Abstract
A key step towards a chemical picture of enzyme catalysis was taken in 1913, when Leonor Michaelis and Maud Menten published their studies of sucrose hydrolysis by invertase. Based on a novel experimental design and a mathematical model, their work offered a quantitative view of biochemical kinetics well before the protein nature of enzymes was established and complexes with substrates could be detected. Michaelis-Menten kinetics provides a solid framework for enzyme kinetics in vitro, but what about kinetics in cells, where enzymes can be highly regulated and participate in a multitude of interactions? We discuss this question using the Extracellular Signal Regulated Kinase (ERK) as a model of an important enzyme for which we have crystal structures, quantitative in vitro assays, and a vast list of binding partners. Despite great progress, we still cannot quantitatively predict how the rates of ERK-dependent reactions respond to genetic and pharmacological perturbations. Achieving this goal, which is important from both fundamental and practical standpoints, requires measuring the rates of enzyme reactions in their native environment and interpreting these measurements using simple but realistic mathematical models, the two elements which served as the cornerstones for the seminal 1913 paper.
Introduction
One hundred years ago, Leonor Michaelis and Maud Menten published their landmark paper on enzyme kinetics, in which they studied how a two-ring sugar – sucrose – is hydrolyzed by a yeast-derived enzyme – invertase, so named because hydrolysis changes optical rotation from positive for sucrose to negative for the mixture of fructose and glucose (Fig. 1a) (1, 2). The choice of this chemical reaction can be traced back to Louis Pasteur, a founding father of microbiology who made many remarkable discoveries, but was convinced that enzyme reactions require the presence of living organisms that provide a vital force, irreducible to laws of physics and chemistry. By 1913, this view had been losing ground, largely due to the work of Eduard Buchner, who demonstrated fermentation in the absence of live cells. This reinforced the view that enzymes can be understood using the principles of chemistry, at that time, still an emerging discipline, with Emil Fischer as one of the leading figures, famous for his synthesis of natural products, including sugars (3).
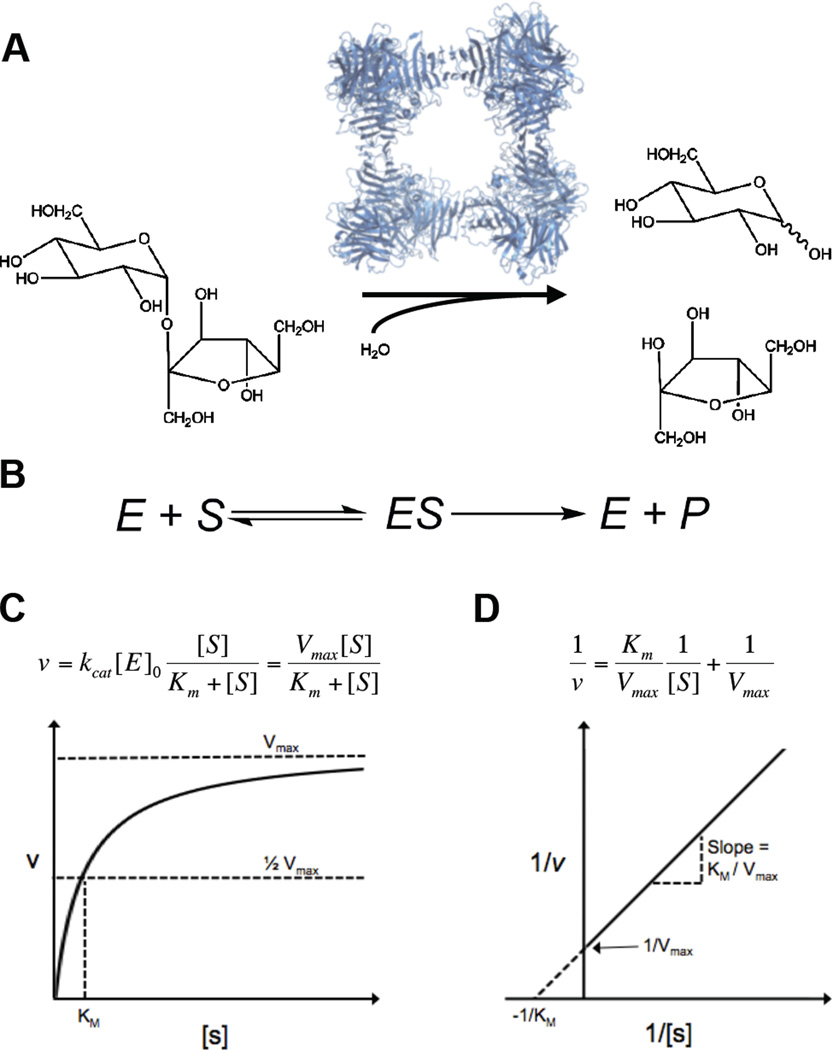
Figure 1.
The Michaelis-Menten model of enzyme kinetics. (A) Yeast invertase and the hydrolysis of sucrose to glucose (top) and fructose (bottom). Structure of S. cerevisiae invertase drawn from PDB file 4EQV (81). (B) The model proposed by Michaelis and Menten, wherein an enzyme and a substrate bind reversibly to form a complex, which is converted to a product and the enzyme. (C) The Michaelis-Menten equation and its graphical representation. Plotting the initial rate vs. substrate concentration enables the determination of both Vmax and Km. (D) Lineweaver Burk representation of the Michaelis-Menten equation. Taking the reciprocal of both sides of the Michaelis-Menten equation yields a linear relationship. Plotting the reciprocal of the initial rate vs. the reciprocal of substrate concentration allows the determination of Km and Vmax from the y and×intercept and the slope of the line.
Working with synthetic sugars and different types of yeast enzyme preparations, Fischer concluded that enzyme catalysis requires shape complementarity between enzymes and their substrates and put forward his famous “lock-and-key” model of enzyme action. Michaelis and Menten’s approach to evaluating Fischer’s model was based on formal chemical kinetics, which is standard today but had been only a couple of decades old in the beginning of the 20th century. In this approach, one postulates a mechanism and derives from it an algebraic equation for the overall reaction rate as a function of reaction conditions, such as reactant concentrations. Fitting the derived equation to rates measured over a range of conditions can be used to assess the validity of the mechanism (4).
The first application of kinetic approach to enzymes is attributed to Victor Henri, whose dissertation, published in 1903, contains the now familiar mechanism in which reversible formation of a complex precedes its irreversible decomposition into enzyme and product (Fig. 1b) (3, 4). However, analysis of Henri’s data was complicated by product inhibition, which was significant at high substrate conversions in his experiments. Michaelis and Menten worked at low conversions and measured initial rates of reaction, which allowed them to neglect product inhibition and simplified kinetic analysis. Their analysis revealed that the rate of reaction is accurately described by a simple formula, linear at small substrate concentrations and approaching a constant value when substrate concentrations are high (Fig. 1c,d) (5). The fact that one formula fit the data over wide range of substrate concentrations was clearly consistent with Fischer’s idea and Henri's mechanism. A rigorous proof of this mechanism, based on direct observation of enzyme-substrate complexes, appeared only decades later, after the protein nature of enzymes was established (6, 7). Nevertheless, the clarity of the paper made it an instant classic and ensured that kinetic approach was rapidly and successfully applied to other enzymes.
The groundbreaking studies on invertase took the first steps towards establishing a chemical picture of a constitutively active enzyme that processes a single substrate. But things are much more complex inside cells, where enzymes can be highly regulated and need to work on multiple substrates. Our understanding of such systems is still incomplete, despite great advances in conceptual analysis of intracellular processes and their experimental analysis. Here, we use the Extracellular Signal-Regulated Kinase1/2 (in the rest of the text, just ERK) as a model to highlight some of the most important aspects of enzyme kinetics in cells. Some of them, such as substrate selectivity, can be addressed by studies with a small number of purified components. Other features, such as spatial control of enzyme activity, require studies in vivo. The general concepts developed by Michaelis and Menten still hold, but they must be applied in the context of enzyme networks and complex intracellular environments.
The paper is organized as follows. After providing a brief history of ERK, we discuss how it interacts with substrates, highlighting modifications to the model envisioned by Fischer. After that, we summarize recent efforts to identify ERK substrates. Finally, we discuss network-level effects, which require considering the joint effects of ERK regulators and substrates.
A brief history of ERK
ERK was discovered in studies of protein phosphorylation, in the context of cell stimulation by growth factors, which act through receptor tyrosine kinases (see (8, 9) for review). The first protein shown to undergo phosphorylation in response to growth factors was the ribosomal protein S6. Since this protein is phosphorylated on serines, it cannot be a direct substrate of tyrosine kinases. This led to the identification of the protein Ser/Thr kinases RSK and S6K, which are again activated by phosphorylation on serines and threonines. The kinase responsible for the phosphorylation of RSK was discovered by Sturgill and Ray (10), and the corresponding gene was subsequently cloned by Cobb and collaborators (11, 12). Although it was initially known by several distinct acronyms, the enzyme’s name settled on the Extracellular Signal-Regulated Kinase (ERK).
ERK became the founding member of an important class of Mitogen Activated Protein Kinases (MAPKs), serine/threonine kinases which control a wide range of processes in adult and developing cells (13). Enzymatic activity of ERK requires its phosphorylation on Tyr and Thr within the activation loop of the kinase (14). The dual specificity protein kinase responsible for this activation was identified (15, 16) and cloned (17, 18) shortly after the cloning of ERK itself, and was termed MAP Kinase/ERK Kinase (MEK). The reverse modification, which dephosphorylates and deactivates ERK, is accomplished by phosphatases, which were identified relatively early in ERK research (19, 20).
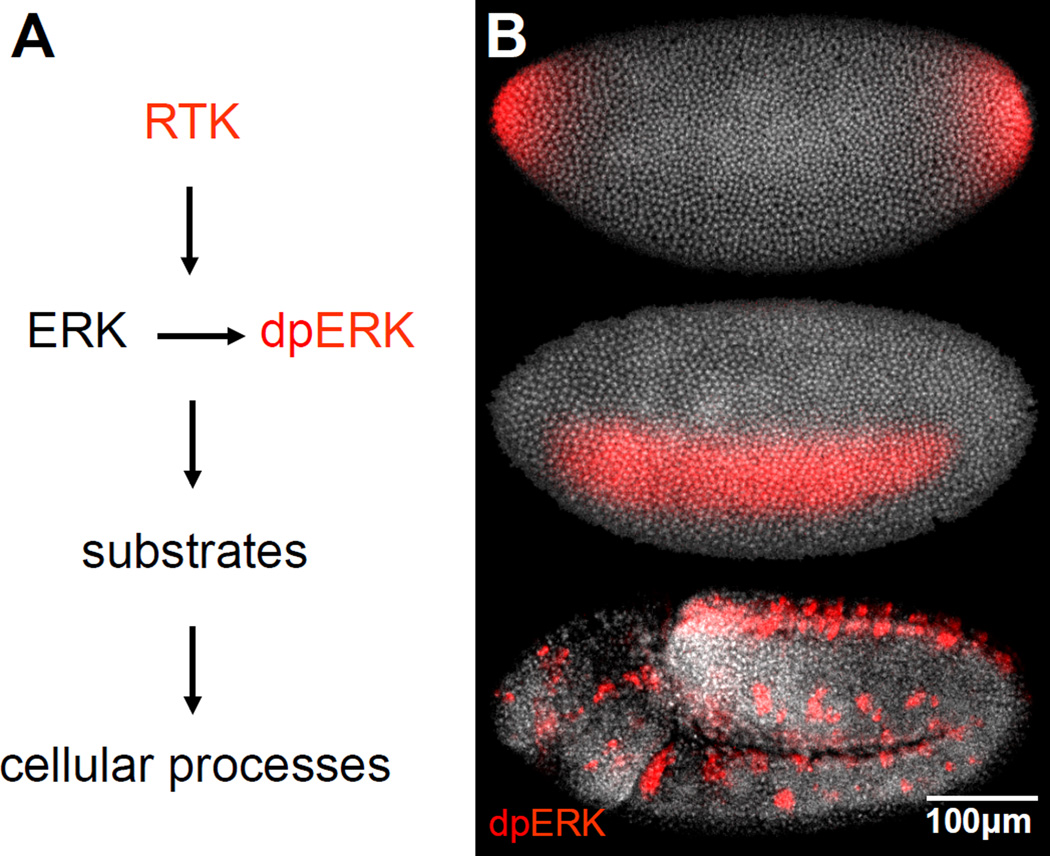
In the beginning of the 1990’s, studies in model genetic organisms, most notably Drosophila, began to complement experiments in cultured cells and established that ERK plays a key role in embryogenesis (21). In vivo effects of ERK were initially studied by observing consequences of ERK mutations on morphological structures, such as the faceted Drosophila eye (22). A critical new tool emerged in 1997, with the development of an antibody that recognizes the active (dually phosphorylated) form of the enzyme (dpERK) (Fig. 2, 4a) (23). This antibody revealed intricate spatiotemporal patterns of ERK activation in vivo (24, 25). The emergence of these patterns and analysis of their effects on processes such as gene expression and morphogenesis is a subject of intensive research in different model systems, from gonad development in worms to segmentation in vertebrates (26, 27).
Figure 2.
Patterns of ERK activation in organisms. (A) ERK activation requires its phosphorylation by MEK. Active ERK controls cellular processes by phosphorylating multiple substrates. (B) Active ERK (red) at three different time points in Drosophila embryos. Active ERK is first detected at the embryonic poles, where it specifies the nonsegmented terminal regions of the future larva. Within the next 30 minutes, ERK is activated in a lateral domain corresponding to the presumptive neurogenic ectoderm. After gastrulation, ERK is active along the ventral midline and in tracheal pits.
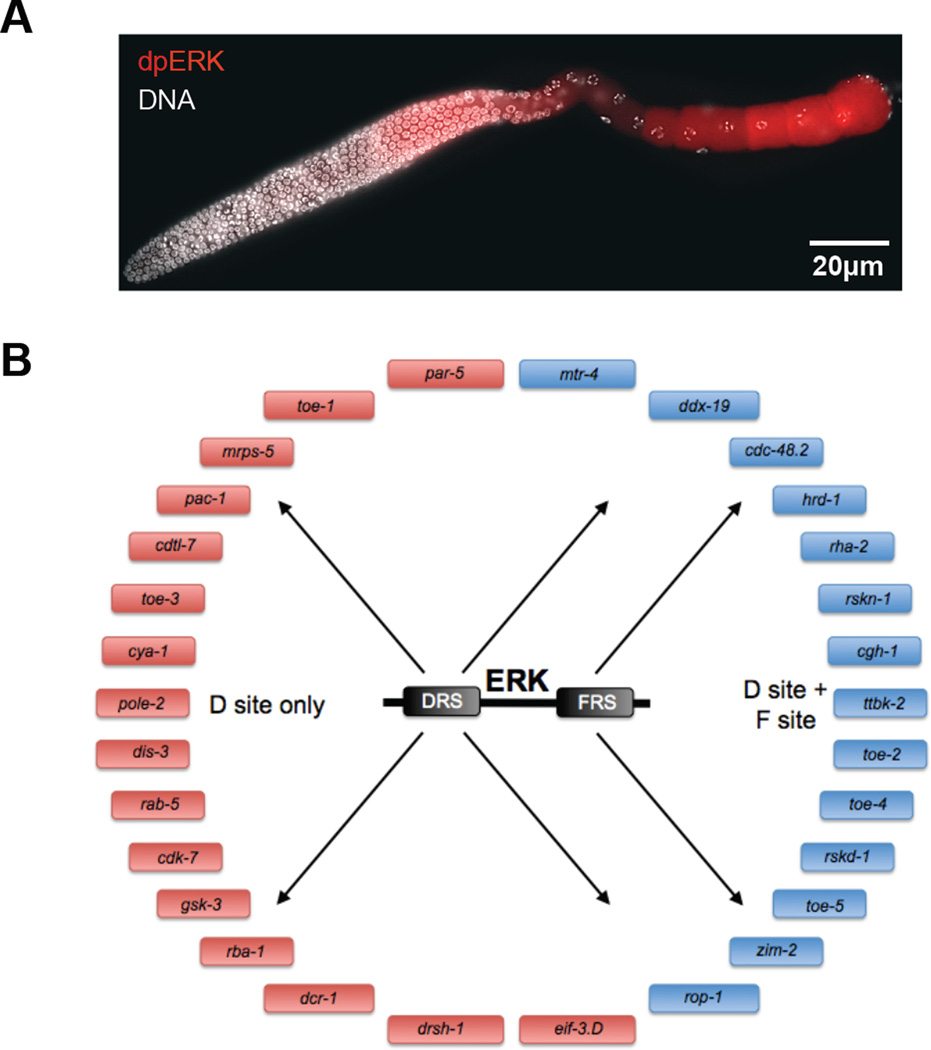
Figure 4.
Identification of ERK substrates in C. elegans. (A) Dissected C. elegans hermaphrodite germ line from wild type animals oriented from left to right, stained for dpERK (red) and DNA (white). Wild type germ lines reveal two zones of ERK activation, zone 1 and 2, with brief down regulation of active ERK in the loop region. (B) Schematic representation of ERK substrates identified in C. elegans by searching the genome for ERK docking site motifs and screening putative substrates in vivo.
Over time, multiple substrates that connect ERK activation to different aspects of cell biology have been identified and their sensitivity to ERK dissected at the molecular level (28–30). These studies revealed the importance of docking interactions, which use distinct parts of the ERK molecule, bringing it together with a wide range of binding partners (31, 32). The first structures of ERK, active or inactive (33, 34), and in complexes with some of its regulators and substrates appeared over the past two decades, providing important insights into the mechanisms of ERK activation and specificity.
The first proof of association between deregulated ERK activation and human disease appeared in 1994 (35). Subsequent work established that deregulation can result from gain of function mutations in the signaling cascade that links RTKs to ERK. In particular, gain of function mutations in enzymes that activate ERK were identified in a broad spectrum human cancers and developmental abnormalities (36). Strong associations between ERK signaling and human diseases continue to drive studies of the ERK cascade at multiple levels of organization, from structures of proteins to multicellular patterns in tissues.
The emerging picture is complex, involving numerous components and layers of regulation (13, 37). Mathematical modeling, which was an integral part of the Michaelis and Menten paper, provides a sensible way of dealing with this complexity (38–41). Starting from 1996, mathematical models have been used to integrate data from a wide range of experimental approaches and predict how ERK activity responds to genetic and pharmacological perturbations. All of these models use the Michaelis-Menten description of enzyme kinetics as a building block of more complex networks. Since this description was established based on studies with isolated components, it is reasonable to ask whether it is appropriate for enzymes in cells. Addressing this question is the main goal of this essay.
Quantitative analysis of ERK-substrate interactions
Michaelis and Menten studied an enzyme that is highly specific. Indeed, invertase only acts on sucrose. Such specificity is fairly common for enzymes with small substrates that bury themselves in clefts where catalysis occurs. Thus, the formation of the enzyme-substrate complex is closely coupled to catalysis. However, other enzymes – especially those that act on proteins – behave differently and have multiple substrates. ERK provides a vivid example of an enzyme with broad specificity and is believed to have over 250 substrates (42–44). Protein substrates are modified at specific positions that interact with the active site. However, the large size of these substrates enables binding between the substrate and enzyme at positions that are distal to the site of catalysis. These docking site interactions play an important role in targeting the activity of enzymes to their substrates.
While ERK phosphorylation is targeted to specific sites – a serine or a threonine followed by a proline ((S/T)P) – these sites are quite common, occurring in approximately 80% of all proteins (45). Clearly, catalytic site targeting is insufficient to guide ERK to substrates. Instead, specific docking domains on ERK and its substrates are used to enhance targeting. There are two well-characterized docking domains on ERK termed the D Recruitment Site (DRS) (also called CD domain), which interacts with D site motifs (also called DEJL motifs) on ERK-interacting proteins, and the F Recruitment Site (FRS), which interacts with proteins containing an F site (also called FXF motif, DEF motif) (46–50) (Fig. 3a–c).
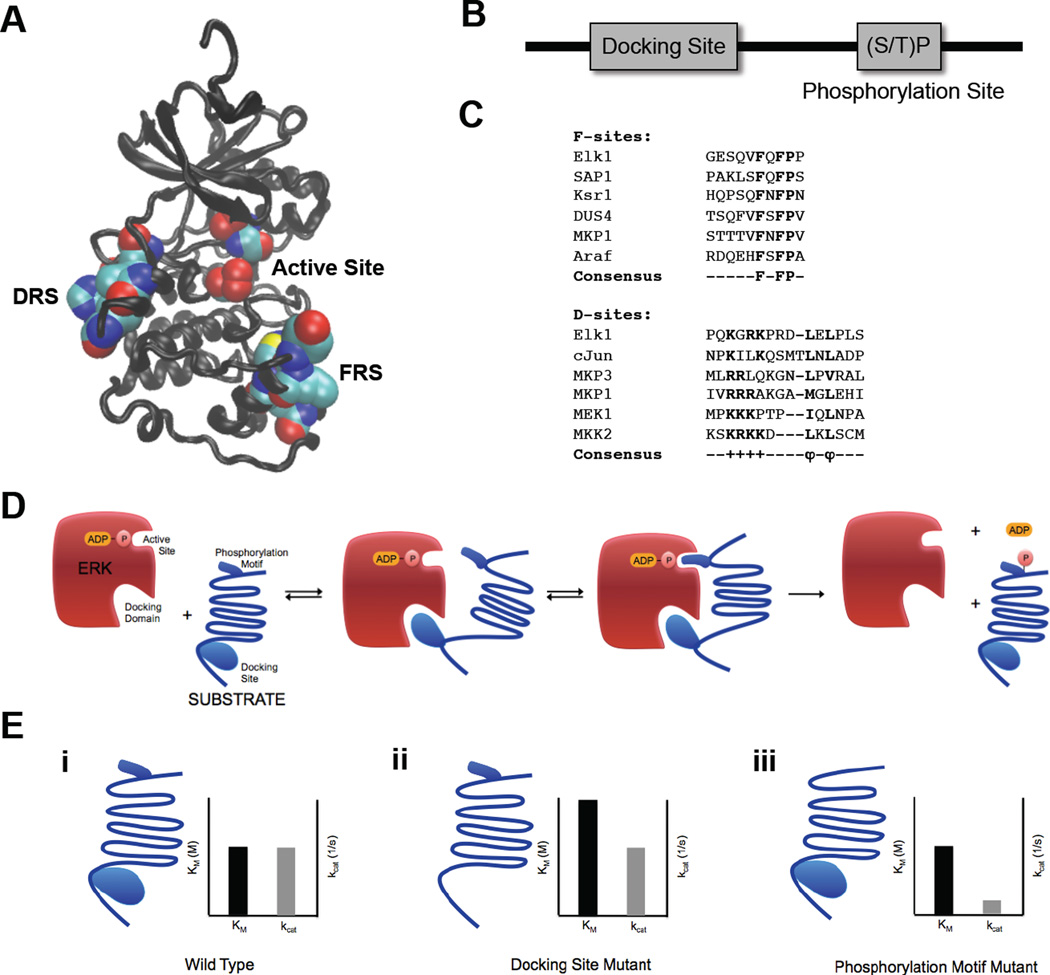
Figure 3.
ERK2 docking domains and ERK2 substrate docking sites. (A) ERK2 structure with docking domains – DRS domain (left) and FRS domain (right) – and active site (middle) in space filling representation. Structure drawn from PDB file 1ERK (33) (B) Schematic representation of ERK2 substrate sequence containing a docking site and a (S/T)P phosphorylation site. (C) Alignment of F-site and D-Site sequences from multiple ERK2 interacting substrates and regulators. (D) Schematic representation of ERK2 interactions with substrate docking site and (S/T)P phosphorylation motif. (E) Effects of mutations on substrate docking site and phosphorylation site and insights into molecular mechanisms of ERK2 catalysis. (i) Wild type substrate and associated Michaelis-Menten parameters. (ii) Substrate with docking site mutations and associated Michaelis-Menten parameters. Mutating the docking site significantly increases the Km but has little effect on kcat. (iii) Substrate with phosphorylation motif mutations and associated Michaelis-Menten parameters. Mutating the phosphorylation motif significantly decreases the kcat but has little effect on Km.
Docking sites do not only increase the specificity of the ERK-substrate interaction, they often increase efficiency of ERK-mediated phosphorylation (50). This may seem counter-intuitive; tight docking interactions could theoretically reduce substrate turnover if ERK substrates are not released promptly after phosphorylation. It appears that many ERK-substrate interactions are destabilized after phosphorylation (51) and others may be sufficiently weak, allowing ERK to dissociate from a phosphorylated product after catalysis has occurred.
Because the formation of the enzyme-substrate complex employs interactions distal to the active site, the formation of the complex can be functionally separated from catalysis. This can be clearly seen in the Michaelis-Menten parameters extracted from studies of ERK-dependent phosphorylation in vitro. For example, Ets1, a transcription factor phosphorylated by ERK, contains both an ERK binding site and a distal phosphorylation site (TP). Phosphorylation of this substrate was studied by separating and quantifying the role of the binding site and phosphorylation motif (52). Interestingly, mutations to each of these sites have a distinct effect on ERK-mediated phosphorylation. Mutating the proline in the TP site to different amino acids has a significant effect on the rate of phosphorylation, kcat, despite the relatively weak effect these mutations have on equilibrium dissociation constants. On the other hand, mutations in the docking site do not have a significant effect on kcat, but lead to much weaker binding to ERK and thus increase the dissociation constant and Km (Fig. 3d,e).
Overall, mutations to both the phosphorylation motif and the docking site affect the ability of Ets1 and ERK to form a catalytically relevant complex, but in very different ways. The binding site increases the local concentration of the TP motif near the catalytic site, but has little effect on catalysis. On the other hand, the TP sequence is critical for positioning residues for the transfer of a phosphoryl group to Ets1, but does not have a significant effect on the strength of the ERK-Ets1 binding (Fig. 3d,e). Thus, studies of enzyme catalysis using modern techniques such as directed mutagenesis and kinase activity and binding assays revealed mechanisms that could never have been imagined in 1913. Yet, we are still able to interpret and model these mechanisms using the language of the 1913 paper.
Identification of ERK substrates in cultured cells and model organisms
Elucidating the parameters that describe ERK activity in a test tube provides valuable information, but it says nothing about which substrates it is acting on at any given time and place in a cell or an organism. This question goes beyond the scope of studies enabled by the Michaelis-Menten model, which focuses on one enzyme and one substrate. However, knowing what makes a protein an ERK substrate – such as sequences of binding and phosphorylation sites – and the advent of post-genomic techniques have enabled us to begin to piece together the puzzle (26, 43, 53–59). We highlight proteomics and bioinformatics as two approaches to identify ERK substrates in living systems.
The proteomic approach aims to find proteins whose interactions with ERK or phosphorylation state change upon ERK activation. A number of studies have used two-dimensional gel electrophoresis to separate proteins by mass and isoelectric point in cells with and without initiation of ERK activity. Proteins that have been phosphorylated will shift positions on a gel, and characterization by mass spectrometry enables the identification of the proteins that display this behavior (54, 55). These studies provide lists of the ERK pathway targets and can identify its novel physiological functions. For example, Kosako et al. used this approach to identify a component of the nuclear pore as a new ERK substrate and proposed that ERK regulates nucleocytoplasmic transport (43). Clearly, intracellular proteins phosphorylated in response to ERK activation are not necessarily ERK substrates. Indeed, their phosphorylation may be induced by enzymes controlled by the ERK pathway. A different set of techniques aims to identify direct binding partners of ERK (53, 56–59). In particular, von Kriegsheim and colleagues assessed the relative amounts of proteins that coimmunoprecipitate with ERK from cells which had been exposed to ERK activation for different periods of time (57). This study revealed the highly dynamic nature of ERK interactome and its sensitivity to the level of pathway activation.
Knowledge about the molecular-level mechanisms of enzyme-substrate interactions and genome sequencing made it possible to combine bioinformatics with genetics and biochemistry to identify ERK substrates in vivo. In a recent study of ERK effects in C. elegans germline development (Fig. 4a), consensus sequences of binding and phosphorylation sites were used to computationally search for ERK substrates in the C. elegans genome (26). A list of substrate candidates was generated by searching the proteome for ERK docking sites in the vicinity of phosphorylation motifs. Candidate substrates were then screened in vivo, using RNAi knockdown to identify which are responsible for translating ERK signaling into multiple aspects of germline development. In this way, a list of 161 candidates predicted by bioinformatics led to 37 proteins that regulate ERK-dependent biological processes in vivo. Finally, in vitro phosphorylation assays confirmed that many of these proteins are true ERK substrates (Fig. 4b). Thus, in this combined bioinformatic, genetic, and biochemical approach ERK substrates were not only identified, but tied to specific cellular and developmental processes, such as translational control.
These studies reveal that analysis of the biological effects of ERK activity requires simultaneous consideration of multiple substrates. To model the enzymatic activity of ERK in cells, even under highly simplified assumptions, we need in vivo concentrations of ERK and its interactors, as well as rates of association, dissociation and catalysis. Currently, we have very little of this information for most ERK substrates. Furthermore, the identification of multiple substrates and processes regulated by ERK raises a number of questions that did not apply to the single substrate system studied by Michaelis and Menten. One of these questions is related to ERK substrate competition.
ERK substrate competition
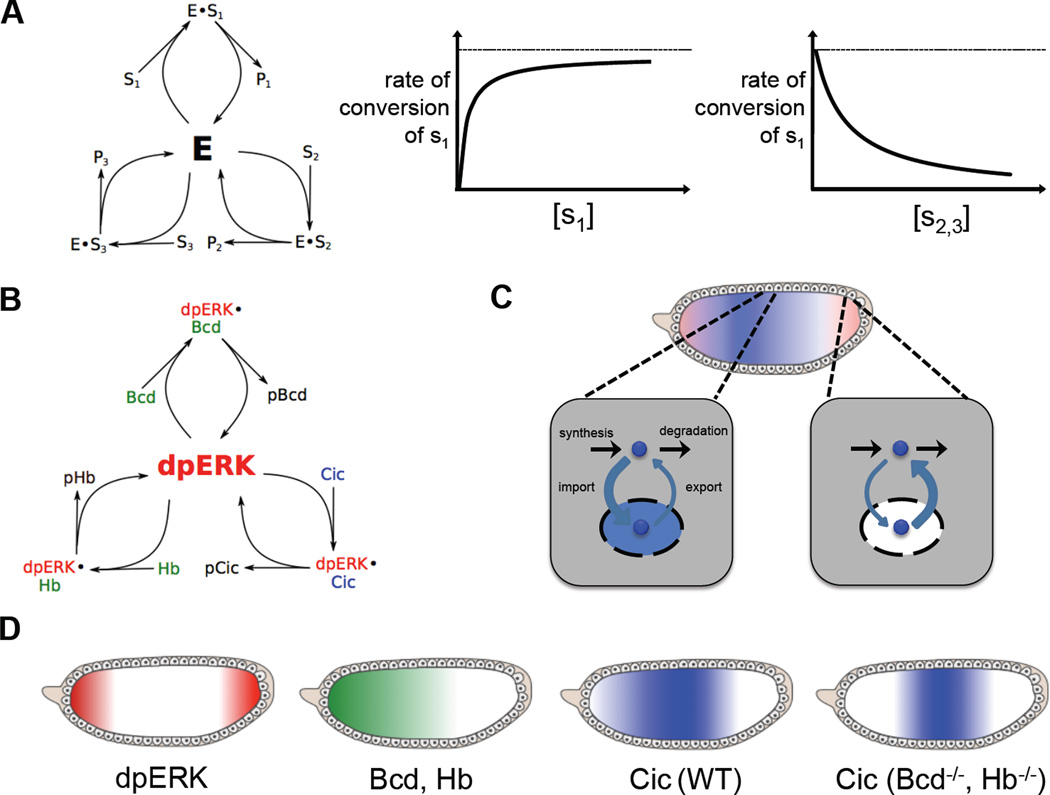
How does ERK multiplex between its multiple substrates at a given time point (Fig. 5a,b)? One can imagine two different scenarios. In the first scenario, active ERK is present in excess of its substrates. In this case, the enzyme is mostly free, and its substrates can be viewed as independent sensors of ERK activity which do not affect each other. Genetic removal or overexpression of any given substrate will not affect the extent to which ERK modifies other substrates present in the cell. In this regime, the enzyme can be readily recruited to modify new substrates.
Figure 5.
Substrate Competition in the ERK pathway. (A) Substrate competition and its effects on enzyme kinetics. When multiple substrates compete for the activity of a single enzyme, the rate of conversion of a single substrate decreases as the concentration of competing substrates increases (B) ERK substrate competition in the Drosophila embryo. Three substrates – Bicoid (Bcd), Capicua (Cic) and Hunchback (Hb) – compete for ERK activity. (C) ERK-mediated downregulation of Cic. At the embryonic poles, where ERK is active, Cic is exported out of the nucleus into the cytoplasm, where it is degraded. (D) Distribution of ERK and its substrates in the Drosophila embryo in the presence and absence of substrate competition. ERK is doubly phosphorylated and activated at the poles of the embryo. Two substrates – Bcd and Hb – are present only at the anterior pole of the embryo. In wild type embryos, Cic is downregulated at the poles unevenly, with a higher concentration at the anterior pole. However, when the anteriorly located ERK substrates Bcd and Hb are removed, Cic is downregulated more evenly, indicating that ERK’s ability to phosphorylate and downregulate Cic is inhibited by the presence of these other ERK substrates at the anterior pole.
In a second scenario, active ERK is saturated by substrates. What happens when the expression levels of different substrates are perturbed in this regime? A simple mathematical model, where different substrates of ERK act as competitive inhibitors of each other, shows that the effect depends on substrate concentrations and their relative affinities for the enzyme (Fig. 5a,b). When ERK deals with a large number of substrates, each of which amounts to a small fraction of its interaction partners, substrates are independent of each other, just as in the case when ERK is present in excess. On the other hand, when a substrate constitutes a significant fraction of ERK interaction partners, its knockdown or overexpression can strongly influence the effects of ERK on other substrates.
Which of the two scenarios reflects the situation in cells? At this point, when we are only beginning to identify ERK substrates in different cell types and have no reliable estimates of their concentrations and relative affinities for ERK, the answer to this question is still unknown. Some insights can be provided by studies in model organisms, such as Drosophila, where multiple ERK substrates have been identified and several methods exist for perturbing their expression in vivo. A recent study of ERK signaling in the early Drosophila embryo suggests that competition between ERK substrates is appreciable in magnitude and functionally significant (60).
In this system, ERK is activated in a localized pattern, with pronounced peaks at the anterior and posterior poles (Fig. 2b). One of the ERK the substrates in this system is a transcriptional repressor Capicua (Cic), which is excluded from nuclei and degraded in the cytoplasm in response to phosphorylation by ERK (Fig. 5c) (61). The spatial pattern of Cic downregulation is highly asymmetric: Cic is downregulated much more strongly at the posterior pole. Genetic experiments established that this asymmetry reflects ERK substrate competition. Specifically, at least two ERK substrates, the transcription factors Bicoid and Hunchback, are localized to the anterior pole where they act as competitive inhibitors of ERK-dependent downregulation of Cic. Accordingly, genetic removal of these substrates from the embryo increases the level of Cic downregulation at the anterior pole and makes the pattern of Cic downregulation symmetric (Fig. 5d) (60).
These observations are consistent with a simple Mass Action model, where ERK substrates inhibit each other competitively (Fig. 5a,b). These competitive interactions can spread through a larger network that controls gene regulation in the embryo and subdivides it into different tissue types (62). Thus, at least in this particular system, it appears that competition between ERK substrates is appreciable and plays a role in determining the biological effects of ERK activation. These observations motivate multiple lines of inquiry. First, it remains to be determined whether the observed competition between ERK substrates implies that they use the same docking site. Second, it is important to determine whether substrate competition is appreciable in other biological contexts. If this turns out to be the case, we might need to rethink the interpretation of experiments based on genetic perturbations of individual substrates.
Enzyme Regulation and Enzyme Networks
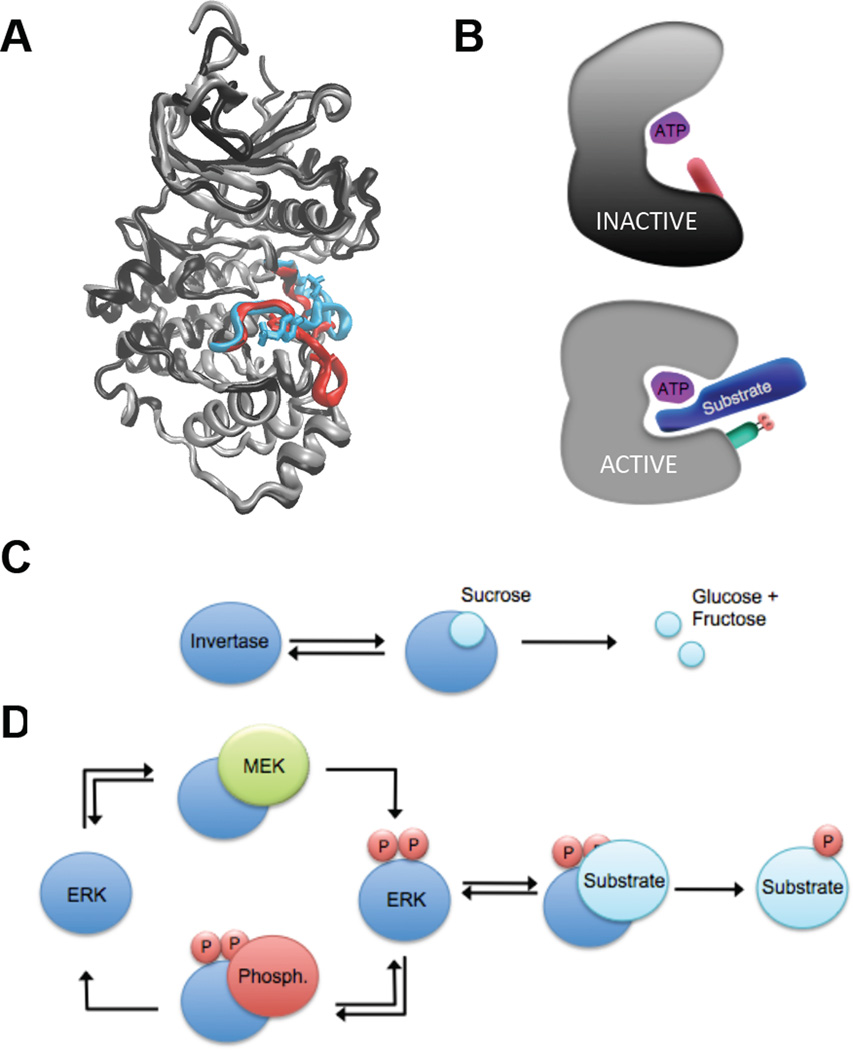
In cells, ERK cycles between a catalytically active and inactive state. How can an enzyme’s activity be turned on and off? The answer to this question can be explained by studying the three-dimensional enzyme structure in atomic detail, a line of inquiry that could not have been imagined in 1913. ERK is a substrate of activating and deactivating enzymes. The mechanism of this regulation is the addition of a phosphoryl group by MEK to two positions on the ERK activation loop, which causes a conformational rearrangement (34, 63, 64). The phosphorylation lip moves considerably and the two lobes of the protein rotate closer to one another. This forms a pocket allowing the proper binding and orientation of substrate residues such that ERK can transfer a phosphoryl group from ATP to the target residue. This conformational change is reversed when ERK is dephosphorylated by one of a number of phosphatases (Fig. 6a,b) (65). Note that the protein-protein interactions responsible for ERK regulation share similarities to those involved in ERK-mediated catalysis. MEK and phosphatases often use the same docking sites to modify ERK’s activity that ERK uses to phosphorylate its substrates (50). Thus, docking domains can be thought of as nodes for the multiple reactions that ERK is involved in.
Figure 6.
Regulation of ERK activity. (A) Superimposition of inactive (black and red) and active (grey and cyan) ERK2 highlighting the activation loop and phosphorylated residues. Structures of inactive and active ERK2 drawn from PDB files 1ERK and 2ERK, respectively (33, 34). (B) Schematic representation of structural changes upon ERK activation and the effects on ATP/Substrate binding and orientation. Upon activation, the N terminal and C terminal lobes of ERK rotate toward one another and residues is the active site are repositioned, allowing proper binding and orientation of substrates for catalysis. (C) A classical single enzyme/substrate network, exemplified by the system used by Michaelis and Menten – invertase-mediated hydrolysis of sucrose. (D) A simplified ERK network involving multiple individual enzyme/substrate interactions – ERK activation by MEK, ERK inactivation by a phosphatase, and ERK-mediated phosphorylation of one substrate.
In order to predict the enzymatic activity of ERK in cells, we must consider its phosphorylation and activation by MEK, ERK-mediated phosphorylation of its many substrates, and deactivation by regulatory phosphatases (Fig. 6c,d). Each of these individual interactions can be modeled by the Michaelis-Menten mechanism, but to fully capture the behavior of ERK, these modules must be coupled in a model that describes a complex network. Molecular level perturbations can ripple through such networks and lead to systems level responses that could not be predicted by looking at individual reactions (66, 67).
One example of the surprising effects of network-level interactions arose from the search for mutants of ERK with increased activity. The sevenmaker gain of function mutant of ERK was discovered in a genetic screen performed in Drosophila (68, 69). The mutation was found to lie in one of the ERK docking domains, the DRS. However, the identification of the location of this mutation raised as many questions as it resolved. The DRS interacts with D sites on MEK, substrates and deactivators alike, which raises the question: why would a mutation in the DRS necessarily lead to increased ERK activity?
It is thought that the sevenmaker mutation has a detrimental effect on ERK activation, deactivation, and catalysis individually. However, increased activity may arise from an imbalance of this effect on different ERK-interacting proteins. It is possible that deactivation of ERK by phosphatases is more sensitive to disruption of the DRS than activation by kinases or catalysis by ERK (69, 70). Thus, even if all binding interactions involving the DRS are negatively impacted by the sevenmaker mutation, if the disruption is more pronounced in ERK phosphatases relative to ERK’s other binding partners, the ratio of active to inactive ERK increases. Thus, in vivo effects of the sevenmaker mutation cannot be reduced to binary interactions and must be considered only in the context of a network.
Discussion
Analysis of enzyme kinetics in cells, especially for highly regulated enzymes with multiple substrates, requires analysis of enzyme networks. And just as Michaelis and Menten based their work on measurements of reaction rates and mathematical models of reaction progress, we need mathematical models and experimental tools appropriate for networks. Are we there yet?
Most of the existing models of cell biochemistry assume that enzymes, substrates, and complexes are perfectly mixed. This might be true in vitro, but not in cells, where reaction medium is highly crowded. Recent work on ERK activation by MEK reveals that crowding has a strong effect on reaction kinetics (71). When the level of mixing is high, MEK phosphorylates ERK following a distributive mechanism, whereby ERK is phosphorylated on two different sites, in two distinct enzyme-substrate encounters, which can involve two different MEK molecules. But when diffusion is slowed down by crowding, the molecule that phosphorylated ERK for the first time binds to it with high probability before mixing with other molecules in the medium and carries out the second phosphorylation. In this regime, a distributive mechanism appears as processive, whereby sequential modifications of the substrate require just a single enzyme-substrate encounter.
This change in the apparent kinetics leads to a qualitative change in the dynamics of the enzymatic cycle that controls ERK phosphorylation. When ERK phosphorylation follows a distributive mechanism, the cycle can have multiple steady states (72), whereas the cycle in which ERK phosphorylation is processive is always monostable (73). This effect can be readily captured by computational models that describe ensembles of individual enzymes and substrates, but not by conventional chemical kinetics. Thus, some aspects of enzyme kinetics in cells need models that differ from those used by Michaelis and Menten.
A typical model of ERK dynamics provides information about biochemical modification and subcellular locations of multiple species. While only a small fraction of these species can be measured by current experimental techniques, new tools of cellular biochemistry, including quantitative proteomics, promise to expand the list of species and reactions that can be monitored in the same sample (59, 74, 75). Furthermore, most of the current models of enzyme kinetics in cells have been formulated based on data that neglect spatial organization of cellular processes. However, rapid development of live imaging techniques can provide data about spatial distribution of network components and reaction rates (76). In parallel, advances in understanding the mechanisms by which ERK interacts with substrates have led to the development of live reporters of its enzymatic activity (77–79).
Overall, when it comes to enzyme kinetics in cells, we are still far from the experimental and conceptual standards established by the 1913 paper. Given the complexity of these processes and the fact that some of the key molecular players have been discovered only recently, this should not come as a surprise. At the same time, progress of experimental techniques for monitoring different aspects of enzyme kinetics in cells and our increasing ability to perturb enzyme networks will inevitably lead to more advanced physicochemical models (80). Armed with these models, we will be better equipped to predict how these networks respond to genetic, environmental and pharmacological perturbations.
Acknowledgements
This work was supported by R01GM086537 from the National Institute of General Medical Sciences. S.Y.S. thanks Swathi Arur, Kevin Dalby, Ze’ev Paroush, Alexey Veraksa and Benny Shlio for helpful discussions and Bomyi Lim and Henry Mattingly for assistance with figure preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Johnson KA, Goody RS. The Original Michaelis Constant: Translation of the 1913 Michaelis–Menten Paper. Biochemistry. 2011;50(39):8264–8269. doi: 10.1021/bi201284u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michaelis L, Menten ML. Die kinetik der invertinwirkung. Biochem. Z. 1913;49:333–369. [Google Scholar]
- 3.Fruton JS. Proteins, Enzymes, Genes: The Interplay of Chemistry and Biology. Yale University Press; 1999. [Google Scholar]
- 4.Cornish-Bowden A. The origins of enzyme kinetics. FEBS letters. 2013;587(17):2725–2730. doi: 10.1016/j.febslet.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Johnson KA. A century of enzyme kinetic analysis, 1913 to 2013. FEBS letters. 2013;587(17):2753–2766. doi: 10.1016/j.febslet.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chance B. The Kinetics of the Enzyme-Substrate Compound of Peroxidase. Journal of Biological Chemistry. 1943;151(2):553–577. [Google Scholar]
- 7.Gunawardena J. Some lessons about models from Michaelis and Menten. Molecular Biology of the Cell. 2012;23(4):517–519. doi: 10.1091/mbc.E11-07-0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avruch J. MAP kinase pathways: The first twenty years. Biochim Biophys Acta. 2007;1773(8):1150–1160. doi: 10.1016/j.bbamcr.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seger R, Krebs EG. The MAPK signaling cascade. FASEB J. 1995;9(9):726–735. [PubMed] [Google Scholar]
- 10.Sturgill TW, Ray LB, Erikson E, Maller JL. Insulin-stimulated MAP-2 kinase phosphorylates and activates ribosomal protein S6 kinase II. Nature. 1988;334:715–718. doi: 10.1038/334715a0. [DOI] [PubMed] [Google Scholar]
- 11.Boulton TG, et al. An insulin-stimulated protein kinase similar to yeast kinases involved in cell cycle control. Science. 1990;249(4964):64–67. doi: 10.1126/science.2164259. [DOI] [PubMed] [Google Scholar]
- 12.Boulton TG, et al. ERK's: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell. 1991;65:663–675. doi: 10.1016/0092-8674(91)90098-j. [DOI] [PubMed] [Google Scholar]
- 13.Keshet Y, Seger R. The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. Methods Mol Biol. 2010;661:3–38. doi: 10.1007/978-1-60761-795-2_1. [DOI] [PubMed] [Google Scholar]
- 14.Anderson NG, Maller JL, Tonks NK, Sturgill TW. Requirement for integration of signals from two distinct phosphorylation pathways for activation of MAP kinase. Nature. 1990;143:651–653. doi: 10.1038/343651a0. [DOI] [PubMed] [Google Scholar]
- 15.Ahn NG, et al. Multiple components in an epidermal growth factor-stimulated protein kinase cascade. In vitro activation of myelin basic protein/microtubule-associated protein-2 kinase. J. Biol. Chem. 1991;266:4220–4227. [PubMed] [Google Scholar]
- 16.Gomez N, Cohen P. Dissection of the protein kinase cascade by which nerve growth factor activates MAP kinases. Nature. 1991;353:170–173. doi: 10.1038/353170a0. [DOI] [PubMed] [Google Scholar]
- 17.Crews CM, Alessandrini A, Erikson RL. The primary structure of MEK, a protein kinase that phosphorylates the ERK gene product. Science. 1992;258:478–480. doi: 10.1126/science.1411546. [DOI] [PubMed] [Google Scholar]
- 18.Seger R, et al. Human T-cell Map kinase kinases are related to yeast signal transduction kinases. J. Biol. Chem. 1992;267:25628–25631. [PubMed] [Google Scholar]
- 19.Alessi DR, et al. Inactivation of p42 MAP kinase by protein phosphatase 2A and a protein tyrosine phosphatase, but not CL100, in various cell lines. Curr Biol. 1995;5(3):283–295. doi: 10.1016/s0960-9822(95)00059-5. [DOI] [PubMed] [Google Scholar]
- 20.Sun H, Charles CH, Lau LF, Tonks NK. MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell. 1993;75(3):487–493. doi: 10.1016/0092-8674(93)90383-2. [DOI] [PubMed] [Google Scholar]
- 21.Biggs WHd, Zipursky SL. Primary structure, expression, and signal-dependent tyrosine phosphorylation of a Drosophila homolog of extracellular signal-regulated kinase [published erratum appears in Proc Natl Acad Sci U S A 1993 Jul 1;90(13):6377] Proc. Natl. Acad. Sci. USA. 1992;89(14):6295–6299. doi: 10.1073/pnas.89.14.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Neill EM, Rebay I, Tjian R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78(1):137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- 23.Yung Y, et al. Detection of ERK activation by a novel monoclonal antibody. FEBS Lett. 1997;408(3):292–296. doi: 10.1016/s0014-5793(97)00442-0. [DOI] [PubMed] [Google Scholar]
- 24.Gabay L, Seger R, Shilo BZ. In situ activation pattern of Drosophila EGF receptor pathway during development. Science. 1997;277(5329):1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- 25.Gabay L, Seger R, Shilo BZ. MAP kinase in situ activation atlas during Drosophila embryogenesis. Development. 1997;124(18):3535–3541. doi: 10.1242/dev.124.18.3535. [DOI] [PubMed] [Google Scholar]
- 26.Arur S, et al. Multiple ERK substrates execute single biological processes in Caenorhabditis elegans germ-line development. Proceedings of the National Academy of Sciences. 2009;106(12):4776–4781. doi: 10.1073/pnas.0812285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delfini M-C, Dubrulle J, Malapert P, Chal J, Pourquié O. Control of the segmentation process by graded MAPK/ERK activation in the chick embryo. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(32):11343–11348. doi: 10.1073/pnas.0502933102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gille H, Sharrocks AD, Shaw PE. Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature. 1992;358(6385):414–417. doi: 10.1038/358414a0. [DOI] [PubMed] [Google Scholar]
- 29.Seth A, Alvarez E, Gupta S, Davis RJ. A phosphorylation site located in the NH2-terminal domain of c-Myc increases transactivation of gene expression. The Journal of biological chemistry. 1991;266(35):23521–23524. [PubMed] [Google Scholar]
- 30.Pulverer BJ, Kyriakis JM, Avruch J, Nikolakaki E, Woodgett JR. Phosphorylation of c-jun mediated by MAP kinases. Nature. 1991;353(6345):670–674. doi: 10.1038/353670a0. [DOI] [PubMed] [Google Scholar]
- 31.Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 1999;13(2):163–175. [PMC free article] [PubMed] [Google Scholar]
- 32.Sharrocks AD, Yang SH, Galanis A. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem Sci. 2000;25(9):448–453. doi: 10.1016/s0968-0004(00)01627-3. [DOI] [PubMed] [Google Scholar]
- 33.Zhang F, Strand A, Robbins D, Cobb MH, Goldsmith JG. Atomic structure of the MAP kinase ERK2 at 2.3 Å resolution. Nature. 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 34.Canagarajah BJ, Khokhlatchev A, Cobb MH, Goldsmith EJ. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell. 1997;90(5):859–869. doi: 10.1016/s0092-8674(00)80351-7. [DOI] [PubMed] [Google Scholar]
- 35.Mansour SJ, et al. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 36.Plotnikov A, Zehorai E, Procaccia S, Seger R. The MAPK cascades: Signaling components, nuclear roles and mechanisms of nuclear translocation. Biochim Biophys Acta. 2011;1813(9):1619–1633. doi: 10.1016/j.bbamcr.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 37.Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat Rev Mol Cell Biol. 2005;6(11):827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 38.Ferrell JE., Jr Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem Sci. 1996;21(12):460–466. doi: 10.1016/s0968-0004(96)20026-x. [DOI] [PubMed] [Google Scholar]
- 39.Huang CY, Ferrell JE., Jr Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc Natl Acad Sci U S A. 1996;93(19):10078–10083. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bluthgen N, Legewie S. Systems analysis of MAPK signal transduction. Essays in biochemistry. 2008;45:95–107. doi: 10.1042/BSE0450095. [DOI] [PubMed] [Google Scholar]
- 41.Fritsche-Guenther R, et al. Strong negative feedback from Erk to Raf confers robustness to MAPK signalling. Molecular systems biology. 2011;7 doi: 10.1038/msb.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carlson SM, et al. Large-Scale Discovery of ERK2 Substrates Identifies ERK-Mediated Transcriptional Regulation by ETV3. Sci. Signal. 2011;4(196):rs11-. doi: 10.1126/scisignal.2002010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kosako H, et al. Phosphoproteomics reveals new ERK MAP kinase targets and links ERK to nucleoporin-mediated nuclear transport. Nat Struct Mol Biol. 2009;16(10):1026–1035. doi: 10.1038/nsmb.1656. [DOI] [PubMed] [Google Scholar]
- 44.Yoon S, Seger R. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth factors (Chur, Switzerland) 2006;24(1):21–44. doi: 10.1080/02699050500284218. [DOI] [PubMed] [Google Scholar]
- 45.Bardwell L. Mechanisms of MAPK signalling specificity. Biochemical Society Transactions. 2006;34:837–841. doi: 10.1042/BST0340837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang S-H, Yates PR, Whitmarsh AJ, Davis RJ, Sharrocks AD. The Elk-1 ETS-Domain Transcription Factor Contains a Mitogen-Activated Protein Kinase Targeting Motif. Molecular and Cellular Biology. 1998;18(2):710–720. doi: 10.1128/mcb.18.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes & Development. 1999;13(2):163–175. [PMC free article] [PubMed] [Google Scholar]
- 48.Lee T, et al. Docking Motif Interactions in MAP Kinases Revealed by Hydrogen Exchange Mass Spectrometry. Molecular cell. 2004;14(1):43–55. doi: 10.1016/s1097-2765(04)00161-3. [DOI] [PubMed] [Google Scholar]
- 49.Tanoue T, Maeda R, Adachi M, Nishida E. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 2001;20(3):466–479. doi: 10.1093/emboj/20.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol. 2000;2(2):110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 51.Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 1997;16(8):1909–1920. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rainey MA, Callaway K, Barnes R, Wilson B, Dalby KN. Proximity-Induced Catalysis by the Protein Kinase ERK2. Journal of the American Chemical Society. 2005;127(30):10494–10495. doi: 10.1021/ja052915p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bandyopadhyay S, et al. A human MAP kinase interactome. Nat Meth. 2010;7(10):801–805. doi: 10.1038/nmeth.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lewis TS, et al. Identification of Novel MAP Kinase Pathway Signaling Targets by Functional Proteomics and Mass Spectrometry. Molecular cell. 2000;6(6):1343–1354. doi: 10.1016/s1097-2765(00)00132-5. [DOI] [PubMed] [Google Scholar]
- 55.Lovrić J, Dammeier S, Kieser A, Mischak H, Kolch W. Activated Raf Induces the Hyperphosphorylation of Stathmin and the Reorganization of the Microtubule Network. Journal of Biological Chemistry. 1998;273(35):22848–22855. [PubMed] [Google Scholar]
- 56.Vinayagam A, et al. A Directed Protein Interaction Network for Investigating Intracellular Signal Transduction. Sci. Signal. 2011;4(189):rs8-. doi: 10.1126/scisignal.2001699. [DOI] [PubMed] [Google Scholar]
- 57.von Kriegsheim A, et al. Cell fate decisions are specified by the dynamic ERK interactome. Nat Cell Biol. 2009;11(12):1458–1464. doi: 10.1038/ncb1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whisenant TC, et al. Computational Prediction and Experimental Verification of New MAP Kinase Docking Sites and Substrates Including Gli Transcription Factors. PLoS computational biology. 2010;6(8):e1000908. doi: 10.1371/journal.pcbi.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Friedman AA, et al. Proteomic and Functional Genomic Landscape of Receptor Tyrosine Kinase and Ras to Extracellular Signal-Regulated Kinase Signaling. Sci. Signal. 2011;4(196):rs10-. doi: 10.1126/scisignal.2002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim Y, et al. MAPK Substrate Competition Integrates Patterning Signals in the Drosophila Embryo. Current biology : CB. 2010;20(5):446–451. doi: 10.1016/j.cub.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grimm O, et al. Torso RTK controls Capicua degradation by changing its subcellular localization. Development. 2012;139(21):3962–3968. doi: 10.1242/dev.084327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim Y, et al. Gene Regulation by MAPK Substrate Competition. Developmental cell. 2011;20(6):880–887. doi: 10.1016/j.devcel.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robbins DJ, et al. Regulation and properties of extracellular signal-regulated protein kinases 1 and 2 in vitro. The Journal of biological chemistry. 1993;268(7):5097–5106. [PubMed] [Google Scholar]
- 64.Payne DM, et al. Identification of the regulatory phosphorylation sites in pp42/mitogen-activated protein kinase (MAP kinase) EMBO J. 1991;10(4):885–892. doi: 10.1002/j.1460-2075.1991.tb08021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anderson NG, Maller JL, Tonks NK, Sturgill TW. Requirement for integration of signals from two distinct phosphorylation pathways for activation of MAP kinase. Nature. 1990;343(6259):651–653. doi: 10.1038/343651a0. [DOI] [PubMed] [Google Scholar]
- 66.Kiel C, Serrano L. Cell Type-Specific Importance of Ras-c-Raf Complex Association Rate Constants for MAPK Signaling. Sci. Signal. 2009;2(81):ra38-. doi: 10.1126/scisignal.2000397. [DOI] [PubMed] [Google Scholar]
- 67.Kim Y, et al. Substrate-dependent control of MAPK phosphorylation in vivo. Molecular systems biology. 2011;7:467. doi: 10.1038/msb.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brunner D, et al. A gain-of-function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signaling pathways. Cell. 1994;76(5):875–888. doi: 10.1016/0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 69.Oellers N, Hafen E. Biochemical characterization of rolledSem, an activated form of Drosophila mitogen-activated protein kinase. The Journal of biological chemistry. 1996;271(40):24939–24944. doi: 10.1074/jbc.271.40.24939. [DOI] [PubMed] [Google Scholar]
- 70.Chu Y, Solski PA, Khosravi-Far R, Der CJ, Kelly K. The mitogen-activated protein kinase phosphatases PAC1, MKP-1, and MKP-2 have unique substrate specificities and reduced activity in vivo toward the ERK2 sevenmaker mutation. The Journal of biological chemistry. 1996;271(11):6497–6501. doi: 10.1074/jbc.271.11.6497. [DOI] [PubMed] [Google Scholar]
- 71.Aoki K, Yamada M, Kunida K, Yasuda S, Matsuda M. Processive phosphorylation of ERK MAP kinase in mammalian cells. Proceedings of the National Academy of Sciences. 2011;108(31):12675–12680. doi: 10.1073/pnas.1104030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Markevich NI, Hoek JB, Kholodenko BN. Signaling switches and bistability arising from multisite phosphorylation in protein kinase cascades. The Journal of cell biology. 2004;164(3):353–359. doi: 10.1083/jcb.200308060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Takahashi K, Tanase-Nicola S, ten Wolde PR. Spatio-temporal correlations can drastically change the response of a MAPK pathway. Proc Natl Acad Sci U S A. 2010;107(6):2473–2478. doi: 10.1073/pnas.0906885107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prabakaran S, et al. Comparative analysis of Erk phosphorylation suggests a mixed strategy for measuring phospho-form distributions. Molecular systems biology. 2011;7:482. doi: 10.1038/msb.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Friedman A, Perrimon N. A functional RNAi screen for regulators of receptor tyrosine kinase and ERK signalling. Nature. 2006;444(7116):230–234. doi: 10.1038/nature05280. [DOI] [PubMed] [Google Scholar]
- 76.Fujioka A, et al. Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. The Journal of biological chemistry. 2006;281(13):8917–8926. doi: 10.1074/jbc.M509344200. [DOI] [PubMed] [Google Scholar]
- 77.Fritz RD, et al. A Versatile Toolkit to Produce Sensitive FRET Biosensors to Visualize Signaling in Time and Space. Science signaling. 2013;6(285):rs12. doi: 10.1126/scisignal.2004135. [DOI] [PubMed] [Google Scholar]
- 78.Tomida T, Oda S, Takekawa M, Iino Y, Saito H. The temporal pattern of stimulation determines the extent and duration of MAPK activation in a Caenorhabditis elegans sensory neuron. Science signaling. 2012;5(246):ra76. doi: 10.1126/scisignal.2002983. [DOI] [PubMed] [Google Scholar]
- 79.Harvey CD, et al. A genetically encoded fluorescent sensor of ERK activity. Proceedings of the National Academy of Sciences. 2008;105(49):19264–19269. doi: 10.1073/pnas.0804598105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kiel C, Serrano L. Challenges ahead in signal transduction: MAPK as an example. Current opinion in biotechnology. 2012;23(3):305–314. doi: 10.1016/j.copbio.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 81.Sainz-Polo MA, et al. Three-dimensional structure of Saccharomyces invertase: role of a non-catalytic domain in oligomerization and substrate specificity. The Journal of biological chemistry. 2013;288(14):9755–9766. doi: 10.1074/jbc.M112.446435. [DOI] [PMC free article] [PubMed] [Google Scholar]