and fluorescent in situ hybridization
We grew primary human foreskin fibroblasts (ATCC CRL 2097) or HeLa cells in Dulbecco's modified eagle's medium with glutamax (DMEM, Life Technologies) supplemented with penicillin/streptomycin and 10% fetal bovine serum. We enriched for G0/G1 phase cells through a double-thymidine block (2mM thymidine in medium) procedure, which arrested cells at the beginning of S phase. We released the cells and let them go through S, G2, M, G1, S, G2, M, and then fixed them when they were in G1. We let the cells go through over one complete cell cycle to minimize any potential transcriptional or structural effects due to the block itself. To fix the cells, we followed the protocol of Raj et al. Nat. Meth. 2008. Briefly, we fixed the cells for 10 minutes at room temperature using 4% formaldehyde/10% formalin in 1x phosphate buffered saline solution (PBS), followed by two rinses in 1x PBS, after which we permeabilized the cells with 70% EtOH and stored at 4°C at least overnight.
To perform fluorescence in situ hybridization (FISH), we again followed the procedure of Raj et al. Nature Methods 2008 with some minor modifications. We prewashed with a wash buffer containing 10% formamide and 2x saline-sodium citrate (SSC), then hybridized by adding the appropriate amount and type of probe (described later) in a buffer containing 10% formamide, 2x SSC and 10% dextran sulfate (W/V). We empirically determined the optimal concentration of each probe, which in most cases was roughly equivalent to the concentrations used in Raj et al. Nature Methods 2008. We hybridized our samples overnight in a humidified chamber kept at 37°C, then washed twice for 30 minutes with wash buffer at 37°C (adding DAPI at a concentration of 50 ng/mL in the second wash), and then imaged in 2x SSC as described below.
In the case of the experiments involving Actinomycin D, we incubated HeLa cells in 2μg/mL of Actinomycin D (Sigma) for 0, 30, 60, and 120 minutes (as described in Supplementary fig. 3), after which we fixed the cells and performed FISH. We made sure to thoroughly mix the Actinomycin D into the medium before adding it to avoid spatial inhomogeneity in the activity of the drug.
For the RNase experiments, we fixed and permeabilized the cells as just outlined, after which we aspirated the 70% EtOH, washed once with 1x PBS, then added 1x PBS with 10μg/mL of RNase A (Sigma). We incubated the fixed cells at 37C for 30 minutes, washed with 1x PBS, and then proceeded with FISH as outlined above. As a control, we performed the exact same procedure on cells in a neighboring well, but didn't add RNase A to the 1x PBS for the incubation (as described in Supplementary fig. 2).
Imaging
We imaged all our samples on a Nikon Ti-E inverted fluorescence microscope using a 100x Plan-Apo objective (numerical aperture of 1.43) and a cooled CCD camera (Pixis 1024B from Princeton Instruments). We sequentially acquired three-dimensional stacks of fluorescent images in 6 different fluorescent channels using filter sets for DAPI, Atto 488, Cy3, Alexa 594, Atto 647N, and Atto 700. Our exposure times were roughly 2-3 seconds for most of the dyes except for DAPI (which we exposed for ~100ms) and Atto 700 (~5 seconds, due to somewhat weaker illumination on our apparatus). The spacing between consecutive planes in our stacks is 0.3 μm. The filter sets we used were 31000v2 (Chroma), 41028 (Chroma), SP102v1 (Chroma), a custom set from Omega as described in ref. 4, SP104v2 (Chroma), and SP105 (Chroma) for DAPI, Atto 488, Cy3, Alexa 594, Atto 647N, and Atto 700, respectively.
Image analysis
Once we acquired our images, we put them through an image analysis pipeline made up of custom semi-automated spot recognition software we wrote in MATLAB with the following series of steps:
We first identified candidate spots in the three-dimensional image by filtering the image with a Laplacian of Gaussian filter, and taking the top 300 spots as candidates. In some cases, we also chose cells to analyze based on phase in the cell-cycle. In those cases, we chose cells that had little or no Cyclin A2 mRNAs. Our experiments in Supplementary fig. 11 validate this approach.
For each candidate, we then fit the candidate to a Laplacian of Gaussian intensity profile, thereby giving us precise estimates of the center, width, and intensity of the spot.
Based on histograms of the intensities and widths, we manually selected a subset of the spots with qualities (uniform width, higher intensity) that were higher than background. This is similar in spirit to the procedure described in Raj et al. Nature Methods 2008, in which the experimenter chose a threshold to separate legitimate RNA spots from background spots. In this case, we erred on the side of including spots that may be background, because our multicolor scheme for spot assignment provided us another means by which to discard background spots.
Once we had selected the spots, we then ran software that found the fiducial markers (in this case, probes in all 5 RNA colors targeting SUZ12 mRNA, which are present at an abundance of roughly 20-50 clear cytoplasmic spots per cell). In this manner, we could measure the displacements between different fluorescence channels in each cell individually. We then applied these shifts to align the computationally identified spots between the different fluorescence channels.
After alignment, we then ran software that looked for colocalized spots corresponding to the particular pseudocoloring scheme we chose for the introns we targeted. We estimate that our software is roughly 75% accurate in assigning colocalized spots to particular genes at this stage.
We then went through a manual correction process in which we corrected mistakes the software made in identifying spots. Common issues were failure to detect dim (but clearly present) signals in one of the fluorescent channels and resolving two spatially close fluorescent spots that the laplacian of gaussian filtering and candidate identification steps had labeled as a single spot.
Once we had correctly annotated the introns of the gene loci we had labeled, we then examined cells manually to separate out individual chromosomes. We would discard cells in which the chromosomes overlapped since this made it difficult to assign gene spots to particular chromosomes.
In order to determine the distance of the chromosome from the nuclear periphery, we first determined the average position of the spots of the chromosome in x and y and then found the Euclidean distance between this point and the nuclear periphery as outlined by our DAPI stain.
Characterization of error rate
In order to gain some sense of the rate of false positives, we performed a hybridization in foreskin fibroblasts in which we left out 10 of the 20 genes comprising our iceFISH assay (randomly chosen by another member of the lab), and proceeded with our spot identification procedure as usual (Supplementary fig. 10). We found that our rate of false identification was very low, with the vast majority (97%) of spots we assigned corresponding to genes which we had targeted in our assay.
We also probed a set of 2 genes (RPS19, TOMM40) one at a time with oligonucleotides labeled with a single dye rather than the combination of 2 or 3 dyes used in our pseudocoloring strategy. Our aim was to determine to what extent our pseudocoloring strategy would result in false negatives in spot identification. We found that the spot per chromosome frequencies measured with a singly-colored probe alone were 0.56, 0.27, while the spot frequencies measured by pseudocoloring were 0.57, 0.25, respectively, in a total of 30 cells. Although statistical effects preclude a definitive statement, our results are consistent with our pseudocoloring strategy correctly identifying virtually all spots detectable by RNA FISH targeting introns.
Probe design
We designed 20 base oligonucleotide probes against introns using custom FISH design software (http://www.biosearchtech.com/stellarisdesigner/). Where possible, we tried to design 16 oligonucleotides targeting the first intron of the gene. We ordered the oligonucleotides from Biosearch Technologies (Novato, CA), who synthesized the oligonucleotides with amine groups attached to the 3’ end. We coupled these 3’ ends to various organic dyes (including Atto 488 (Atto-Tec), Cy3 (GE), Alexa 594 (Invitrogen), Atto 647N (Atto-Tec), and Atto 700 (Atto-Tec)) as indicated in the text and in Supplementary Table 1. We purified the probes by HPLC4.
Karyotyping of HeLa cells
We performed G-band analysis (karyotyping) on metaphase spreads of our HeLa cells following standard procedures. This indicated that our cells contained two intact copies of chromosome 19 and a full third copy of chromosome 19 split into two fragments and fused to other chromosomes (Supplementary fig 12). One fragment includes the first half of the chromosome 19 p-arm and is fused to a large portion of chromosome 6. The second fragment is the remaining portion of chromosome 19 (half the p-arm through the centromere and entire q-arm), which is fused to the q-arm of chromosome 13. In order to conclusively demonstrate that chromosome 19 was split in this particular way, we performed a DNA FISH analysis on the same metaphase spreads that we performed the G-band analysis on. We used probes targeting loci within the 19p13 and 19q13 regions on chromosome 19, each labeled with a different fluorophore (Abbott Molecular). The results confirm the results of the G-band analysis. We performed this analysis on 10 cells, each of which showed the same genetic abnormalities, indicating that the cells do not vary much in this particular characteristic from cell to cell.
Click-iT EdU analysis of cell cycle progression
In order to demonstrate that Cyclin A2 mRNA was an accurate marker of position in the cell cycle, we used the Click-iT EdU Alexa Fluor 594 Imaging kit (Invitrogen), which incorporates a targetable chemical into newly replicated DNA. In this case, we incubated foreskin fibroblasts with the 10μM Click-iT EdU reagent for 5 minutes before fixing the cells. We performed our FISH protocol on these cells using a Cyclin A2 mRNA Cy3 probe and after hybridization and wash steps followed the instructions provided with the kit for fluorescently labeling the incorporated EdU. We ultimately did not elect to use the Click-iT EdU kit directly in most of our experiments (and instead opted to use Cyclin A2) because we found that performing the Click-iT EdU procedure interfered with our nascent RNA FISH detection, most likely either due to interference with transcription itself or by making our spot detection less reliable because of additional washing steps associated with the Click-iT procedure.
DNA FISH
We performed DNA FISH with BAC probes from Empire Genomics, using their reference hybridization protocol. In the human foreskin fibroblast cells we applied pairs of fluorescently labeled BAC clones from the human RPCI-11 library targeting human chromosome 19 at positions 2.8-4.5 Mb (268O21), 39.0-39.5 Mb (31D10), or 52.5-52.7 Mb (43N16). We denatured the DNA by immersing the cells in 70% formamide, 2X SSC buffer at 80°C for 5 minutes, and then transferred to series of ethanol steps increasing to 70, 85, and then 100% ethanol. We added 10μL of BAC probes to the air dried sample, applied a coverslip, and incubated overnight in humidified slide chamber. The next day we washed the sample with 0.4X SSC at 73°C for 2 minutes, removed the coverslip, transferred to room temperature 2X SSC for 1 minute, then to 10μL of 2X SSC with DAPI at 50 ng/mL, and applied a new coverslip. We performed imaging similar to our iceFISH probes with dye pairs Red 5-ROX and TAMRA.
Combined DNA/RNA FISH
We performed a sequential DNA/RNA FISH in HeLa cells by first performing DNA FISH using BAC clones as and then performing RNA FISH, both by following the protocols outlined above. We found that both the bright exonic transcription sites and the intron spots were considerably brighter than single mRNA spots, thus showing that the RNA probes were not simply targeting the DNA directly. We compared the location of SLC1A5 exonic (Alexa 594) and intronic (ATTO647N) RNA to the location of DNA FISH probes using BAC clones RP11-687M15 (TAMRA).
Statistical analysis
In Figure 3, we looked for deviations from independence in the transcriptional frequencies of all pairs of genes we examined. We performed the Fisher Exact Test on all 2×2 tables generated by counting the number of chromosomes where gene A or B was transcriptionally active vs. inactive. We reported the two-sided p-value corresponding to the chance of obtaining a similar deviation from independence via random chance, with a smaller p-value corresponding to a more significant result. In Figure 3, we show the results we obtained by analyzing a dataset consisting of the combination of two independent biological replicates; we also performed the analysis on each individual biological replicate, as shown in Supplementary figs. 18 & 19. Note that we have not applied a multiple hypothesis correction in our presentation of the p-values; however, our results would remain statistically significant if we applied the crude correction of just multiplying our p-values by 190, which is the number of pairs of genes we examined. We chose to convey the information in this manner because the number of hypotheses tested depends on the particular question being asked of the data. For instance, if one decides that, based on the human foreskin fibroblast data, one wanted to focus on interactions between RPS19 and ZNF444, then the p-values for the specific hypothesis comparing these two genes in, say, HeLa cells, would not be subjected to this same correction. We leave such interpretative matters to the reader.
We also report the correlation coefficient between RPS19 and ZNF444; although it is a somewhat imperfect measure of the lack of independence for this sort of data, it has the advantage of being familiar to many researchers. We obtained standard errors for the correlation coefficient by bootstrapping.
In Figure 2, we obtained p-values for the difference in transcriptional frequency between the copy of the gene on the t(13;19) (or t(6;19)) chromosome and the copies of the gene on the normal copies of chromosome 19 by rejecting the null hypothesis in which the frequency of transcription was the same for all three copies. We did this by computationally generating the probability density function for the difference in transcriptional frequencies between two sets chosen to match our experimental data in size under the null hypothesis that the frequency is the same for both sets, and then directly calculated the probability of finding our observed difference by chance.
Supplementary Material
Acknowledgements
We thank Biosearch Technologies (esp. Marc Beal and Ron Cook) for providing many of the reagents used in the iceFISH assay and thank Jennifer Morrissette for her lab's assistance in performing the cytogenetic analysis. We also thank Long Cai, Hyun Youk, Dale Muzzey, and Jeff Gore for insightful comments, as well as Hedia Maamar, Olivia Padovan-Merhar and other members of the Raj lab for useful discussions, especially Gautham Nair for discussions about statistics. We thank the lab of Phillip Sharp (MIT) for providing the HeLa cells. We gratefully acknowledge a University of Pennsylvania URF pilot grant, the NIH Director's New Innovator Award (1DP2OD008514) and a Burroughs-Wellcome Fund Career Award at the Scientific Interface for supporting our work.
Footnotes
Author contributions:
MJL and AR conceived the project, performed the analyses and wrote the paper. MJL performed the experiments.
References
- 1.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 2.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 3.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 4.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fremeau RT, Lundblad JR, Pritchett DB, Wilcox JN, Roberts JL. Regulation of pro-opiomelanocortin gene transcription in individual cell nuclei. Science. 1986;234:1265–1269. doi: 10.1126/science.3775385. [DOI] [PubMed] [Google Scholar]
- 6.Gribnau J, et al. Chromatin interaction mechanism of transcriptional control in vivo. EMBO J. 1998;17:6020–6027. doi: 10.1093/emboj/17.20.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xing Y, Johnson CV, Dobner PR, Lawrence JB. Higher level organization of individual gene transcription and RNA splicing. Science. 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- 8.Vargas DY, et al. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell. 2011;147:1054–1065. doi: 10.1016/j.cell.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 10.Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr Biol. 2006;16:1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suter DM, et al. Mammalian genes are transcribed with widely different bursting kinetics. Science. 2011;332:472–474. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- 13.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 14.Lubeck E, Cai L. Single-cell systems biology by super-resolution imaging and combinatorial labeling. Nat Methods. 2012;9:743–748. doi: 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mateos-Langerak J, et al. Spatially confined folding of chromatin in the interphase nucleus. Proc Natl Acad Sci USA. 2009;106:3812–3817. doi: 10.1073/pnas.0809501106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eward KL, Van Ert MN, Thornton M, Helmstetter CE. Cyclin mRNA stability does not vary during the cell cycle. Cell Cycle. 2004;3:1057–1061. [PubMed] [Google Scholar]
- 17.Macville M, et al. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res. 1999;59:141–150. [PubMed] [Google Scholar]
- 18.Harewood L, et al. The effect of translocation-induced nuclear reorganization on gene expression. Genome Res. 2010 doi: 10.1101/gr.103622.109. doi:10.1101/gr.103622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 20.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.