Abstract
Finding mucosal abnormalities (e.g., erythema, blood, ulcer, erosion, and polyp) is one of the most essential tasks during endoscopy video review. Since these abnormalities typically appear in a small number of frames (around 5% of the total frame number), automated detection of frames with an abnormality can save physician’s time significantly. In this paper, we propose a new multi-texture analysis method that effectively discerns images showing mucosal abnormalities from the ones without any abnormality since most abnormalities in endoscopy images have textures that are clearly distinguishable from normal textures using an advanced image texture analysis method. The method uses a “texton histogram” of an image block as features. The histogram captures the distribution of different “textons” representing various textures in an endoscopy image. The textons are representative response vectors of an application of a combination of Leung and Malik (LM) filter bank (i.e., a set of image filters) and a set of Local Binary Patterns on the image. Our experimental results indicate that the proposed method achieves 92% recall and 91.8% specificity on wireless capsule endoscopy (WCE) images and 91% recall and 90.8% specificity on colonoscopy images.
Keywords: Wireless capsule endoscopy, Colonoscopy, Filter bank, Local binary pattern, Texton, Texton dictionary
1. Introduction
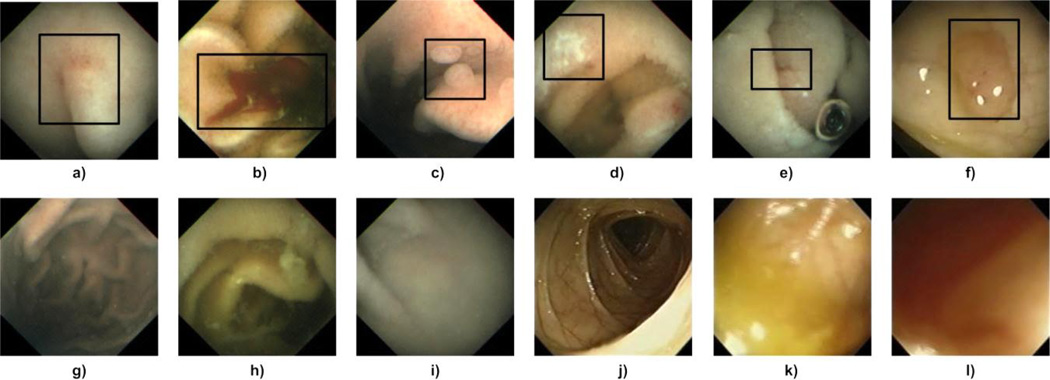
Endoscopy was introduced to examine the interior of a hollow organ or cavity of the body by directly inserting an endoscope inside the body. It has shown tremendous success in examination of gastrointestinal (GI) tract. Currently, there are various types of endoscopy available, namely, colonoscopy, bronchoscopy, gastroscopy, push enteroscopy, wireless capsule endoscopy (WCE), and upper endoscopy. The main aim of an endoscopy procedure is to find abnormal lesions (abnormalities) in the inspected human organ. Important abnormalities include erythema, blood (bleeding), ulcer, erosion, and polyp. We define that a frame showing an abnormality is an abnormal image, and a frame not showing any abnormality is a normal image. Fig. 1 shows examples of abnormal and normal images of WCE and colonoscopy. Our work focuses on abnormal image detection of two main endoscopy procedures: WCE and colonoscopy, which is briefly summarized as follows.
Fig. 1.
Sample WCE and colonoscopy abnormal and normal images. (a) WCE erythema, (b) WCE blood, (c) WCE polyp, (d) WCE ulcer, (e) WCE erosion, (f) colonoscopy polyp, (g) – (i) WCE normal images, and (j) – (l) colonoscopy normal images. In abnormal images, an abnormal area is manually marked inside a rectangle.
Most types of endoscopy have limited capability in viewing the small intestine. To address this problem, wireless capsule endoscopy (WCE), a device that integrates wireless transmission with image and video technology, was first developed in 2000 [1–3]. Physicians use WCE to examine the entire small intestine non-invasively. A typical WCE capsule is 11 mm in diameter and 25 mm in length. The front of the capsule has an optical dome where white light emitting diodes (LEDs) illuminate the luminal surface of the gut, and a micro camera sends images via wireless transmission to a wireless receiver worn by a patient. The capsule also contains a small battery that can last up to 8 ~ 11 hours. Once the capsule is swallowed, it passes naturally through the digestive system taking thousands of images (typically around 50,000 images) for about 8 hours. These images are transmitted to a receiver worn on the patient's belt. An expert later reviews these images to check whether any abnormal lesions are present. Although WCE is a technical breakthrough, the video viewing and interpretation time takes up to one hour even for an experienced gastroenterologist because of the large number of frames. This limits its general applications and incurs a considerable amount of healthcare costs. Thus, there is significant interest in the medical imaging community to discover a reliable and automated method for distinguishing abnormal images from normal images in a WCE video. Important abnormal lesions (abnormalities) in WCE are blood (bleeding), ulceration, erosion, erythema, and polyp. Details about these abnormalities can be found in [1–3]. Fig. 1 (a-e) show five abnormal images from WCE videos.
Colonoscopy is a preferred screening modality for prevention of colorectal cancer---the second leading cause of cancer-related deaths in the USA [4]. As the name implies, colorectal cancers are malignant tumors that develop in the colon and the rectum. The survival rate is higher if the cancer is found and treated early before metastasis to lymph nodes or other organs occur. The major abnormality in a colonoscopy video is the polyp. Fig. 1(f) shows a colorectal polyp image from a colonoscopy video. Early detection of polyps is one of the most important goals of colonoscopy. Recent evidence suggests that there are numbers of polyps that are not detected during colonoscopy [4]. To address this issue, we propose a texture-based polyp detection method.
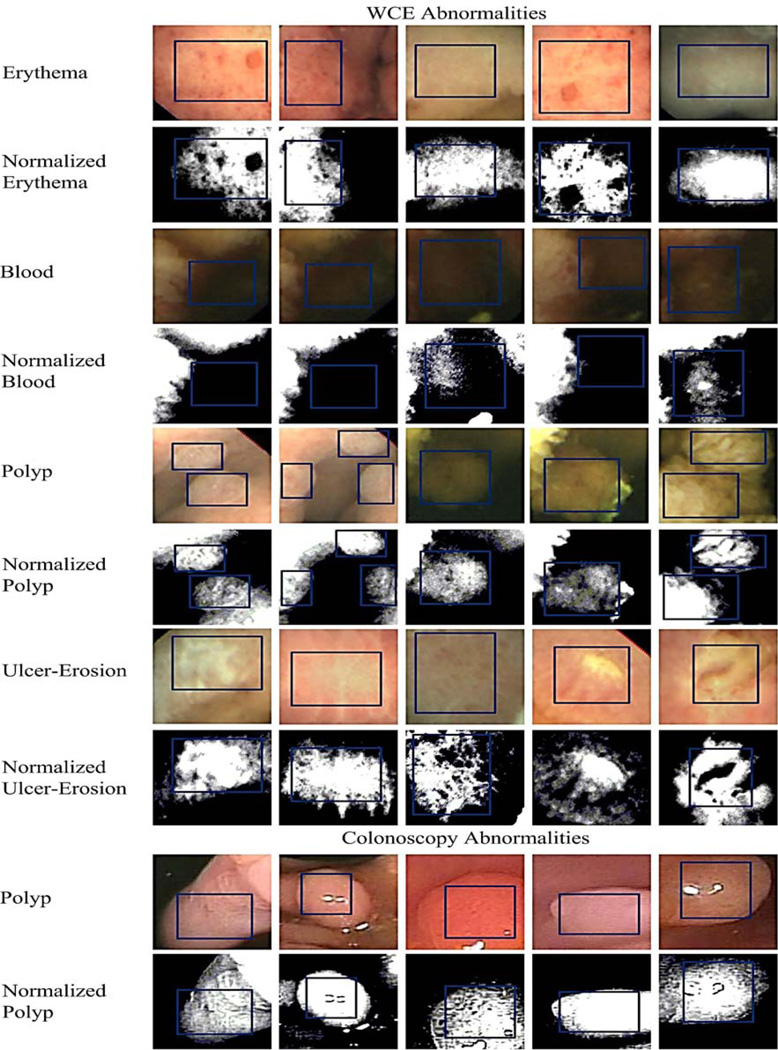
Distinguishing abnormal images from normal images in endoscopy is very challenging since endoscopy images are hampered by various artifacts. The aim of this work is to propose one method which can detect multiple abnormalities present in endoscopy images. The intended abnormalities include erythema, blood, polyps, ulcer and erosion. It is clear that erythema, polyps, ulcer and erosion do not have a dominant color. Therefore, when developing multiple abnormality detection methods color based approaches are not reliable. It is apparent that blood, erythema, ulcer and erosion do not possess a regular shape. Thus the shape-based approaches are not dependable when designing a multiple abnormality detection method. It has been shown that all these abnormalities can be successfully detected using a good texture analysis method [16, 18, 20–22, 27, 28]. Most abnormalities in endoscopy images have unique textures that are clearly distinguishable from normal textures, which make texture-based detection a preferred option for detecting multiple abnormalities. Fig.3 in Section 3.1 gives a clear picture about the inconsistent color and shape information and clear texture information of the endoscopy abnormalities. After all the abnormal frames are identified from an endoscopy video, finding a particular abnormality is not very time-consuming. Thus, a method to detect multiple abnormal frames is desirable.
Fig. 3.
Sample abnormal image textures from WCE and colonoscopy. For every abnormal texture, top row shows the original image texture and bottom row shows the normalized image textures. Blue rectangles point to the exact abnormal texture region. For training stage 1 (i.e., texton generation), we extract the regions shown using blue rectangles.
We design a multi-texture analysis method to detect multiple abnormalities. Over the years, numerous methods have been proposed to represent image textures. They include co-occurrence matrix, wavelets, local binary pattern, and many others [5]. Among them, texton-based texture analysis, first proposed in [6], is one of the most recent image texture analysis approaches. In general, the definition of a texton is vague. Theoretically, a texton is defined as a fundamental microstructure in natural images, and considered as the atoms of pre-attentive human visual perception [6]. However, in practice, a texton of a particular texture is a set of feature values that are most likely to occur after applying a feature detection method. Each type of texture has its own set of textons. Moreover, texton-based texture analysis supports classification of multiple textures with very high accuracy as shown in [7–11].
Here, we propose a multi-texture analysis method based on textons to discern abnormal images from normal images in endoscopy videos. It was found that the best way to generate textons is to use a set of filters such as Gaussian filters [7–11]. This set of filters is called a “filter bank”; each filter in the filter bank can extract a different texture feature of an image. By applying a filter bank to an image, a mixture of texture features such as directionality, orientation, line-likeness, and repetitiveness could be extracted. For the texton generation, the most commonly used filter banks are Leung-Malik (LM), Schmid (S), Maximum Responses (MR), or Gabor (G) [7–11]. After performing an extensive study, we found that using a new combination of Leung and Malik (LM) filter bank and a set of local binary patterns [12] can represent textures of endoscopy images more accurately compared to other filter banks. We call this hybrid model as the “LM-LBP” filter bank. In our method, a texton of a particular WCE or colonoscopy texture is the set of feature values generated by the LM-LBP filter bank, which are most likely to occur for only that texture. We combine all textons from all texture types (i.e., both abnormal and normal), and build a model called “texton dictionary.” We classify an unseen image as abnormal or normal by analyzing its distribution of textons (which is called “texton histogram”) with the texton dictionary.
Our experimental results on wireless capsule endoscopy (WCE) videos indicate that the proposed method achieves 92% recall and 91.8% specificity. Corresponding values for colonoscopy videos are 91% and 90.8%, respectively. Our contributions are as follows.
We have shown that the textures of endoscopy images can be represented as textons using a filter bank. We have proposed a new filter bank (i.e., LM-LBP) which is a combination of the LM filter bank and four local binary patterns.
This new filter bank works well for multiple abnormality detection in endoscopy images. This is significant since most endoscopy abnormality detection methods consider only one or two abnormalities in their schemes. With the capability of filter banks in detecting many different texture features, and with the capability of textons and texton histograms in representing these features, our proposed method can detect multiple abnormalities.
We have provided an extensive experimental study to show the effectiveness of the proposed LM-LBP filter bank by comparing it with other existing filter banks and related methods for endoscopy abnormal detection.
The remainder of this paper is organized as follows. Related works on wireless capsule endoscopy and colonoscopy are discussed in Section 2. Our proposed method for abnormality detection is described in Section 3. We discuss our experimental results in Section 4. Finally, Section 5 presents concluding remarks.
2. Related Works
The WCE and colonoscopy video processing has been one of the active research areas in medical image processing recently. The related works on WCE can be divided into three main categories: (1) video preprocessing such as image enhancement, frame reduction, and so on, (2) detection of abnormalities such as bleeding and ulceration, and (3) segmentation of WCE video into smaller segments corresponding to different types of the organs. Most of the research works in WCE aim at developing automated algorithms for abnormality detection using either color or texture analyses. Coimbra and Cunha proposed a method to detect multiple abnormalities such as blood, ulcer, or polyp, using MPEG-7 visual descriptors [14]. Penna et al. implemented a blood detection algorithm based on color modeling, edge masking, and RX detection [15]. In [16, 17], Li and Meng proposed individual methods to identify ulcers, polyps, and tumors. The ulcer detection method uses a curvelet transform, local binary pattern, and a multilayer perceptron neural network [16]. Polyp detection method in [17] is based on MEPG-7 region-based shape descriptors, Zernike moments, and a multilayer perceptron neural network for classification. Wavelet-based local binary pattern for texture feature extraction and Support Vector Machine (SVM) for classification are used in [18] for small bowel tumor detection. Two methods by Karargyris and Bourbakis for detection of ulcers and polyps are based on HSV color information and log-Gabor filter [19, 20]. In [21], the authors use statistical texture descriptors based on color curvelet covariance to identify small bowel tumors. A multi-texture abnormal detection method based on the bag of visual word approach is proposed in [22] to detect blood, polyp and ulcer images. The texture features are generated using Gabor filters and the classification is performed using the support vector machine classifier. In [23] a set of multiple abnormalities is detected using color histograms, color statistics and local binary patterns as features and combined patch classifiers as the classifier. Table 1 (first 10 rows) gives a summary of the detected abnormalities, features used, dataset used, classifiers used and the performance of each of the above method. The main limitation we see is that most of the WCE abnormality detection methods in Table 1 are focused on detecting only one type of abnormality [15–21], and the reported performances of multiple abnormality detection methods in [14], [22] and [23] are somewhat low. For instance, the given recall values of [14], [22] and [23] are 58%, 77% and 76%, respectively.
Table 1.
Analysis of related works (W – A work on WCE and C – A work on colonoscopy)
| Work | Detected Abnormalities | Features | Dataset | Classifiers | Average Performance |
|---|---|---|---|---|---|
| W: Coimbra et al.[14] | Blood, ulcer, polyp | MPEG-7 visual descriptors |
400 images from each texture |
Their own classifier |
Recall= 58% |
| W: Penna et al. [15] | Blood | Color features | 341 bleeding and 770 normal images |
No classifiers | Recall = 92% Specificity = 88% |
| W: Li and Meng [16] | Ulcer | Curvelet transformation and local binary pattern |
100 images | Multilayer perceptron neural network and support vector machines |
Recall = 93% Specificity = 91% |
| W: Li et el [17] | Polyp | Zernike moments and MEPG-7 region-based shape descriptor |
150 polyp and 150 normal images |
Multilayer perceptron neural network |
Recall = 90% Specificity = 83% |
| W: Li and Meng [18] | Ulcer | Wavelet based local binary pattern |
150 polyp and 150 normal images |
Support vector machines |
Recall = 97% Specificity = 96% |
| W: Karagysis et al. [19] | Ulcer | Log Gabor features | 20 ulcer and 30 normal images |
Support vector machines |
Recall = 75% Specificity = 73% |
| W: Karagysis et al.[20] | Polyp | Log Gabor features | 10 ulcer and 40 normal images |
Support vector machines |
Recall = 100% Specificity = 68% |
| W: Barbosa et al. [21] | Small bowel tumors | Statistical texture descriptors |
200 ulcer and 400 normal images |
Multilayer perceptron neural network |
Recall = 97% Specificity = 97% |
| W: Hwang [22] | Blood, ulcer, polyp | Gabor filters and color statistics |
50 blood, 50 polyp, 50 ulcer and 100 normal images |
Support vector machines |
Recall = 77% Specificity =91 % |
| W: Zhang et al. [23] | Hyperplasia, ulcer, anabrosis, oncoides, umbilication, infiltration, cabrities and bleeding |
Color histograms, color statistics and local binary patterns |
2949 images | Combined patch classifier |
Recall = 76% Specificity = 77% |
| C: Hwang et al. [25, 26] | Polyp | Shape features | 815 polyp and 7806 normal images |
No classifiers | Not given |
| C: Alexandre et al. [27] | Polyp | Color wavelet covariance and local binary pattern |
35 images | Lib-SVM | Area under the curve = 94% |
| C: Cheng et al. [28] | Polyp | Co-occurrence matrix and color texture |
74 images | Support vector machines |
Recall = 86% |
| C: Park et al. [29] | Polyp | Spatial and temporal features |
35 videos | Conditional random field |
Recall = 89% |
| C: Wang et al. [30] | Polyp | Edge cross-section based features |
247 training images and 1266 test images |
Decision tree and Support vector machines | Recall = 81% with falsely detected regions = 0.32 |
The related works on colonoscopy video processing could be divided into two main categories: (1) quality analysis of colonoscopy procedures and (2) detection of abnormalities such as polyps and ulcers. In [24], quality metrics were introduced to measure the quality of colonoscopy procedures. Most recent methods for automatic colorectal polyp detection are based on shape, color and/or texture. In [25] and [26], Hwang et al. proposed a polyp detection method based on a marker-controlled watershed algorithm and elliptical shape features. Because this technique relies on ellipse approximation, it can miss polyps in which their shapes are not close to ellipse. A texture-based polyp detection method [27] uses region color, local binary pattern (LBP), and SVM for classification. In [28], performance comparison was conducted to determine the best features among LBP, opponent color-LBP, wavelet energy, color wavelet energy, and wavelet correlation signatures for colonoscopy polyp detection using the same videos as training and testing data. The limitations of the methods in [25–28] are a high false alarm ratio and very slow detection speed. In [29], a framework for polyp detection in colonoscopy is proposed using a combination of spatial and temporal features. Spatial features include both texture and shape features which are represented as eigenvectors. They also incorporate some other features such as the number and size of the edges and the distance between the specular reflections and edges. A conditional random field (CRF) model is used for the classification. Although polyps can be detected more accurately when both shape and texture features are combined as outlined in [29], the method can be computationally complex since more features (i.e., texture and shape) need to be calculated. Also, this method cannot be used for multiple abnormality detection as shape is not common for other abnormalities such as ulcers. The work in [30] presents a polyp detection method based on edge cross section profiles (ECSPs). This method computes features by performing derivative functions on the edge cross section profiles. The feature values are fed into the decision tree and support vector machine classifiers to distinguish polyp images from non-polyp images. Although this technique detects various types of polyps with a higher accuracy it cannot be applicable to multiple abnormality detection as abnormalities such as ulcer do not have unique edge cross section profiles. Other research works on polyp detection in colonoscopy videos can be found in [31]. Detected abnormalities, features used, dataset used, classifiers used and the performance of each of the above polyp detection methods are listed in Table 1 (last 5 rows). Although most of the methods have provided very good classification performance they are specifically designed only to detect colorectal polyps.
In summary, most of the aforementioned abnormality detection methods focused on detecting only one type of abnormality, or the overall accuracies for detecting multiple abnormalities are somewhat low. The proposed method detects multiple abnormalities with a higher accuracy.
3. Proposed Method
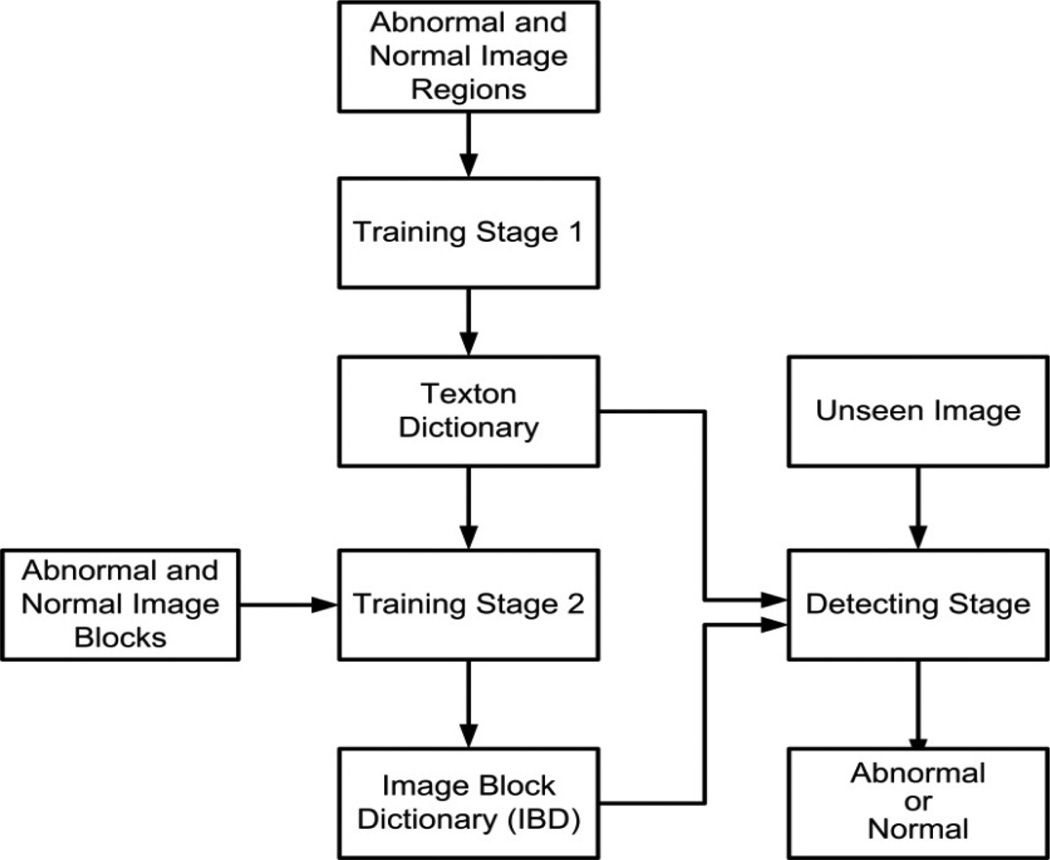
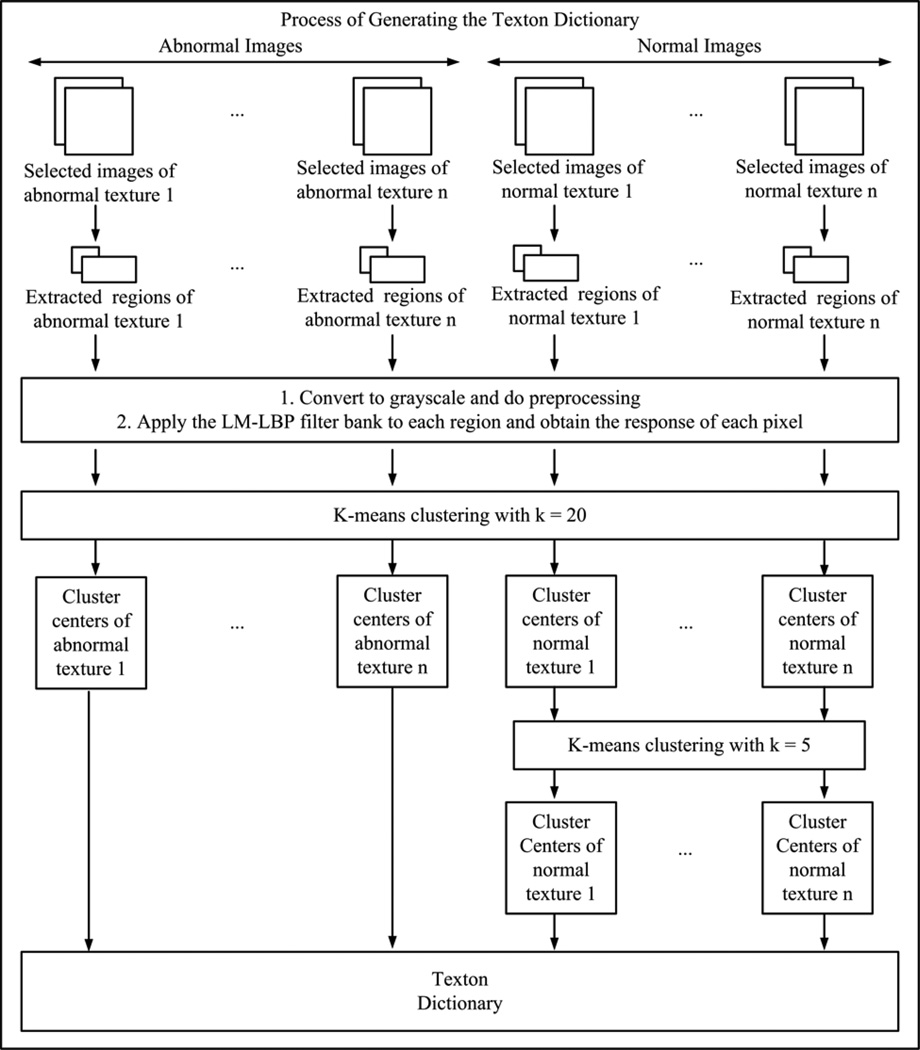
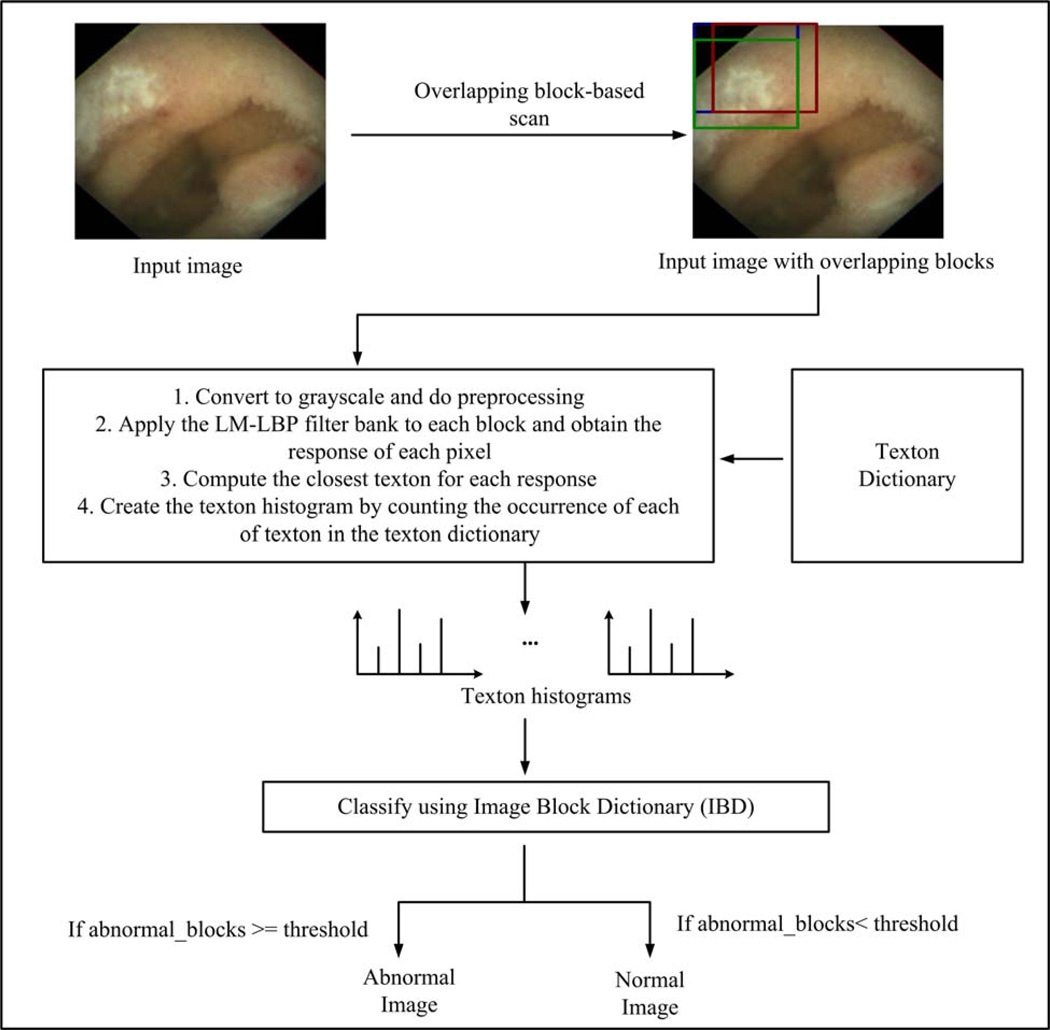
The proposed method consists of two training stages (Training stage 1 and Training stage 2), and a detecting stage. Fig. 2 shows a summary of the proposed method. Training stage 1 finds textons to represent WCE and colonoscopy textures using the proposed LM-LBP filter bank. The input for this stage is a set of image regions from each WCE and colonoscopy texture where each image region is covered entirely by one type of texture. The output of Training stage 1 is the collection of all textons from all types of textures of WCE and colonoscopy (i.e., both abnormal and normal). We call this collection the “texton dictionary.” In Training stage 2, we build an “image block dictionary (IBD)” classifier. The inputs for Training stage 2 are the texton dictionary and a significant number of sample image blocks (128 by 128 pixels) extracted from abnormal and normal textures from WCE and colonoscopy images. IBD is a trained classifier with k-nearest neighbor (KNN) method, which classifies an image block (128 by 128 pixels) as abnormal or normal based on its texton distribution. In the detecting stage, we evaluate an unseen image to determine whether it is abnormal or normal by scanning the image in both row wise and column wise. The inputs for the detecting stage are the unseen images, the texton dictionary from Training stage 1, and the IBD from Training stage 2. The output of the detecting stage is a label of an unseen image, which can be either abnormal or normal. Since our classification is a multi-class classification, the abnormal label is referred by the name of the abnormality. That is, for WCE, abnormal label can be one of erythema, blood, polyp, and ulcer-erosion. For colonoscopy the abnormal label can be polyp. In the following subsections, these three stages are discussed in detail.
Fig. 2.
An overview of the proposed abnormal image detection method.
3.1. WCE and Colonoscopy Textures
Before presenting our proposed method, we discuss the WCE and colonoscopy textures that are considered in designing the method. In WCE, we consider erythema, blood, ulcer, erosion, and polyp as abnormal textures. We observe that the ulcer and erosion show a similar kind of texture, so we combine ulcer and erosion as a one type of texture named UE. In colonoscopy, polyp is considered as the only abnormal texture, and other abnormal textures will be added later. Analyzing the normal textures is very complex due to their diversities. Fortunately, when we closely analyze the normal textures, we can categorize them into a handful of texture types. After analyzing these textures, WCE normal textures are divided into 10 types, and colonoscopy normal textures are divided into 8 types. Table 2 shows all textures types used to develop the proposed method.
Table 2.
Normal and abnormal textures of WCE and colonoscopy.
| WCE | Colonoscopy | ||
|---|---|---|---|
| Normal Texture | Abnormal Texture | Normal Texture | Abnormal Texture |
| Before Stomach | Erythema | Blurry | Polyp |
| Stomach Type I | Blood | Colon with Folds | |
| Stomach Type II | Polyp | Colon with Blood vessels | |
| Stomach Type III | UE (Ulcer-Erosion) | Colon without Blood vessels | |
| Small Intestine Type I | Colon with Lumen | ||
| Small Intestine Type II | Stool | ||
| Colon Type I | Specular Reflection | ||
| Colon Type II | Water | ||
| Stool | |||
| Specular Reflection | |||
In Fig. 3, we show five sample image blocks (texture area is marked using blue rectangles) for each abnormal texture of WCE and colonoscopy. We do not show normal textures to avoid having a very large figure. For each abnormal texture in Fig.3, the first row shows the original image block and the second row shows the same image block which is normalized using for example, z-score normalization (see Section 3.2.2). This step is done to show that one abnormality can have different combinations of colors and textures. In material textures we can highlight one type of arrangement of pixels as line-likeness, checkerboard and so on. But, it is important to realize that it is not possible to highlight one type of arrangement of pixels for endoscopy textures. The texture of blood is more or less uniform. The proposed method has detected the blood images in our dataset with a very high accuracy. Our assumption is that there is a texture in blood which cannot be seen by the human eyes. Sometimes the erythema texture can be seen as a thin cloud or salt and pepper noise. Polyp texture is similar to a surface of a caterpillar (animal) or some kind of a fungus. Texture of ulcer and erosion appear as a thick cloud of pixels. All the above descriptions are what can be observed by the human eyes. Fig. 3 shows all the above mentioned texture properties using the original and their corresponding normalized image areas for each abnormal image texture. For instance, the area of blood in normalized images is seen as black except the images in the 3rd and the 5th columns that have a different texture. Erythema and UE textures can be seen as clouds. The both WCE and colonoscopy polyp texture stands out from other textures showing fungus like texture. Also within one abnormal texture slight variations of textures can be seen. In the proposed method, we can represent these slight variations with textons. Moreover, Fig. 3 illustrates that it is very hard to find common shape or dominant color information from these abnormalities.
3.2. Training Stage 1: Generation of Textons and Texton Dictionary
To generate textons first we collect multiple image regions for each texture presented in Table 2. Since each image region contains one type of texture, the regions may not have the same size. Extracted image regions are similar to the ones marked using blue rectangles in Fig. 3. Within one texture, we observe slight variations due to illumination changes. We make sure that all variations are included in the extracted image regions for each texture.
3.2.1. LM-LBP Filter Bank
As mentioned in Section 1, the most commonly used filter banks for texton generation are Leung-Malik (LM), Schmid (S), Maximum Responses (MR), or Gabor (G) [7–11]. Our study focuses on the LM filter bank since it contains a variety of Gaussian filters. We conducted an extensive experimental study to demonstrate that the Gaussian filters can represent the endoscopy image textures more effectively than other filters. This study is presented in Section 4.
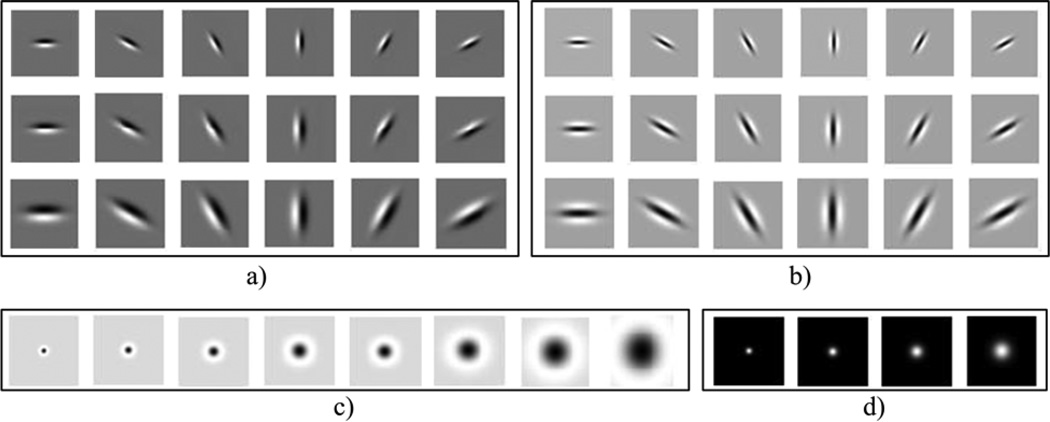
The LM filter bank consists of 48 different Gaussian filters. Fig. 4 shows the 48 filters in the LM filter bank. 18 first-order derivative of Gaussian filters in three different scales and six different orientations (See Fig. 4(a)), and another 18 second-order derivative of Gaussian filters in three different scales and six different orientations (See Fig. 4(b)) give 36 filters in total. The three scales are and six orientations are (0°, 30°, 60°,90°, 120°,150°). The remaining filters are 8 Laplacian of Gaussian (LoG) filters with , Considering P equally spaced pixels on a circular neighborhood with radius R, the local binary pattern (LBP) of the central pixel is defined as given in Equation (1) [12]. 12) (See Fig. 4(c)) and 4 other Gaussian filters with (See Fig. 4(d)). When an image is convoluted with the LM filter bank, we obtain 48 responses from each pixel.
| (1) |
Fig. 4.
Filters in the LM filter bank. a) 18 first derivative of Gaussian filters with three scales and six orientations, b) 18 second derivative of Gaussian filters with three scales and six orientations, c) 8 Laplacian of Gaussian filters, and d) 4 Gaussian filters.
In Equation (1), gc and gp are gray values of the central pixel (x, y) and the pth pixel (xp, yp) on the circular neighborhood; s is a function used to calculate the binary values. The location of the pth pixel, (xp, yp) is calculated using the formula (-Rsin(2πp/P), Rcos(2πp/P)). If the location is not the center of the pixel, the gray value is approximated using interpolation as mentioned in [12]. We experimentally found that a set of four LBPs with (P = 4, R = 1.0), (P = 8, R = 1.0), (P = 12, R = 1.5), and (P = 16, R = 2.0) shown in Fig. 5 best represents the WCE and colonoscopy textures with the highest accuracy for abnormal image detection. In fact, we do not observe any significant change in accuracy with R = 2.0 or larger. It makes sense that the pixels too far away from their center do not contribute much to the pattern of the center pixel. LBP responses are converted to values in the range between 0 and 1 (i.e., normalization) to make it consistent with the responses from filters in the LM filter bank. We combine the LM filter bank and the set of four LBPs to form the LM-LBP filter bank. By applying the LM-LBP filter bank to a pixel we get 52 filter responses (48 from LM and 4 from LBP).
Fig. 5.
Four LBPs with different P and R values. In a), b), c) and d), (P, R) values are (4, 1.0), (8, 1.0), (12, 1.5), and (16, 2.0), respectively.
3.2.2. Preprocessing and Filter Response Generation
Since various illumination and/or lighting effects of the camera distort endoscopy images, preprocessing is performed on the image regions before applying the LM-LBP filter bank to them. Due to illumination changes we can observe different textures of the same image tissue captured from different locations. Sometimes we see similar texture from different tissues located at different locations also due to illumination changes. However, once the illumination changes are removed, we can obtain a unique texture for each type of abnormal and normal textures. In the preprocessing step, we convert each region into grayscale and perform the z-score normalization by normalizing the image region to have zero mean and unit standard deviation. This is to ensure that we obtain exact textural information rather than color variations in the pixels. Our experimental results confirm that the proposed method has overcome this problem up to some extent. In Fig. 3, images in the second row of each abnormality show the preprocessed image blocks for each texture. After the preprocessing step, each region is convoluted with each filter in the LM-LBP filter bank. We arrange all the 52 responses into one vector of 52 dimensions, and call it a “response vector”. Each resulting filter response is normalized using the Webber's formula [7], which is defined in Equation (2). In Equation (2), F(x) and L(x) are the filter response and its L1-norm value, respectively. This step ensures that when calculating Euclidean distance (i.e., k-means clustering) the scaling and the range of each filter response are the same. The total number of response vectors in each texture is equal to the total number of pixels in all image regions of that particular texture.
| (2) |
3.2.3. Texton Generation Using K-means Clustering
We generate the textons as follows. Fig. 6 shows the process of generating the texton dictionary step by step. We cluster the response vectors from each texture using k-means clustering [7, 13] with the aim of finding a set of cluster centers This is because, in general a texton is a cluster center from the k-means clustering of response vectors. For an abnormal texture, we apply k-means clustering by setting k = 20 (based on our experiments). The resulting cluster centers are called “textons.” That means “textons” are representative response vectors of each texture. We apply k-means clustering to the normal texture in two steps. The purpose of the two-step approach is to reduce the dimension of the feature vectors by reducing the number of textons for the normal textures, which speeds up the overall procedure (to be described in Section 3.3). In Step 1, k-means clustering is applied to each type of normal texture by setting k = 20. This produces 200 textons for WCE normal textures and 160 textons for colonoscopy normal textures since WCE normal textures have 10 different types, and colonoscopy normal textures have 8 different types. In Step 2, k-means clustering is applied with k = 5 on the cluster centers obtained in Step 1. Overall, we generate 20 textons from each abnormal texture and 5 textons from each normal texture for both WCE and colonoscopy. The collection of all textons from all texture types (i.e., both abnormal and normal) is called the “texton dictionary.” Basically, the texton dictionary contains a set of representative response vectors that are generated by applying the LM-LBP filter bank to a particular WCE or colonoscopy image pixel. The WCE texton dictionary has 130 textons (80 from 4 abnormal textures and 50 from 10 normal textures), and the colonoscopy texton dictionary has 60 textons (20 from the polyp texture and 40 from 8 normal textures). In this way, any endoscopy image textures can be represented using the texton dictionary.
Fig. 6.
A summary of the training stage 1: Textons and their dictionary generation.
3.3. Training Stage 2: Generating the Image Block Dictionary (IBD)
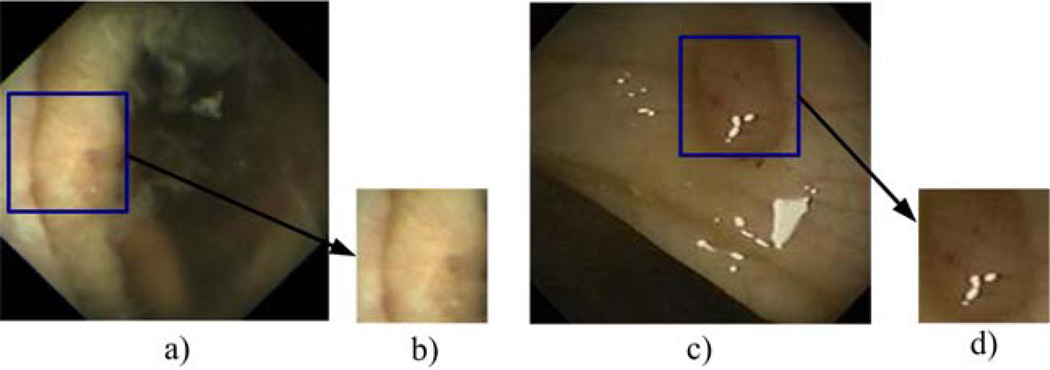
First, we extract a significant number of image blocks (of size 128 by 128 pixels) of abnormal and normal textures from WCE and colonoscopy images. All extracted image blocks were verified by the domain experts. We use fixed size image blocks instead of regions segmented using a region segmentation algorithm because it is simple to implement and fast to compute, and we do not need to concern about the accuracy of region segmentation. We initially tried segmentation algorithms such as Watershed [5] which did not provide a proper segmentation result. We experimentally found that the image block size of 128 by 128 pixels provides the best accuracy for our proposed method. Each selected image block contains a particular abnormal or normal texture. We make sure that the set of image blocks extracted for each texture include examples from all different illumination changes, viewing angles, and patients in our dataset. Since we extract the image blocks so that they contain as many abnormal areas as it does not matter whether the image blocks are overlapping or non-overlapping. Fig. 7 shows how an abnormal image block is extracted from an image to include as much abnormal area as possible. The normal image blocks are similarly obtained.
Fig. 7.
Two examples of abnormal image block extraction. a) An image with ulcer in WCE, b) an image block (128 by 128 pixels) of ulcer from a), c) an image with polyp in colonoscopy, and d) an image block (128 by 128 pixels) of polyp from c).
3.3.1. Texton Histogram Generation
We preprocess each image block and generate filter responses by applying the LM-LBP filter bank as mentioned in Section 3.2.2. For instance, we get 16,384 response vectors from an image block of 128 by 128 pixels. Using these response vectors, we build a histogram for each image block, which is called “texton histogram”. A texton histogram of an image block is a representation of the number of occurrences of the textons in the texton dictionary generated in Training stage 1. The number of bins in the histogram is equal to the number of textons in the texton dictionary. It shows how the texture of the image block is formed using the textons. In most cases multiple textures can be seen in the endoscopy tissue of the image block. Hence, to represent the texture of an image block accurately, we require textons corresponding to all abnormal and normal textures. This is the motive behind the fact that we include all textons from abnormal and normal textures in the texton dictionary created in Training stage 1 and use them as bins in the texton histogram.
A texton histogram for an image block of WCE has 130 bins, and a texton histogram for an image block of colonoscopy has 60 bins. The texton histograms are normalized to have values from 0 to 1. To build a texton histogram, we map each response vector to the closest matching texton in the texton dictionary, and increase the count of the bin corresponding to the matching texton. To decide the closest matching texton of a response vector, we calculate the Euclidean distance between the response vector and each texton in the texton dictionary. We find a texton with the minimum Euclidean distance as given in Equation (3). In Equation (3), RV = (RV1, …, RV52) is the response vector, Ti = (Ti1, … , Ti52) is the ith texton in the texton dictionary. The value of i varies from 1 to 130 for WCE, and from 1 to 60 for colonoscopy.
| (3) |
From this process, we get a texton histogram for each image block, and the total number of texton histograms is equal to the number of selected image blocks for that texture. In this way, a group of histograms that represents a particular texture could be derived.
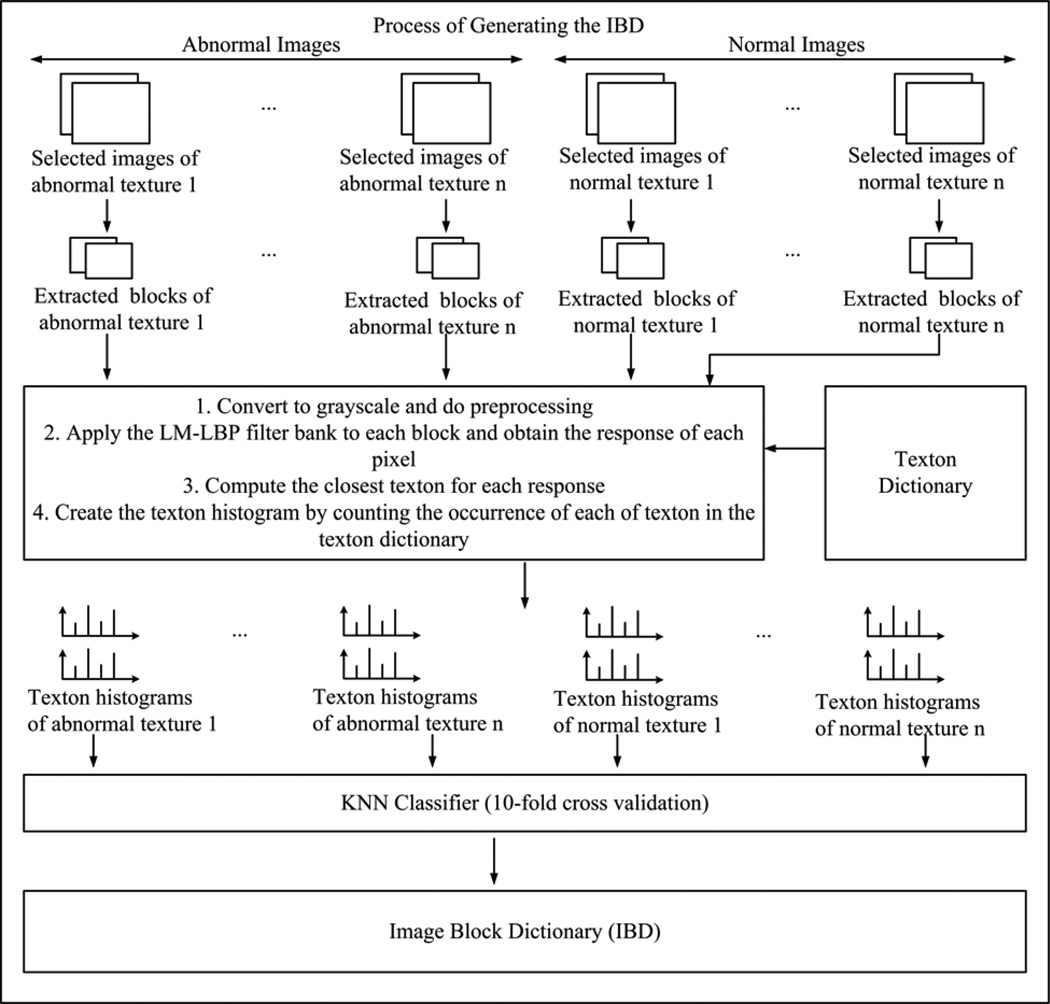
3.3.2. Image Block Dictionary (IBD) Generation
In the next step, we train a classifier that can distinguish abnormal image blocks from normal ones. The process of IBD generation is shown in Fig. 8 step by step. The texton histograms of image blocks are considered as feature vectors for the classifier. The dimension of a feature vector is equivalent to the number of textons in the texton dictionary. Using the trained classifier, previously unseen image blocks are evaluated. We tested five of the most popular classification algorithms used in machine learning [13]: K-nearest neighbor (KNN), Support vector machines (SVM), Decision tree (DT), Naïve Bayes, and Artificial neural networks to find the best one for our domain. We omitted the details about these classifiers due to space constraints, which can be found in [13]. In order to train a classifier, we used the 10-fold cross validation [13]. Experimentally, we found that the KNN classifier performed better than the other four classifiers. In the KNN classifier, the distance between texton histograms is calculated by employing the chi-square distance as given in Equation (4). In Equation (4), THi = (THi1, …, THiN) and THj = (THj1, …, THjN), are two texton histograms. The values of N are 130 and 60 for WCE and colonoscopy, respectively.
| (4) |
We call this trained classifier “Image Block Dictionary (IBD).” Image block dictionary is a trained classifier which can classify image blocks. It basically contains a set of texton histograms that represents the textures of abnormal and normal image blocks. IBD classifies an input image block using these representative texton histograms. The set of texton histograms acts like a dictionary or a database. Hence we named this model as the image block dictionary.
Fig. 8.
A summary of the training stage 2: Generating the IBD.
Basically, IBD decides whether an image block is abnormal or normal. An IBD has three components: a filter bank to generate the response vectors, image block size, and a classifier. Hence, we represent an IBD with three parameters, for example, IBD (filter bank, block size, classifier).
3.4. Detecting Stage: Evaluating Images for Abnormal Detection
In this stage, we evaluate an unseen image to determine whether it is abnormal or normal. The image is scanned both row and column wise using image blocks having a predefined overlap with the previous scan as shown in Fig. 9 (red and green squares). We experimentally verified that with overlapping image blocks the proposed method performs better compared to non-overlapping image blocks. Texton histogram of each resulted block from the scanning is evaluated with the IBD. Thus, the texton histograms of the image blocks are used as the input to the trained KNN classifier. The number of blocks detected as abnormal is counted during the scanning. If the abnormal block count of an image is more than a certain threshold value, the image is categorized as an abnormal image. For instance, if 4 blocks are detected as erythema, then the input image is labeled as ‘erythema’. Otherwise, the image is categorized as a normal image. This threshold value is decided to compensate the accuracy of block evaluation. Theoretically, if at least one block is detected as abnormal, then image should be labeled as abnormal. However, since it is very hard to achieve 100% accuracy for image block evaluation, the threshold value could be more than one. We decide this threshold value experimentally, which will be discussed in the following section.
Fig. 9.
A summary of the detecting stage.
4. Experimental Results and Discussion
In this section, we assess the effectiveness of the proposed abnormal image detection technique. Two separate experiments were carried out for WCE and colonoscopy videos and summarized as follows.
We compare the results from the proposed LM-LBP filter bank with those from three other filter banks: LM, MR8, and Gabor (G).
We also compare the results from the proposed LM-LBP filter bank with textons generated only using the set of four LBPs discussed in Section 3.2 and with the textons generated using image patch exemplars [9, 31]. Image patch exemplars are a very good candidate for texton generation as described in [9]. We named the method as the ‘local neighborhood method’. To generate textons, the local neighborhood method considers the 3 × 3 local neighborhoods around each pixel as the response vectors [32] (see Section 3). We tested larger neighborhood such as 5×5 and 7×7 pixels. For 5×5 local neighborhoods we observed 1–2% improvement in the performance compared to 3×3 local neighborhoods. However, for 7×7 local neighborhoods we saw a 2–3% decrease in performance compared to 3×3 local neighborhoods. We call the local neighborhood based method as “LN filter bank”, and the local binary pattern based method as “LBP filter bank” for easier reference. For both LN and LBP filter banks the filter responses are normalized to have values from 0 to 1.
To verify that IBDs based on image blocks of 128 by 128 pixels perform better than the other sizes of image blocks, we tested our method on two different image block sizes: 64 by 64 and 128 by 128 pixels.
To show that IBDs based on the KNN classifier outperform IBDs based on other classifiers, we present the results for KNN, SVM, and Decision Tree (DT) classifier based IBDs. Due to the space limitations we have only provided the results of KNN, SVM and DT classifiers. KNN, SVM and DT are the top three classifiers in terms of performance. Experiments with KNN classifier were performed with different values of “K”, which is the number of nearest neighbors. Since K=1 produced the best classification accuracy, we reported the performance measures only for K=1. Sequential minimization optimization (SMO) and C-SVM based implementation [13] were used for the SVM classifier, in which the Gaussian radial basis function (RBF) kernel [13] was used as the kernel function. We obtained the best results when the cost parameter (C) was 1. The reported performance values for SVM classifier were obtained when using the set of parameters which gave the best classification performance for SVM classifier. We tested the SVM classifier with the polynomial, normalized polynomial and radial basis function (RBF) kernels. We concluded that the RBF kernel provides the best performance. Similarly we tuned the cost parameter c and found that c = 1 gave the best performance. For the decision tree classifier we used the J48 based decision tree. Three classifiers were tested using the 10-fold cross validation [13]. As mentioned in Section 3.3.2, texton histograms were used as the input to a classifier. Thus the 10-fold cross validation was used on these texton histograms by creating 10 different training and testing combinations.
To verify the effectiveness of the filters in the LM-LBP filter bank and the size of the texton dictionary, we show the performance of various combinations of filters exists within the LM-LBP filter bank.
To show the soundness of selecting four original LBPs in the LM-LBP filter bank, we compare the results with other variants of LBPs.
For Training stage 1, we created texton dictionaries with LM, MR8, G, LN, LBP, and LM-LBP filter banks by applying the algorithm mentioned in Section 3.2. For Training stage 2, we generated 36 IBDs by using the algorithm mentioned in Section 3.3 and by varying the filter bank type, image block size, and the classifier. All combinations of six filter banks, two block sizes, and three classifiers give us 36 different IBDs as outlined in Table 3. The same filter bank used for the texton generation in Training stage 1 is used to generate the IBDs in Training stage 2. We show that the proposed method works best with the combination of LM-LBP filter bank, 128 by 128 (pixels) image blocks, and KNN classifier (i.e., IBD (LM-LBP, 128, KNN) (No. 34)).
Table 3.
Generated image block dictionaries(IBDs).
| No. | IBD | No. | IBD | No. | IBD | No. | IBD |
|---|---|---|---|---|---|---|---|
| 1 | IBD(LM, 64, KNN) | 10 | IBD(LN, 64, KNN) | 19 | IBD(LM, 128, KNN) | 28 | IBD(LN, 128, KNN) |
| 2 | IBD(LM, 64, SVM) | 11 | IBD(LN, 64, SVM) | 20 | IBD(LM, 128, SVM) | 29 | IBD(LN, 128, SVM) |
| 3 | IBD(LM, 64, DT) | 12 | IBD(LN, 64, DT) | 21 | IBD(LM, 128, DT) | 30 | IBD(LN, 128, DT) |
| 4 | IBD(MR8, 64, KNN) | 13 | IBD(LBP, 64, KNN) | 22 | IBD(MR8, 128, KNN) | 31 | IBD(LBP, 128, KNN) |
| 5 | IBD(MR8, 64, SVM) | 14 | IBD(LBP, 64, SVM) | 23 | IBD(MR8, 128, SVM) | 32 | IBD(LBP, 128, SVM) |
| 6 | IBD(MR8, 64, DT) | 15 | IBD(LBP, 64, DT) | 24 | IBD(MR8, 128, DT) | 33 | IBD(LBP, 128, DT) |
| 7 | IBD(G, 64, KNN) | 16 | IBD(LM-LBP, 64, KNN) | 25 | IBD(G, 128, KNN) | 34 | IBD(LM-LBP,128, KNN) |
| 8 | IBD(G, 64, SVM) | 17 | IBD(LM-LBP, 64, SVM) | 26 | IBD(G, 128, SVM) | 35 | IBD(LM-LBP, 128,S VM) |
| 9 | IBD(G, 64, DT) | 18 | IBD(LM-LBP, 64, DT) | 27 | IBD(G, 128, DT) | 36 | IBD(LM-LBP, 128, DT) |
We present our results using commonly used performance metrics: recall (R), specificity (S), precision (P), and accuracy (A). Table 4 shows the definitions of True Positive (TP), False Positive (FP), False Negative (FN), and True Negative (TN). Recall (R) is the percentage of correctly classified positive instances and it is defined as TP/(TP+FN). Precision (P) computed from TP/(TP+FP) is the percentage of correctly classified positive instances from the predicted positives; specificity (S) is the percentage of correctly classified negative instances and it is calculated from TN/(TN+FN). The accuracy (A) which is equal to (TP+TN)/(TP+FP+FN+TN) is the number of correctly classified instances. All experiments were performed on a computer with Intel(R) Core i7 2600K, 64-bit, 3.40 GHz processor and 8 GB memory. The programming environment was Matlab R2012b. SVM and decision tree classifiers were tested using the corresponding implementations in the WEKA package [33].
Table 4.
Accuracy metrics.
| Actual Texture | Predicted Texture |
|
|---|---|---|
| Positive | Negative | |
| Positive | TP | FN |
| Negative | FP | TN |
The remainder of the experimental section is organized as follows. Experiments performed with WCE videos are described in the Subsection 4.1, and experiments performed with colonoscopy videos are described in the Subsection 4.2. In both Subsections 4.1 and 4.2, we present the performance of IBDs (i.e., Training stages). Then, we present the performance of the abnormal image detection (i.e., Detecting stage). In Section 4.3, we provide experimental evidence to show selection of filters in the LM-LBP filter bank and the size of the texton dictionary.
4.1. Experiments with WCE Videos
We extracted a set of images from five real WCE videos for each abnormal and normal textures outlined in Table 2. All WCE images are generated from the MicroCam capsule from IntroMedic. The resolution of an image is 320×320 pixels. Our domain expert verified the ground truth of the WCE images. We made sure that as many as possible texture variations are included in both abnormal and normal images extracted. From each texture, a portion of the image set was used for the training (Stage 1 and Stage 2), and the remaining portion was used for the detecting stage. The number of images used for the two training stages and the detecting stage are given in Columns 2 and 3 of Table 5. The number of image regions extracted for Training stage 1 (i.e., texton generation) and the number of image blocks extracted for Training stage 2 (i.e., IBD generation) are given in Columns 4–6 of Table 5. Both image regions for Training stage 1 and image blocks for Training stage 2 were extracted from the set of training images listed in Column 2 of Table 5. We made sure that the extracted regions for Training stage 1 listed in column 4 of Table 5 include as many variations as possible for each WCE texture as mentioned in Section 3.2. For instance, 50 examples for erythema regions include five different textures. Moreover, we ensured that extracted image blocks for training stage listed in column 5 and column 6 of Table 5 contain as many as possible variations of abnormal and normal areas as mentioned in Section 3.3. Using the image regions in Training stage 1 and the image blocks in Training stage 2, we built the 36 IBDs listed in Table 3 for WCE. We used the same image set for both training stage 1 and 2 since they are two steps of one continuous training process. Input from the training stage 1 (i.e., texton dictionary) is used to build the texton histogram in the training stage 2. The main goal of the whole training process is to create a set of histograms which can accurately represent abnormal and normal image blocks. The best way of generating textons to be used in the texton histograms is to generate them from the same image set.
Table 5.
Images, image regions, and image blocks used for the WCE experiment.
| Texture | # of Images for Training Stage 1 and Stage 2 |
# of Images for Detecting Stage |
# of Image Regions for Training Stage 1 |
# of Image Blocks (64 by 64) for Training Stage 2 |
# of Image Blocks (128 by 128) for Training Stage 2 |
|---|---|---|---|---|---|
| Erythema | 100 | 25 | 50 | 200 | 100 |
| Blood | 100 | 25 | 50 | 200 | 100 |
| Polyp | 100 | 25 | 60 | 200 | 100 |
| UE | 150 | 25 | 70 | 300 | 150 |
| 800 | 400 | 400 | 1600 | 800 | |
| Normal | (80 from each of 10 normal textures) |
(40 from each of 10 normal textures) |
(40 from each of 10 normal textures) |
(160 from each of 10 normal textures) |
(80 from each of 10 normal textures) |
4.1.1. WCE IBD Performance analysis
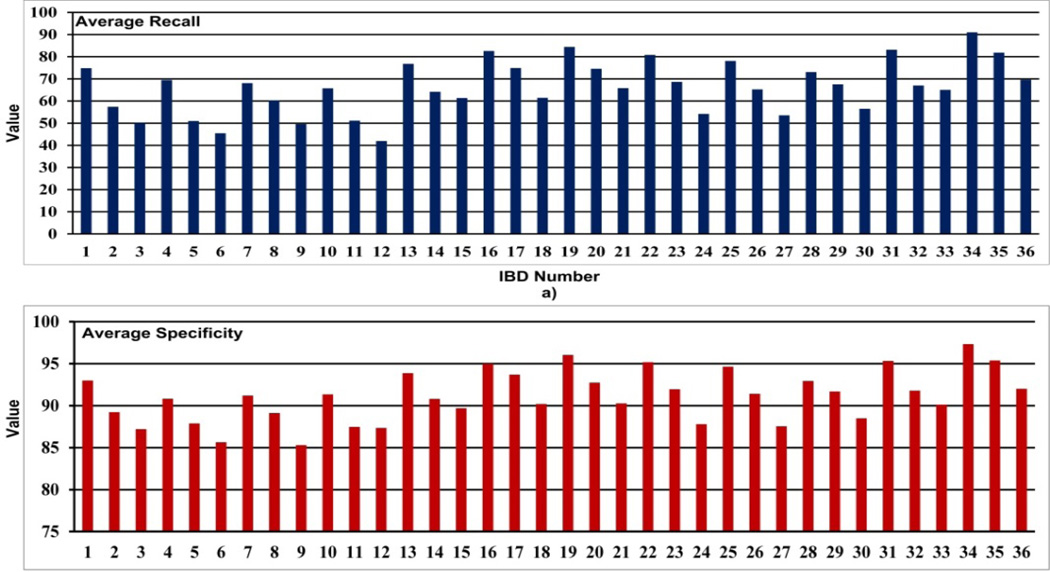
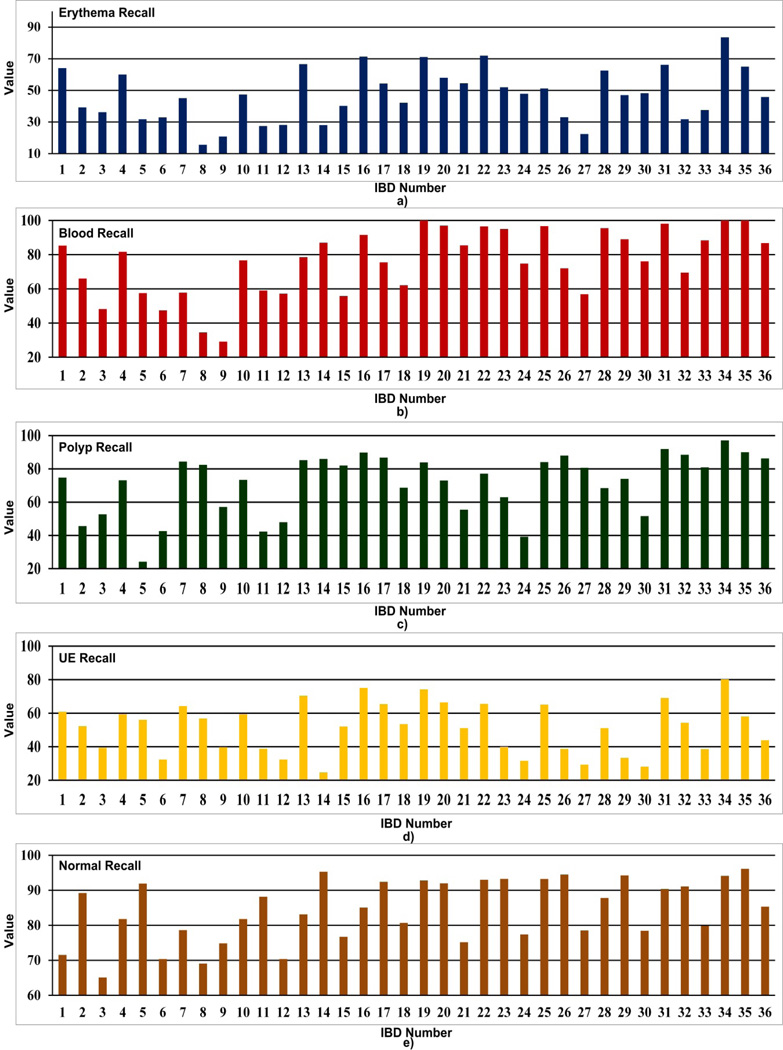
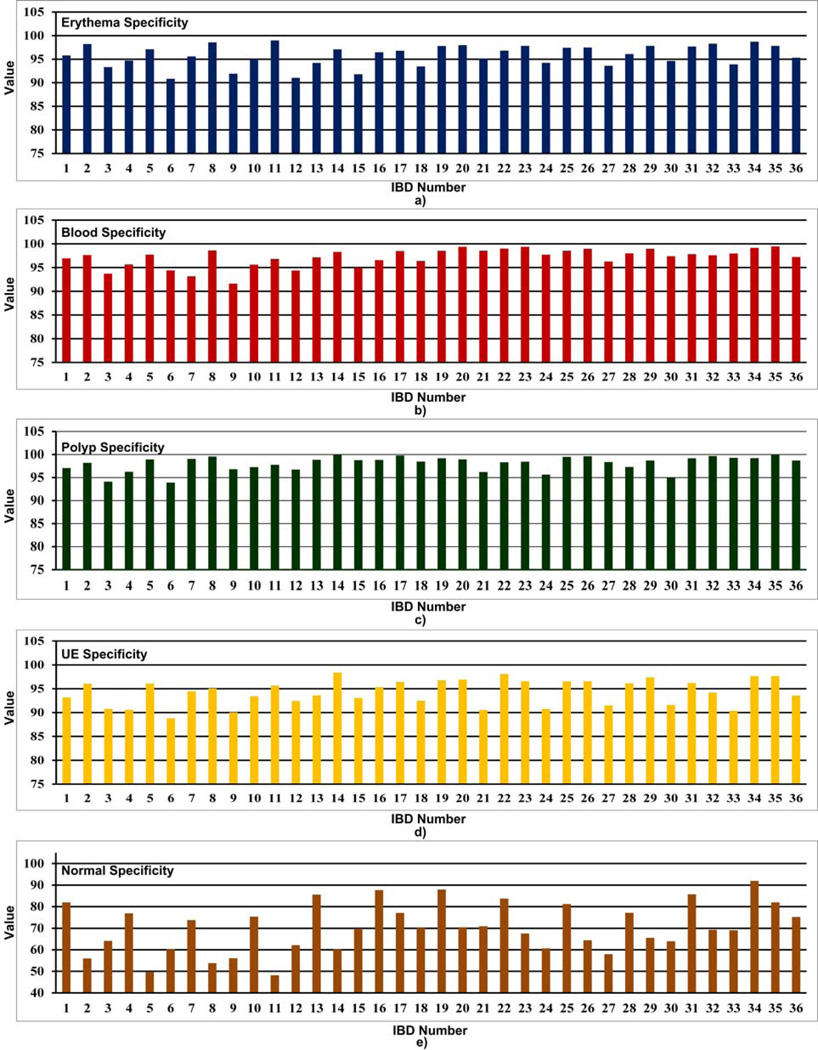
Average performance of the four abnormal (i.e., erythema, blood, polyp and UE) and other normal textures of the 36 IBDs are given in Table 6. Fig. 10 gives a graphical comparison of the average recall and specificity values of each IBD. Fig. 11 and Fig. 12 show comparisons of the recall and specificity values of each IBD for four abnormal and other normal textures.
Table 6.
Average performance of the 36 WCE IBDs.
| IBD No. | P | R | S | A | IBD No. | P | R | S | A | IBD No. | P | R | S | A |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 71.1 | 74.8 | 93.0 | 91.0 | 13 | 74.0 | 76.8 | 93.9 | 91.8 | 25 | 80.8 | 78.1 | 94.7 | 94.4 |
| 2 | 70.9 | 57.4 | 89.2 | 89.1 | 14 | 75.0 | 64.2 | 90.8 | 92.0 | 26 | 75.5 | 65.2 | 91.4 | 92.2 |
| 3 | 48.6 | 50.1 | 87.2 | 84.0 | 15 | 61.4 | 61.4 | 89.7 | 87.3 | 27 | 54.3 | 53.5 | 87.5 | 86.6 |
| 4 | 63.6 | 69.4 | 90.8 | 88.1 | 16 | 78.5 | 82.6 | 95.0 | 93.4 | 28 | 71.8 | 73.1 | 92.9 | 92.2 |
| 5 | 66.8 | 51.0 | 88.0 | 88.0 | 17 | 81.1 | 74.9 | 93.7 | 93.2 | 29 | 76.6 | 67.5 | 91.7 | 92.5 |
| 6 | 44.1 | 45.5 | 85.6 | 82.0 | 18 | 63.9 | 61.4 | 90.2 | 88.2 | 30 | 54.6 | 56.5 | 88.5 | 87.1 |
| 7 | 66.6 | 65.7 | 91.2 | 89.3 | 19 | 83.7 | 84.4 | 96.1 | 95.5 | 31 | 81.1 | 83.1 | 95.3 | 94.7 |
| 8 | 71.6 | 56.1 | 89.1 | 89.4 | 20 | 79.9 | 72.4 | 92.7 | 93.3 | 32 | 75.6 | 67.0 | 91.8 | 91.3 |
| 9 | 43.0 | 42.0 | 85.3 | 82.1 | 21 | 61.6 | 62.7 | 90.3 | 88.8 | 33 | 64.5 | 65.1 | 90.1 | 88.9 |
| 10 | 65.9 | 68.1 | 91.4 | 89.3 | 22 | 81.8 | 80.8 | 95.2 | 94.8 | 34 | 89.0 | 91.0 | 97.3 | 96.9 |
| 11 | 66.8 | 51.1 | 87.5 | 87.5 | 23 | 76.7 | 68.7 | 92.0 | 92.5 | 35 | 86.6 | 81.8 | 95.4 | 95.6 |
| 12 | 86.8 | 49.6 | 87.4 | 84.6 | 24 | 53.9 | 54.2 | 87.8 | 86.5 | 36 | 68.0 | 69.6 | 92.0 | 91.0 |
Fig. 10.
a) Average recall and b) average specificity analysis of 36 WCE IBDs.
Fig. 11.
Comparison of recall values of a) erythema, b) blood, c) polyp, d) UE and e) other normal textures for 36 WCE IBDs.
Fig. 12.
Comparison of specificity values of a) erythema, b) blood, c) polyp, d) UE and e) other normal textures for 36 WCE IBDs.
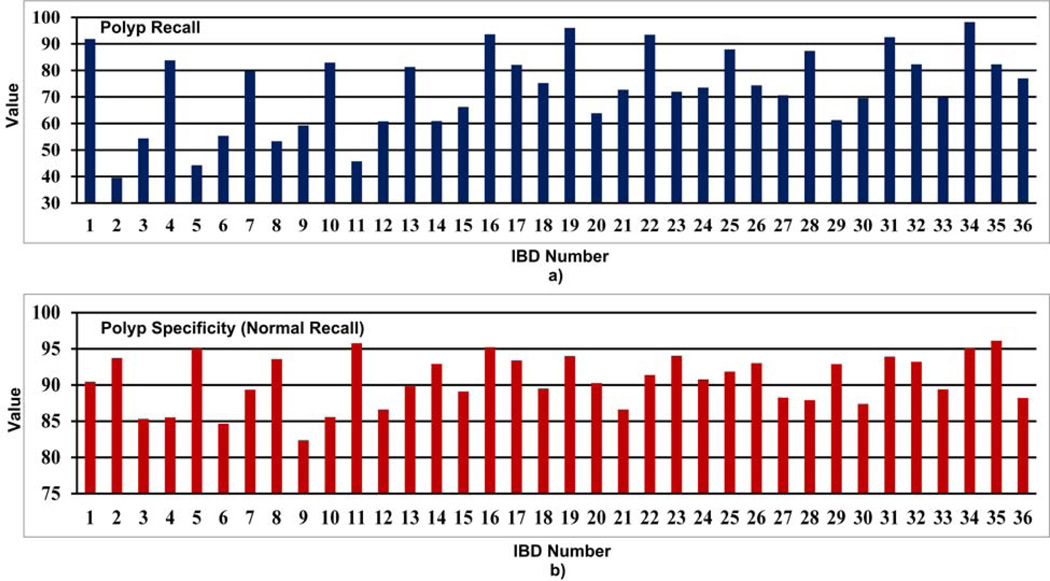
When analyzing Table 6 and Fig. 10, we see that IBD No. 34 (LM-LBP, 128, KNN) shows the highest average recall of 91.0% and the highest average specificity of 97.3%. IBD No. 19 (LM, 128, KNN) and IBD No. 31 (LBP, 128, KNN) perform with average recall values of 84.4% and 83.1%, and average specificity of 96.1% and 95.3%, respectively, which are very close to IBD No. 34 (LM-LBP, 128, KNN). It is apparent that when LM filter bank and a set of LBPs are combined to form the LM-LBP filter bank, we can see a significant improvement in the recall and the specificity. Fig. 11 shows that the IBD No. 34 (LM-LBP, 128, KNN) provides the highest recall values for all abnormal textures, and a very high recall value of 94.1% for other normal textures. Note that the IBD No. 34 (LMLBP, 128, KNN) provides 100% recall for blood, which is the same with IBD No. 19 (LM, 128, KNN) and IBD No. 35 (LM-LBP, 128, SVM). As seen in Fig. 12, the specificity values of most of the IBDs are above 80%. Except for normal textures, the specificity values of the IBD No. 34 (LM-LBP, 128, KNN) for other textures are not the highest but very close to the highest. Its specificity values for four abnormal textures and other normal textures are 98.7%, 99.2%, 99.2%, 97.6%, and 92.0%, respectively, which are good enough even if they are not the highest. However, the average specificity of the four abnormal and other normal textures (i.e., 97.3%) is the highest of all 36 IBDs. These prove that the IBD No. 34 (LM-LBP, 128, KNN) classifies the 128 by 128 image blocks with better recall values while maintaining very high specificity values for all types of textures compared to other IBDs. The IBD No. 34 (LM-LBP, 128, KNN) is selected for the WCE abnormal image detecting stage.
LBP and Gabor filters have successfully been used for WCE abnormal detection [16, 22]. In a similar work to the proposed method in [22], Gabor filters are used to generate the bag of visual words (similar to textons). Our experimental results for IBDs based on the Gabor filter banks (i.e., IBDs 7, 8, 9, 25, 26, and 27) for WCE shows that the proposed method outperforms the Gabor filter bank based abnormal detection. When analyzing the LBP based IBDs (i.e., IBDs 7, 8, 9, 25, 26, and 27 in Table 3, Section 4, page 21) we can conclude that our proposed LM-LBP filter bank outperforms the LBP based methods for abnormal detection as well.
4.1.2. WCE Abnormal Image Detection Analysis
We evaluated the detecting stage of the proposed method using our IBD No. 34 (LM-LBP, 128, KNN). We used 100 abnormal images (25 from each texture) and 400 normal images as outlined in Column 3 of Table 5. We intentionally made the dataset imbalance to resemble real world scenario in WCE where the numbers of abnormal and normal images are imbalanced. Each image was evaluated using IBD No. 34 (LM-LBP, 128, KNN) as described in Section 3.4 for abnormal image detection. We experimented with abnormal block thresholds (see Section 3.4) of 1 through 20. Table 7 shows performance comparisons of the detecting stage with abnormal block thresholds of 4, 5, and 6. We found that the threshold value of 5 provides the best image classification performance when both recall and specificity are considered. When abnormal block threshold is 5, the proposed method classifies abnormal and normal images with recall value of 92.0% and a specificity of 91.8%. That means, 8 abnormal images (out of 100) (i.e., false negatives) are missed by our method. This contains 3 erythema, 2 polyp and 3 UE images. Also, there are 33 normal images detected as abnormal images (i.e., false positives). These include 11 erythema, 2 blood, 7 polyp and 13 UE detections. The confusion matrix given in Table 8 shows the detections of each texture. Our WCE abnormal image detection is a multi-class classification problem having erythema, blood, ulcer-erosion, polyp and normal as classes. Hence, in Table 8 we have provided the confusion matrix for those five classes. Overall, the accuracy of erythema and UE image detection is somewhat lower compared to blood and polyp image detection. Despite these false positives and false negatives, a recall of 92.0% and a specificity of 91.8% for abnormal image detection are very high since we detect multiple abnormalities. Normally, the physician’s recommendation is to reduce the miss rate of abnormal images (i.e., true positives) as much as possible while keeping true negatives at a reasonably higher value. Hence, our recall and specificity values are adequate for WCE abnormal image detection.
Table 7.
Evaluation of the WCE abnormal image detection with IBD No. 34 (LM-LBP, 128, KNN).
| Abnormal Block Threshold | P | R | S | A |
|---|---|---|---|---|
| 4 | 52.8 | 94.0 | 79.0 | 82.0 |
| 5 | 73.6 | 92.0 | 91.8 | 91.8 |
| 6 | 79.4 | 85.0 | 94.5 | 92.6 |
Table 8.
Confusion matrix of the WCE abnormal classification for abnormal block threshold = 5.
| Actual/Predicted | Erythema | Blood | Polyp | UE | Normal |
|---|---|---|---|---|---|
| Erythema | 22 | 0 | 0 | 1 | 2 |
| Blood | 0 | 25 | 0 | 0 | 0 |
| Polyp | 0 | 0 | 23 | 0 | 2 |
| UE | 1 | 0 | 0 | 22 | 2 |
| Normal | 11 | 2 | 7 | 13 | 367 |
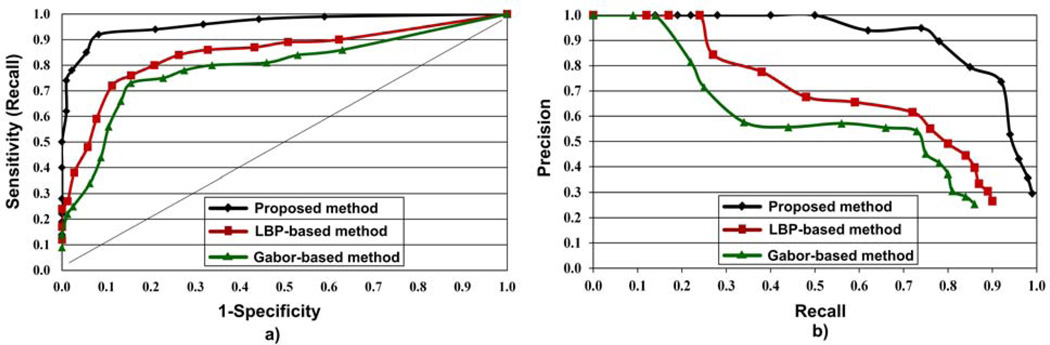
We compared the effectiveness of the proposed method with the LBP and the Gabor filter bank based methods. For the LBP and the Gabor filter bank based methods, full image evaluation was performed with IBD No. 31 (LBP, 128, KNN) and IBD No. 25 (G, 128, KNN), respectively. Fig. 13 depicts the performance comparisons of the three methods using receiver operating characteristic (ROC) and precision-recall plots. ROC and precision-recall plots were generated from the precision, recall, and specificity values obtained by varying the abnormal block thresholds from 1 through 20. Fig. 13 clearly shows the superiority of the proposed method over LBP and Gabor filter bank based methods for WCE image evaluation. According to Fig. 13, ROC and precision-recall plots of the proposed method are better compared to those of the LBP and Gabor filter bank based methods. Moreover from the precision-recall plot of the proposed method in Fig. 13(b), it can be seen that the imbalanced nature of our dataset has not caused a major drop in the performance.
Fig. 13.
Performance comparison of the proposed method with LBP and Gabor filter banks based methods for WCE abnormal detection using a) ROC, and b) precision-recall plots.
4.2. Experiments with Colonoscopy Videos
In this experiment, seven real colonoscopy videos were used to classify images showing polyps from those not showing any polyp. All colonoscopy videos were generated from the Fujinon scope having an image resolution of 720×480 pixels. The same experimental setup used for the WCE experiment was used in this experiment as well (see Section 4.1). Our domain expert verified the ground truth of colonoscopy images. Columns 2 and 3 of Table 9 provide the numbers of images used for the two training stages and the detecting stage. The number of image regions extracted for texton generation (i.e., Training stage 1) and the number of image blocks extracted for IBD generation (i.e., training stage 2) are given in Columns 4–6 in Table 9. The image regions and the image blocks were extracted from the training images (Column 2 in Table 9). We included as many variations as possible of each colonoscopy texture as mentioned in Section 3.1 in the extracted regions listed in Column 4 of Table 9. For instance, the 80 examples for the polyp texture include eight different textures. We ensured that extracted image blocks for IBD generation listed in Column 5 and Column 6 of Table 9 contain as many variations as possible of abnormal and normal areas in the colonoscopy training set as mentioned in Section 3.3. Similar to the WCE experiment, we used the same image set for both training stage 1 and 2. Using the image regions in Training stage 1 and the image blocks in Training stage 2, we built the 36 IBDs listed in Table 3 for colonoscopy abnormal image classification.
Table 9.
Colonoscopy images, image regions, and image blocks.
| Texture | # of Images for Training Stage 1 and Stage 2 |
# of Images for Detecting Stage |
# of Image Regions for Training Stage 1 |
# of Image Blocks (64 by 64) for Training Stage 2 |
# of Image Blocks (128 by 128) for Training Stage 2 |
|---|---|---|---|---|---|
| Polyp | 250 | 100 | 80 | 500 | 250 |
| 800 | 400 | 400 | 1600 | 800 | |
| Normal | (100 from each of 8 normal textures) |
(50 from each of 8 normal textures) |
(50 from each of 8 normal textures) |
(200 from each of 8 normal textures) |
(100 from each of 8 normal textures) |
4.2.1. Colonoscopy IBD Performance Analysis
Performance of each IBD with polyp texture as the positive class is summarized in Table 10. A graphical comparison of the average recall and specificity values of each IBD is shown in Fig. 14. When analyzing Table 10 and Fig. 14, we see that both recall and specificity values are very high for most of the IBDs. We have obtained the best recall value of 98.2% with a very high specificity value of 95.1% for IBD No. 34 (LM-LBP, 128, KNN). In these results, we see almost all trends we observed with the WCE IBDs. LM and LBP filter bank based IBDs (i.e., IBD No. 19 (LM-LBP, 128, KNN) and IBD No. 31 (LM-LBP, 128, KNN)) distinguish abnormal and normal image blocks with very high recall values (96.0% and 92.5%) and specificity values (94.0% and 93.9%). It is clear that when the LM filter bank and a set of LBPs are combined, we see improved recall and the specificity values for colonoscopy abnormal region detection as well. Hence, we used IBD No. 34 (LM-LBP, 128, KNN) in the detecting stage of colonoscopy polyp image classification. In the missed polyp image blocks by IBD No. 34 (LM-LBP, 128, KNN), the polyp region is either partially blurred or partially covered by specular reflection. The partial blurriness and the specular reflections wipe out some of the polyp texture.
Table 10.
Performance of 36 colonoscopy IBDs with polyp texture as the positive class.
| IBD No. | P | R | S | A | IBD No. | P | R | S | A | IBD No. | P | R | S | A |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 76.4 | 91.8 | 90.4 | 90.8 | 13 | 73.0 | 81.3 | 89.9 | 87.7 | 25 | 82.0 | 87.9 | 91.9 | 90.7 |
| 2 | 68.0 | 39.6 | 93.7 | 80.1 | 14 | 74.4 | 60.9 | 92.9 | 84.9 | 26 | 81.8 | 74.4 | 93.0 | 87.5 |
| 3 | 55.6 | 54.4 | 85.3 | 77.5 | 15 | 67.1 | 66.2 | 89.1 | 83.3 | 27 | 71.8 | 70.6 | 88.2 | 83.0 |
| 4 | 66.1 | 83.8 | 85.5 | 85.1 | 16 | 89.2 | 93.6 | 95.2 | 94.7 | 28 | 75.3 | 87.3 | 87.9 | 87.7 |
| 5 | 75.1 | 44.2 | 95.1 | 82.2 | 17 | 84.0 | 82.1 | 93.4 | 90.0 | 29 | 78.5 | 61.3 | 92.9 | 83.5 |
| 6 | 54.9 | 55.4 | 84.7 | 77.3 | 18 | 75.1 | 75.2 | 89.5 | 85.2 | 30 | 70.0 | 69.6 | 87.4 | 82.1 |
| 7 | 71.6 | 79.6 | 89.4 | 86.9 | 19 | 87.1 | 96.0 | 94.0 | 94.6 | 31 | 81.6 | 92.5 | 93.9 | 92.8 |
| 8 | 73.6 | 53.3 | 93.6 | 83.4 | 20 | 73.5 | 63.9 | 90.3 | 82.4 | 32 | 83.6 | 82.3 | 93.2 | 90.0 |
| 9 | 53.1 | 59.2 | 82.4 | 76.5 | 21 | 69.8 | 72.7 | 86.6 | 82.5 | 33 | 69.1 | 69.9 | 89.4 | 84.5 |
| 10 | 66.0 | 83.0 | 85.6 | 84.9 | 22 | 82.1 | 93.5 | 91.4 | 92.0 | 34 | 89.4 | 98.2 | 95.1 | 96.0 |
| 11 | 78.4 | 45.8 | 95.8 | 83.2 | 23 | 83.6 | 71.9 | 94.0 | 87.5 | 35 | 89.8 | 82.3 | 96.1 | 92.0 |
| 12 | 60.5 | 60.8 | 86.6 | 80.1 | 24 | 77.1 | 73.5 | 90.8 | 85.6 | 36 | 73.4 | 77.0 | 88.2 | 84.9 |
Fig. 14.
Comparison of a) recall and b) specificity values of polyp texture for each IBD. Specificity of value of polyp texture is the same as the recall value for normal texture.
4.2.2. Colonoscopy Abnormal Image Detection Analysis
We evaluated the performance of colonoscopy polyp detecting stage using our IBD No. 34 (LM-LBP, 128, KNN). A data set of 500 colonoscopy images were used for the polyp detection in which 100 of them were polyp and 400 of them were normal as given in Column 3 of Table 9. In 100 polyp images there were 8 distinct types of polyps. As in WCE abnormal detection, the dataset was intentionally made to be imbalanced to resemble real world scenario in colonoscopy where the number of abnormal and normal images are imbalanced. Each image was evaluated using IBD No. 34 (LM-LBP, 128, KNN) as described in Section 3.4. We experimented with abnormal block thresholds (see Section 3.4) of 1 through 20, and found that the threshold of 4 provides the best performance. Table 11 shows performance of colonoscopy polyp abnormal image detection with abnormal block thresholds of 3, 4 and 5. The confusion matrix in Table 12 outlines the number of detections of polyp and normal images. In colonoscopy abnormal detection, we have only considered polyp as the abnormal class. Hence, our colonoscopy abnormal detection is a binary classification problem having polyp and normal as classes. Thus Table 12 shows the confusion matrix of the binary classification of polyp and normal classes. We obtained the best recall value of 91.0% and the best specificity value of 90.8% when the abnormal block threshold is 4. Confusion matrix in Table 12 shows the number of missed polyps and normal images by the proposed mehtod. These recall and specificity values prove that our method works for colonoscopy abnormal image detection as well.
Table 11.
Evaluation of colonoscopy abnormal image detection with IBD No. 34 (LM-LBP, 128, KNN).
| Abnormal Block Threshold | P | R | S | A |
|---|---|---|---|---|
| 3 | 50.5 | 95.0 | 76.8 | 80.4 |
| 4 | 71.1 | 91.0 | 90.8 | 90.8 |
| 5 | 79.2 | 84.0 | 94.5 | 92.4 |
Table 12.
Confusion matrix of colonoscopy abnormal image detection for the abnormal block threshold = 4.
| Actual/Predicted | Polyp | Normal |
|---|---|---|
| Polyp | 91 | 9 |
| Normal | 39 | 361 |
Table 13.
Analysis of various filter combinations within the LM-LBP filter bank for WCE and colonoscopy
| Filter Combination | Average Performance for WCE | Average Performance for Colonoscopy | ||||||
|---|---|---|---|---|---|---|---|---|
| P | R | S | A | P | R | S | A | |
| 36 Gaussian filters + four LBPs | 83.9 | 81.7 | 95.2 | 94.8 | 85.2 | 96.1 | 92.9 | 94.0 |
| 18 first-order derivative of Gaussian filters + four LBPs | 78.8 | 80.5 | 94.4 | 93.7 | 85.8 | 95.7 | 93.3 | 94.0 |
| 18 second-order derivative of Gaussian filters + four LBPs | 74.8 | 77.4 | 94.0 | 93.0 | 84.9 | 95.1 | 92.8 | 93.5 |
| 8 Laplacian of Gaussian filters + four LBPs | 69.8 | 67.7 | 91.0 | 90.7 | 83.6 | 93.1 | 92.3 | 92.6 |
| 4 other Gaussian filters + four LBPs | 70.2 | 69.3 | 91.6 | 91.1 | 81.4 | 92.4 | 91.1 | 91.5 |
| All 48 filters + four LBPs | 89.0 | 91.0 | 97.3 | 96.9 | 89.4 | 98.2 | 95.1 | 96.0 |
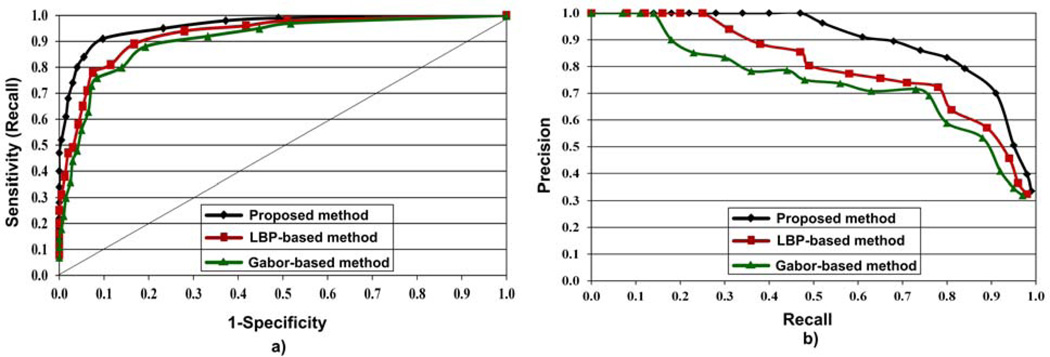
As in the WCE experiment, the performance of the proposed method for colorectal polyp detection was compared with the LBP and Gabor filter bank based methods. IBD No. 31 (LBP, 128, KNN) and IBD No. 25 (G, 128, KNN) were used for colonoscopy full image evaluation for the LBP and Gabor filter bank based methods, respectively. The performances of the three methods are presented as ROC and precision-recall plots in Fig. 15. The resulting precision, recall and specificity values for the abnormal block thresholds from 1 through 20 were used to generate ROC and precision-recall plots. Fig. 15 clearly shows that the performance of the proposed method is better compared to the LBP and the Gabor filter based methods. According to Fig. 15, the proposed method has provided better ROC and precision-recall plots compared to the LBP and Gabor filter bank based methods. Similar to WCE abnormal detection, from the precision-recall graph of the proposed in Fig. 15(b), it can be seen that the imbalanced nature our dataset has not caused a major drop in the colonoscopy abnormal detection performance as well.
Fig. 15.
Performance comparison of the proposed method with LBP and Gabor filter bank based methods for colonoscopy abnormal detection using a) ROC, and b) precision-recall plots.
4.3. Analysis of the Filters in the LM-LBP Filter Bank and the Size of the Texton Dictionary
This section summarizes the experimental study we conducted to show the soundness of the selection of filters in the LM-LBP filter bank and the size of the texton dictionary.
4.3.1. Analysis of filters in the LM-LBP Filter Bank
We tested all filter combinations of LM filter bank with four LBPs to find the best combination of filters for WCE and colonoscopy abnormal image detection. It was found that the best abnormal detection can be obtained when all 48 filters in the LM filter bank and four LBPs are combined. The same trend was seen for both WCE and colonoscopy. We conducted this experiment by creating an IBD with 128×128 image blocks and the KNN classifier for each filter combination. Table 13 outlines the average performance of each of those IBDs for both WCE and colonoscopy. Table 13 clearly shows the necessity to use all filters in the LM filter bank and the four LBPs. In terms of the recall, full LM-LBP filter bank is 10% to 25% and 2% to 6% better compared to other filter combinations for WCE and colonoscopy, respectively. In terms of the specificity, it is 2% to 6% and 2% to 4% better for WCE and colonoscopy, respectively.
Table 14.
Top five most important filters in the LM-LBP filter bank (See Section 3.2.1 for details on each filter)
| WCE | Colonoscopy | ||
|---|---|---|---|
| Filter No. | Description | Filter No. | Description |
| 21 | Second derivative Gaussian filter with and orientation = 60° | 7 | First derivative Gaussian filter with σ = 2 and orientation = 0° |
| 11 | First derivative Gaussian filter with σ = 2 and orientation = 120° | 24 | Second derivative Gaussian filter with and orientation = 150° |
| 42 | Laplacian of Gaussian filter with σ = 6 | 50 | Local binary pattern with P = 8 and R=1 |
| 2 | First derivative Gaussian filter with and orientation = 60° | 30 | Second derivative Gaussian filter with σ = 2 and orientation = 150° |
| 51 | Local binary pattern with P = 12 and R=1.5 | 42 | Laplacian of Gaussian filter with σ = 6 |
We analyzed the importance of each individual filter in the LM-LBP filter bank towards abnormal image detection. For that we removed each individual filter from the 52 filters in the filter bank and created IBDs with the remaining 51 filters. IBDs are created using 128×128 image blocks and the KNN classifier. To analyze the performance we introduce a new measure called ‘g-mean’. The g-mean measure is defined as given in Equation (5).
| (5) |
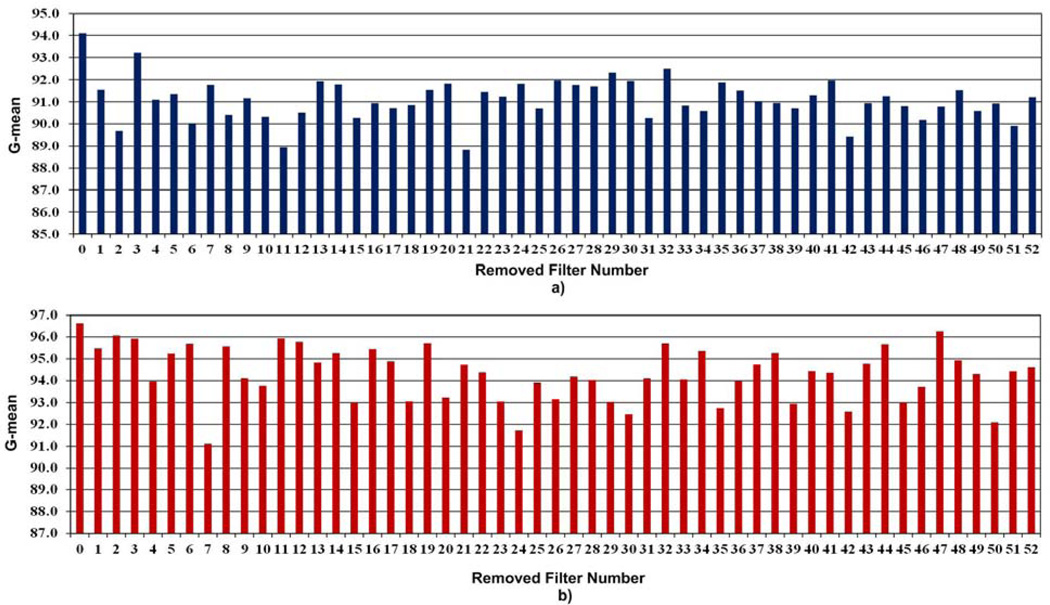
The g-mean value gives a combined performance of recall and specificity. Larger g-mean values imply larger values for both recall and specificity. Column graphs that are shown in Fig. 16 demonstrate the performance in terms of g-mean measure of each of those IBDs for both WCE and colonoscopy. In Fig. 16, lower g-mean indicates a greater significance of the filter to the filter bank. In Fig. 16(a) and 16(b), the column 0 represents the case with no filter is removed. Table 14 summarizes the top 5 most important filters in the LM-LBP filter bank for WCE and colonoscopy abnormal image detection.
Fig. 16.
Analysis of individual filters in the LM-LBP filter bank for a) WCE and b) colonoscopy. X-axis represents the number of the removed filter. Zero represents the case where no filter is removed (i.e., full LM-LBP filter bank).
Several variants of LBPs can be found in the literature. We experimentally verified our selection of LBPs over the other variations of LBPs. Our LBPs are based on the original LBP operator proposed in [12]. We compared the performance of the original LBPs with rotationally invariant LBPs [12], local ternary patterns (LTP) [34], and local derivative patterns (LDP) [35]. We tested each LBP variant by creating IBDs with 128×128 image blocks and the KNN classifier. A bank of four rotationally invariant LBPs with (P = 4, R = 1.0), (P = 8, R = 1.0), (P = 12, R = 1.5), and (P = 16, R = 2.0) was implemented as put forward in [12]. Our LTPs are based on the proposed method in [34] where we created a bank of four LTPs with threshold values of 5, 10, 15 and 20 [34], respectively. Each threshold provides two LTPs as stated in [34] and hence our LTP filter bank has 8 LTPs. A bank of three LDPs with order 1, 2, and 3 was created for the testing using the method proposed in [35]. For that we adopted the code provided in [36]. All three banks of rotationally invariant LBPs, LTPs and LDPs are corresponding to the best performance provided on our dataset. The outcomes from the experiment are presented in Table 15. Table 15 proves our selection of bank of four original LBPs over the other variants of LBPs. The performances of the rotationally invariant LBPs and LTPs are somewhat closer to the original LBPs whereas the performance of LDP is relatively low for both WCE and colonoscopy. Further, the simplicity of original LBPs compared to other three LBP variants helps to achieve a slightly lower computational cost in the proposed method.
Table 15.
Comparison of the original LBPs with other variations of LBPs: rotationally invariant LBP, LTP, and LDP.
| LBP Variant | Average Performance for WCE | Average Performance for Colonoscopy | ||||||
|---|---|---|---|---|---|---|---|---|
| P | R | S | A | P | R | S | A | |
| Original local binary pattern (LBP) | 81.1 | 83.1 | 95.3 | 94.7 | 81.6 | 92.5 | 93.9 | 92.8 |
| Rotationally invariant LBP | 78.1 | 75.7 | 93.7 | 93.5 | 81.1 | 91.1 | 93.2 | 91.5 |
| Local ternary pattern (LTP) | 75.2 | 77.5 | 86.1 | 83.7 | 78.8 | 87.8 | 88.2 | 89.8 |
| Local derivative pattern (LDP) | 57.8 | 55.6 | 82.3 | 76.4 | 68.7 | 71.6 | 84.2 | 81.7 |
4.3.2. Analysis of the Size of the Texton Dictionary
The size of the texton dictionary plays an important role in delivering high abnormal detection performance while keeping the computational cost to a minimum. The number of textons in the texton dictionary is the number of bins in the texton histogram (see Section 3.3.1). As a result when the number of textons in the texton dictionary increases the computational cost also increases. Therefore, we must ensure that the number of textons created for each texture is not that high. As mentioned in Section 3.3.2, we determine the number of textons for each texture using the k-means clustering algorithm. The ‘k’ value (i.e., number of cluster centers) used for k-means clustering is the number of textons from each texture. We investigated the effect of the k value using two separate experiments for abnormal and normal textures. To verify the selection of the ‘k ‘value (i.e., number of textons) for abnormal textures, we conducted one experiment by varying the ‘k’ value for abnormal textures while fixing the ‘k’ value to 5 (in the second stage) (see Section 3.2.3) for normal textures. In the other experiment, we varied the ‘k’ value (in the second stage) for normal textures while keeping the ‘k’ value for abnormal textures to 20. First we generated texton dictionaries for each different ‘k’ value. Using the texton dictionaries we created IBDs with the LM-LBP filter bank, 128×128 image blocks and the KNN classifier. We calculated the g-mean for each of the IBD for both WCE and colonoscopy. Table 16 and 17 show the results of the two experiments. When Table 16 is analyzed, no major improvement in the g-mean value can be seen for ‘k’ values above 20 for both WCE and colonoscopy. Even we can see a drop in the g-mean after k = 40 for WCE and after k = 30 for colonoscopy. Hence we decided to use a ‘k’ value of 20 in the k-means clustering (i.e., number of textons) for each abnormal texture in an attempt to reduce the execution time. Table 17 shows that ‘k’ value for each normal texture can be as low as 5. For the ‘k’ values above 5, we do not see any significant improvement in the g-mean. This is very essential as the size of the texton dictionary gets very big when the ‘k’ value for normal textures increases. For instance, when k = 20 for normal textures, it produces a texton dictionary of size 290 for WCE compared to a texton dictionary of size 130. This would increase the execution time more than twice of the execution time for the texton dictionary with 130 textons.
Table 16.
Effect of the ‘k’ value of the k-means clustering for abnormal textures (i.e., number of textons for each abnormal texture in the texton dictionary). The ‘k’ value is fixed to 5 for normal textures.
| k (number of textons for each abnormal texture) | Size of the texton dictionary (Number of bins in the texton histogram) |
G-mean for WCE | G-mean for Colonoscopy |
|---|---|---|---|
| 10 | WCE-90, Colonoscopy-50 | 89.6 | 94.4 |
| 20 | WCE-130, Colonoscopy-60 | 94.1 | 96.6 |
| 30 | WCE-170, Colonoscopy-70 | 94.4 | 97.3 |
| 40 | WCE-210, Colonoscopy-80 | 94.8 | 96.5 |
| 50 | WCE-250, Colonoscopy-90 | 93.2 | 95.3 |
| 60 | WCE-290, Colonoscopy-100 | 91.1 | 93.8 |
Table 17.
Effect of the k value of the k-means clustering for normal textures (number of textons for each normal texture in the texton dictionary. The ‘k’ value is fixed to 20 for abnormal textures.
| k (number of textons for each normal texture) | Size of the texton dictionary (Number of bins in the texton histogram) |
G-mean for WCE | G-mean for Colonoscopy |
|---|---|---|---|
| 5 | WCE-130, Colonoscopy-60 | 94.1 | 96.6 |
| 10 | WCE-180, Colonoscopy-100 | 94.2 | 97.0 |
| 15 | WCE-230, Colonoscopy-140 | 95.0 | 96.8 |
| 20 | WCE-280, Colonoscopy-180 | 95.6 | 96.7 |
5. Concluding Remarks
Distinguishing abnormal images such as erythema, blood (bleeding), ulcer, erosion, and polyp from normal images in endoscopy videos is a major concern when a gastroenterologist reviews the videos. It is reported that the reviewing takes very long time, which limits its general application and incurs considerable amount of healthcare costs. In this paper, we propose a novel multi-texture analysis method for multiple abnormality detection in endoscopy videos. It is apparent that color and shape based approaches provide very limited support in detecting abnormalities such as erythema, ulcer, erosion, or polyp since they do not possess any unique color range or common shape information. When endoscopy images are closely analyzed, we notice that each abnormality has a unique texture that can be distinguishable from normal ones using an advanced texture analysis method. We found that variety of normal textures can be divided into a handful of textures. We represent these textures as “textons” and propose a new filter bank called LM-LBP to generate the textons. We created a model called Image Block Dictionary (IBD) including a trained KNN classifier, which can classify an image block based on its distribution of textons (i.e., texton histogram). To classify an image, we divide it into blocks. These image blocks are classified as either abnormal or normal blocks using the generated IBD. If the abnormal block count is greater than a certain threshold value, the image is classified as an abnormal image. Otherwise, it is classified as a normal image. The proposed method was tested with two of the most popular endoscopy methods: wireless capsule endoscopy (WCE) and colonoscopy. We found that our IBD (LM-LBP,128, KNN) provides the best classification accuracy for both WCE and colonoscopy videos used in the study. The IBD (LM-LBP, 128, KNN) based abnormal image detection achieves 92.0% recall and 91.8% specificity. Corresponding recall and specificity for colonoscopy videos are 91% and 90.8%, respectively.
The advantage of the proposed method over existing methods is that one scheme finds multiple abnormalities. The LM-LBP filter bank has the ability to generate a unique set of textons for each chosen texture. In this study we have introduced the concept of filter banks for multiple abnormality detection in endoscopy. To verify this, we have presented a broad experimental study which proves the applicability of the proposed LM-LBP filter bank over the other existing filter banks and the related methods. In the future, we will improve the accuracy for ulcer-erosion detection in WCE videos, and detect more abnormalities such as angioectasia (in WCE) and ulcers (in colonoscopy). Moreover improving the computational cost is one of our major future works. The computational cost of the method was not a focus in this work. Most computationally intense part of the proposed method is the convolution of image blocks with the LM-LBP filter bank. We aim to speed up the process by using GPU computing capabilities.
Acknowledgements
This work is partially supported by NSF STTR-Grant No. 0740596, 0956847, National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK DK083745), and the Mayo Clinic. Any opinions, findings, conclusions, or recommendations expressed in this paper are those of authors. They do not necessarily reflect the views of the funding agencies. Johnny Wong, Wallapak Tavanapong, and JungHwan Oh hold positions at EndoMetric Corporation, Ames, IA 50014, U.S.A, a for profit company that markets endoscopy-related software. Johnny Wong, Wallapak Tavanapong, JungHwan Oh, Piet de Groen, and Mayo Clinic own stocks in EndoMetric. Piet de Groen, Johnny Wong, Wallapak Tavanapong, and JungHwan Oh have received royalty payments from EndoMetric.
Biographies

Ruwan Nawarthna received the B.S. degree in Computer Science from the University of Peradeniya, Sri Lanka in 2005, and M.S. and Ph.D. Degrees in Computer Science from the University of North Texas in 2011 and 2013, respectively. Currently, he is working as a research assistant at the Multimedia Information Laboratory, University of North Texas. His research interests include endoscopy image analysis, camera motion analysis and multi-class image texture analysis

JungHwan Oh received M.S. and Ph.D. degrees in computer science from the University of Central Florida in 1997 and 2000 respectively. Dr. Oh worked with the Department of Computer Science and Engineering at University of Texas, Arlington from 2001 to 2006. In 2006, he joined the Department of Computer Science and Engineering at University of North Texas. He has established a medical video research with the collaboration of Iowa State University and Mayo Clinic in 2003. This research focuses on the medical video (colonoscopy, endoscopy, broncoscopy, minimal access surgery, etc.) analyses and mining. As results of these researches, a number of proposals have been granted from National Science Foundation and National Institute of Health. He has organized a number of workshops and Special Sessions on multimedia data processing, and served as a member of program committee and an editor of various international conferences on imaging science, systems, and information technology. He is the author or coauthor of more than 90 journal articles, book chapters, and conference papers.

Jayantha Muthukudage received his M.S. and Ph.D. degrees in Computer Science from the University of North Texas in 2012 and 2013 respectively and his Bachelor’s degree in Computer Science from the University of Peradeniya, Sri Lanka in 2006. His major research interests are endoscopy image analysis, object detection and quality analysis of endoscopy videos. Currently, he is working as a research assistant at the Multimedia Information Laboratory, University of North Texas.

Wallapak Tavanapong received the B.S. degree in Computer Science from Thammasat University, Thailand, in 1992 and the M.S. and Ph.D. degrees in Computer Science from the University of Central Florida in 1995 and 1999, respectively. She joined Iowa State University in 1999 and is currently an Associate Professor and Associate Chair of Computer Science. She is a cofounder and a Chief Technology Officer of EndoMetric Corp., a software company that offers computer-aided technology for endoscopy procedures. She received a National Science Foundation Career grant, the 2006 American College of Gastroenterology Governors Award for Excellence in Clinical Research for “The Best Scientific Paper”, and a US patent on Colonoscopy Video Processing for Quality Metrics Determination. Her current research interests include content-based analysis on multimedia data for healthcare and social media, high performance multimedia computing, medical informatics, and databases. National Science Foundation, Agency for Healthcare Research and Quality, the National Institute of Diabetes and Digestive and Kidney Diseases, Mayo Clinic Rochester, Iowa Department of Economic Development, and EndoMetric Corp., supported her research.

Johnny Wong is Professor & Interim Chair of the Computer Science Department at Iowa State University (ISU). His research interests include Software Systems & Networking, Security & Privacy, and Medical Informatics. Most of his research projects are funded by government agencies and industries, including NSF, DoD, HHS, NIH, Mayo, etc. He is the President/CEO of a startup company EndoMetric Corporation, with software products for Medical Informatics. He is a co-director of the Smart Home Lab in the Department of Computer Science at ISU. He has served as a member of program committee of various international conferences on intelligent & software systems and networking. He was the Program co-Chair of the COMPSAC 2006 and General co-Chair of the COMPSAC 2008 conference, the IEEE Signature Conference on Computers, Software, and Applications. He is a member of the ACM and IEEE Computer Society. He has published over 150 papers in peer reviewed journals and conferences.

Piet C. de Groen, M.D. is a Consultant in Gastroenterology and Hepatology at Mayo Clinic, Professor of Medicine at Mayo Clinic College of Medicine and former Program Director of the Mayo Clinic/IBM Computational Biology Collaboration. He is an NIH-funded clinical investigator and international expert in medical informatics, primary liver cancers and colonoscopy. His endoscopic research is focused on measuring what happens during colonoscopy. Together with collaborators at Iowa State University and the University of North Texas he has created a first-of-a-kind new software system that automatically captures and analyzes “inside-the-patient” video information of medical procedures performed via endoscopy. At present he is studying patient- and endoscopist-specific features, including quality of the colon preparation and effort of the endoscopist to visualize mucosa, remove remaining fecal material and adequately distend the colon. The ultimate goal of his endoscopic research related to colonoscopy is a fully automated quality control system that provides real-time feedback to the endoscopist and insures individualized healthcare by virtually guaranteeing a high quality examination for each patient. Another goal is to extend the technology to other organs examined using endoscopy equipment such as esophagus, stomach, small bowel, bronchi, bladder and joints and combine the information obtained from the optical signal with information derived from molecular tests and other imaging modalities.

Shou-jiang Tang, M.D. graduated from the West China University of Medical Sciences in 1992. After he finished his post-graduate medical training at the Cleveland Clinic Foundation (medical residency), UCLA (gastroenterology fellowship), and University of Toronto (advanced endoscopy fellowship), he joined University of Texas Southwestern Medical Center in Dallas in 2004 as an assistant professor. His research interests include endoscopic research and innovations. Currently, he is the interim Chief, Division of Division of Digestive Diseases, Gastroenterology Fellowship Director, and Director of Endoscopic Research at the University of Mississippi Medical Center. He has published extensively in peer-reviewed clinical journals and book chapters. He holds and applied five patents in endoscopic devices. He is one of CME editors for Gastrointestinal Endoscopy journal and he has served on several American Society of Gastrointestinal Endoscopy Committees (ASGE) committees. He was a recipient of the ASGE Don Wilson Award and served as an ASGE Ambassador to Taiwan in 2012.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Ruwan Nawarathna, Email: RuwanNawarathna@my.unt.edu.
JungHwan Oh, Email: Junghwan.Oh@unt.edu.
Jayantha Muthukudage, Email: MuthukudageKumara@my.unt.edu.
Wallapak Tavanapong, Email: tavanapo@cs.iastate.edu.
Johnny Wong, Email: wong@cs.iastate.edu.
References
- 1.Adler DG, Gostout CJ. State of art: Wireless Capsule Endoscopy. J. Hospital Physician. 2003;39(2):14–17. [Google Scholar]
- 2.Coimbra M, Mackiewicz M, Fisher M, Jamieson C, Scares J, Cunha JPS. Computer vision tools for capsule endoscopy exam analysis. EURASIP Newsletter. 2007;18(2):1–19. [Google Scholar]
- 3.Eliakim R. Video Capsule Endoscopy of the Small Bowel. Current Opinion in Gastroenterology. 2008;24(2):159–163. doi: 10.1097/MOG.0b013e3282f3d946. [DOI] [PubMed] [Google Scholar]
- 4.American Cancer Society, Colorectal Cancer Facts and Figures. American Cancer Society Special Edition. 2005:1–20. [Google Scholar]
- 5.Gonzalez RC, Woods RE. Digital Image Processing. third ed. New Jersy: Prentice Hall; 2008. pp. 776–778. [Google Scholar]
- 6.Julesz B. Texton, the elements of texture perception, and their interactions. Nature. 1981;290(5802):91–97. doi: 10.1038/290091a0. [DOI] [PubMed] [Google Scholar]
- 7.Leung T, Malik J. Representing and recognizing the visual appearance of materials using three-dimensional textons. Computer Vision. 2001;43(1):29–44. [Google Scholar]
- 8.Varma M, Zisserman A. Classifying images of materials: achieving viewpoint and illumination independence. Proceedings of European Conference in Computer Vision. 2002:255–271. [Google Scholar]
- 9.Varma M, Zisserman A. A Statistical approach to texture classification from single images. Comp. Vision: Special Issue on Texture Analysis and Synthesis. 2005;62(1–2):61–81. [Google Scholar]
- 10.Lei Z, Li SZ, Chu Rufeng, Zhu Xiangxin. Face recognition with local gabor textons. Lecture Notes in Computer Science. 2007;4642:49–57. [Google Scholar]
- 11.Liu GH, Yanga J-Y. Image retrieval based on the texton co-occurrence matrix. Pattern Recognition. 2008;41(12):3521–3527. [Google Scholar]
- 12.Ojala T, Pietikainen M, Harwood D. A comparative study of texture measures with classification based on featured distributions. Pattern Recognition. 1996;29(1):51–59. [Google Scholar]
- 13.Duda RO, Hart PE, Stork DG. Pattern Classication. New York: John Wiley and Sons; 2001. pp. 517–581. [Google Scholar]
- 14.Coimbra MT, Cunha JPS. MPEG-7 visual descriptors-contributions for automated feature extraction in Capsule Endoscopy. IEEE Trans. on Circuits and Systems for Video Technology. 2006;16(5):628–637. [Google Scholar]
- 15.Penna B, Tilloy T, Grangettoz M, Magli E, Olmo G. A technique for blood detection in Wireless Capsule Endoscopy images; Proceedings of 17th European Signal Processing Conference (EUSIPCO'09); August 2009; Glasgow, Scotland. pp. 1864–1868. [Google Scholar]
- 16.Li B, Meng MQ-H. Texture analysis for ulcer detection in Capsule Endoscopy images. Image and Vision Computing. 2009;27(9):1336–1342. [Google Scholar]
- 17.Li B, Meng MQ-H, Xu L. A comparative study of shape features for polyp detection in Wireless Capsule Endoscopy images; Proceedings of Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC’09); September 2009; Minneapolis, MN, USA. pp. 373–3734. [DOI] [PubMed] [Google Scholar]
- 18.Li B, Meng MQ-H. Small bowel tumor detection for Wireless Capsule Endoscopy images using textural features and support vector machine; Proceedings of IEEE/RSJ International Conference in Intelligent Robots and Systems (IROS’09); October 2009; St. Louis, MO, USA. pp. 498–503. [Google Scholar]
- 19.Karargyris A, Bourbakis N. Identification of ulcers in Wireless Capsule Endoscopy videos; Proceedings of IEEE International Symposium in Biomedical Imaging: From Nano to Macro (ISBI’09); June–July 2009; Boston, Massachusetts, USA. pp. 554–557. [Google Scholar]
- 20.Karargyris A, Bourbakis N. Identification of polyps in Wireless Capsule Endoscopy videos using Log Gabor Filters; Proceedings of IEEE/NIH Life Science Systems and Applications Workshop (LiSSA’09); April 2009; Bethesda, Maryland, USA. pp. 143–147. [Google Scholar]
- 21.Barbosa DJC, Ramos J, Correia JH, Lima CS. Automatic detection of small bowel tumors in Capsule Endoscopy based on color curvelet covariance statistical texture descriptors; Proceedings of Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC’09); September 2009; Minneapolis, MN, USA. pp. 6683–6686. [DOI] [PubMed] [Google Scholar]
- 22.Hwang S. Bag-of-Visual-Words Approach to Abnormal Image Detection in Wireless Capsule Endoscopy Videos, Advances in Visual Computing. Lecture Notes in Computer Science. 2011;6939:320–327. [Google Scholar]
- 23.Zhang S, Yang W, Wu YL, Yao R, Cheng SD. Abnormal region detection in gastroscopic images by combining classifiers on neighboring patches; International Conference on Machine Learning and Cybernetics (ICMLC’ 09); July 2009; Baoding, Hebei, China. pp. 2374–2379. [Google Scholar]
- 24.Hwang S, Oh J, Lee J, Cao Y, Liu D, Tavanapong W, Wong J, de Groen PC. Automatic measurement of quality metrics for colonoscopy videos; Proceedings of the 13th ACM International Conference on Multimedia (ACM Multimedia’ 2005); November 2005; Hilton, Singapore. pp. 912–921. [Google Scholar]
- 25.Hwang S, Oh J, Tavanapong W, Wong J, de Groen PC. Polyp detection in colonoscopy videos using elliptical shape feature; Proceedings of the IEEE International Conference on Image Processing (ICIP’07); September 2007; San Antonio, TX, USA. pp. 465–468. [Google Scholar]
- 26.Hwang S, Oh J, Tavanapong W, Wong J, de Groen PC. Automatic polyp segmentation for colonoscopy image using Watershed algorithm and ellipse fitting method; Proceedings of SPIE Medical Imaging; February 2007; San Diego, CA, USA. pp. 65141D-1–65141D-12. [Google Scholar]
- 27.Alexandre L, Nobre NN, Casteleiro JC. Color and position versus texture features for endoscopic polyp detection; Proceedings of the International Conference on BioMedical Engineering and Informatics; May 2008; Sanya, China. pp. 38–42. [Google Scholar]
- 28.Cheng DC, Ting WC, Chen YF, Pu Q, Jiang XY. Colorectal polyps detection using texture features and support vector machine; Proceedings of the 3rd international Conference on Advances in Mass Data Analysis of Images and Signals in Medicine, Biotechnology, Chemistry and Food Industry (MDA’08); July 2008; Leipzig, Germany. pp. 62–72. [Google Scholar]
- 29.Park SY, Sargent D, Spofford I, Vosburgh KG, Rahim YA. A colon video analysis framework for polyp detection. IEEE Trans. on Biomedical Engineering. 2012;59(5):1408–1418. doi: 10.1109/TBME.2012.2188397. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Tavanapong W, Wong J, Oh J, de Groen PC. Part-based multi-derivative edge cross-section profiles for polyp detection in colonoscopy. IEEE Journal of Biomedical and Health Informatics. 2013;(99) doi: 10.1109/JBHI.2013.2285230. [DOI] [PubMed] [Google Scholar]
- 31.Ameling S, Wirth S, Paulus D. Methods for polyp detection in colonoscopy video: A review. 14 University of Koblenz-Landau; 2008. [Google Scholar]
- 32.Varma M, Zisserman A. Texture classification: are filter banks necessary?; Proceedings of IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR'03); June 2003; Madison, Wisconsin. pp. 691–698. [Google Scholar]
- 33.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: An update. SIGKDD Explorations. 2009;11(1) [Google Scholar]
- 34.Tan X, Triggs B. Enhanced local texture feature sets for face recognition under difficult lighting conditions. IEEE Transactions on Image Processing. 2010;19(6):1635–1650. doi: 10.1109/TIP.2010.2042645. [DOI] [PubMed] [Google Scholar]
- 35.Zhang B, Gao Y, Zhao S, Liu J. Local derivative pattern versus local binary pattern: face recognition with high-order local pattern descriptor. IEEE Transactions on Image Processing. 2010;19(2):533–544. doi: 10.1109/TIP.2009.2035882. [DOI] [PubMed] [Google Scholar]
- 36.http://www.mathworks.com/matlabcentral/fileexchange/29800-scenesobjects-classificationtoolbox/content/reco_toolbox/html/demo_mlhmsldp_spyr.html.