Abstract
Drug development and preclinical trials are challenging processes and more than 80% to 90% of drug candidates fail to gain approval from the United States Food and Drug Administration. Predictive and efficient tools are required to discover high quality targets and increase the probability of success in the process of new drug development. One such solution to the challenges faced in the development of new drugs and combination therapies is the use of low-cost and experimentally manageable in vivo animal models. Since the 1980’s, scientists have been able to genetically modify the mouse genome by removing or replacing a specific gene, which has improved the identification and validation of target genes of interest. Now genetically engineered mouse models (GEMMs) are widely used and have proved to be a powerful tool in drug discovery processes. This review particularly covers recent fascinating technologies for drug discovery and preclinical trials, targeted transgenesis and RNAi mouse, including application and combination of inducible system. Improvements in technologies and the development of new GEMMs are expected to guide future applications of these models to drug discovery and preclinical trials.
Keywords: Genetically engineered mouse models, Preclinical trials, Drug discovery, Targeted transgenesis, RNAi mouse
INTRODUCTION
Now, more than ever before, drug development and preclinical trials are long and expensive processes. These processes have a high attrition rate and less than 10% of the compounds tested in clinical trials gain approval from the United States Food and Drug Administration (Zambrowicz and Sands, 2003; Sharpless and Depinho, 2006). The cost of launching a new drug exceeds $1 billion. Therefore, technological advances that allow for easy and efficient target identification and validation steps early in the drug discovery process are essential for reducing both the attrition rate and the cost of launching a new drug (Begley and Ellis, 2012). Methods that model the action and predict the efficacy of a drug would improve the overall success rate and provide a reproducible path for drug development.
One solution to the significant challenges faced in the development of new drugs and combination therapies is the use of low-cost, experimentally manageable in vivo animal models (Hansen and Khanna, 2004; Suggitt and Bibby, 2005). Until 1980, mouse models used in drug screening were limited to wild-type or spontaneously mutated models. Many currently used chemotherapeutic agents, including alkylating and other DNA-damaging agents (Esteller et al., 2000), were developed using these animal model systems. Recently, immuno-compromised mice were used for screening anticancer drug candidates (Suggitt and Bibby, 2005; Sharpless and Depinho, 2006). However, large variability existed in the outcomes of these screenings between responses in mice and humans and, as such, these models had a low potential to predict the results of Phase I/II clinical trials (Richmond and Su, 2008).
Genetically engineered mouse models (GEMMs) of human disease could improve drug development (Hansen and Khanna, 2004; Abate-Shen, 2006; Sharpless and Depinho, 2006; Singh and Johnson, 2006; Robles and Varticovski, 2008; Politi and Pao, 2011; Kucherlapati, 2012). Since the 1980’s, several types of GEMMs have been used in drug development and preclinical trials, including transgenic, knockout, and knock-in mouse models (Frese and Tuveson, 2007; Politi and Pao, 2011). Using transgenic technology, extra DNA that encodes the gene of interest can be integrated into the mouse genome; using knockout or knock-in technology, specific regions of the mouse genome can be selectively deleted or modified.
Over the last 30 years, techniques for manipulating the mouse genome and mouse embryos have become increasingly sophisticated. For example, inducible and conditional models, in which genes are turned on or knocked-out temporarily or spatially, have been developed (Lewandoski, 2001; Jonkers and Berns, 2002; Hansen and Khanna, 2004; Frese and Tuveson, 2007; Sun et al., 2007). Knock-in models, in which transgenes are inserted in the specific locus or specific mutations introduced into target genes, have also been developed. The sophisticated mouse models that have been generated through these advances in mouse engineering technologies are useful for drug discovery and preclinical trials.
GEMMs have become an invaluable tool for determining target function, verifying selectivity, and predicting toxicity and drug discovery scientists have developed thousands of mouse models over the last 30 years (Frese and Tuveson, 2007; Kucherlapati, 2012). The primary applications of GEMMs in drug discovery are validating targets, identifying pharmacodynamic markers of drug action, detecting toxicity, and evaluating safety (Bolon, 2004; Sharpless and Depinho, 2006; Politi and Pao, 2011) (Table 1). GEMMs have also been used to validate hypotheses regarding confidence in a new gene’s rationale as a potential therapeutic target.
Table 1.
Examples of GEMMs in drug development processes
| Drug | Drug target (gene) | GEM phenotypes |
|---|---|---|
| Palbociclib (Pfizer) | Cdk4/6 | CDK4 expression is essential for Neu-induced breast tumorigenesis. |
| LEE011 (Novatis) | ||
| Herceptin | ErbB2 (Her2) | Homozygotes for targeted null mutations exhibit cardiac defects. |
| Prozac | Serotonin transporter (5-HTT) | Homozygotes for a targeted null mutation exhibit greatly diminished brain serotonin levels and showed higher anxiety. |
| Paxil | ||
| Zoloft | ||
| Effexor | ||
| Celexa | ||
| Celebrex | Cox2 | KO mice showed reduced inflammation, significant reduction in collagen-induced arthritis, reduced febrile response, and decreased polyp formation |
| Claritin | Histamine H1 receptor (Hrh1) | KO mice showed decreased T- and B-cell response, decreased alertness, and altered activity level. |
| Allegra | ||
| Zyrtec | ||
| Prilosec | H+/K+ ATPase (ATP4A, ATP4B) | Homozygous mutation of ATP4A results in achlorhydria, hypergastrinemia, and abnormalities of the parietal cells. |
| Prevacid | ||
| Takepron | ||
| Pantozol | ||
| Procrit | Erythropoietin (Epo) | Homozygotes for a targeted null mutation exhibit reduced primitive erythropoiesis and die around embryonic day 13 due to impaired fetal liver erythropoiesis |
| Epogen | ||
| Premarin | Estrogen receptor (Esr1, Esr2) | KO mice showed reproductive defects and reduced bone mineral density |
| Evista |
NEW GEMMS IN DRUG DEVELOPMENT AND PRECLINICAL TRIALS
Despite meaningful advantages in the applications of GEMMs, several technical limitations exist in the development of new GEMMs that would be useful in drug development and pre-clinical trials (Beard et al., 2006). In transgenic mice, there is significant heterogeneity among different founders; in knockout mice, the predicted phenotypes are not always readily observed and, occasionally, completely novel or unexpected phenotypes are noted. Another critical problem is the generation period of GEMMs. The timelines required to generate transgenic and knockout mice are quite long and require 12 to 24 months from design to initial experimental cohort.
To address and overcome these obstacles in the generation and application of GEMMs, many mouse geneticists have explored innovative technologies for manipulating the mouse genome and mouse embryos. One such technology is targeted transgenesis, which reduces the cost and accelerates the timeline of GEMM generation (Seibler et al., 2003; Seibler et al., 2005; Beard et al., 2006). This technique also allows the transgene to be stably integrated and expressed in the mouse genome. Using targeted transgenesis, several innovative mouse models have recently become available, including RNA interference (RNAi) and inducible transgenic models, in which specific genes can be regulated (knocked-down or induced) temporally or spatially (Jaisser, 2000; Lewandoski, 2001; Kleinhammer et al., 2011b). These models are expected to strengthen and broaden the application potential of GEMMs in future drug development. Targeted transgenesis will enhance the efficiency and accelerate the generation of GEMMs, and several transgenic and RNAi mouse models will be generated by these methods.
TARGETED TRANSGENESIS IN GEMM GENERATION
Until now, most transgenic mouse models have been based on random transgenesis via pronuclei injection with plasmid-based constructs or transfection of embryonic stem (ES) cells by lentiviral infection (Carmell et al., 2003; Rubinson et al., 2003; Ventura et al., 2004; Coumoul et al., 2005). Pronuclear injection of transgenes results in individual founder lines, each with a unique and irreproducible pattern of transgene expression. Furthermore, random integration of these elements into the genome is problematic because positional-effect variegation can profoundly influence transgene expression and the insertion event itself may disrupt endogenous genes (Beard et al., 2006). Additionally, foreign DNA usually integrates as a linear array, resulting in multiple copies that yield variable levels of gene dosage that may subsequently be silenced by epigenetic mechanisms. As a result, some transgenic mouse models generated by these methods failed to show reproducible gene expression and phenotypes. Lentivirus-based vectors have also been used to introduce transgenes into primary cells, ES cells, and embryos. However, only a fraction of infected cells showed detectable transgene expression at various levels, which indicated that the activity of lentiviral vectors is subject to position-dependent modulation. Overall, previous reports of transgene expression in mice used random transgenesis and required time-consuming and laborious screening of targeted ES cells or mouse lines due to the inherent problem of position-dependent effects on transgene expression.
Targeted transgenesis provides an attractive alternative to random transgenesis by pronuclear injection or lentiviral infection. Transgene integration mediated by recombination at specific sites such as the ROSA26 or COL1A1 loci results in insertion of only a single copy of the transgene (Seibler et al., 2005; Beard et al., 2006; Strathdee et al., 2006). Therefore, this approach avoids the instability issues that are frequently associated with repetitive transgene clusters resulting from pro-nuclear injection or lentiviral infection. Additionally, because transgene insertion occurs at a defined site in the genome, genotyping and identification of homozygous transgenic mice are simple. Single site integration also signifies that targeted transgenes do not independently segregate during breeding. Each transgenic founder is genetically identical, so it is not necessary to breed and characterize multiple lines. Further, targeted transgenesis avoids unpredictable chromosome position effects, which is a challenge of pronuclear and lentiviral transgenics. ROSA26, HPRT, and COL1A1 are mouse genes that are commonly used as host loci for targeted transgene insertion (Beard et al., 2006; Strathdee et al., 2006; Palais et al., 2009).
The ROSA26 locus has become the locus of choice for targeted transgenesis because of the autosomal location, the ubiquitous expression, and the fact that the gene appears not to be subject to epigenetic inactivation (Soriano, 1999; Seibler et al., 2005). The locus was found to be ubiquitously active throughout mouse development in nearly every single cell of the body, so it was identified as being suitable for targeted transgenesis. The open chromatin structure of this locus might promote expression of the transgene or shRNA under the control of an RNA polymerase-dependent promoter. In fact, a single copy of the shRNA transgene inserted into ROSA26 efficiently mediates whole-body gene silencing, with knockdown levels ranging from 80% to 95% in most mouse tissues (Seibler et al., 2005; Seibler et al., 2007).
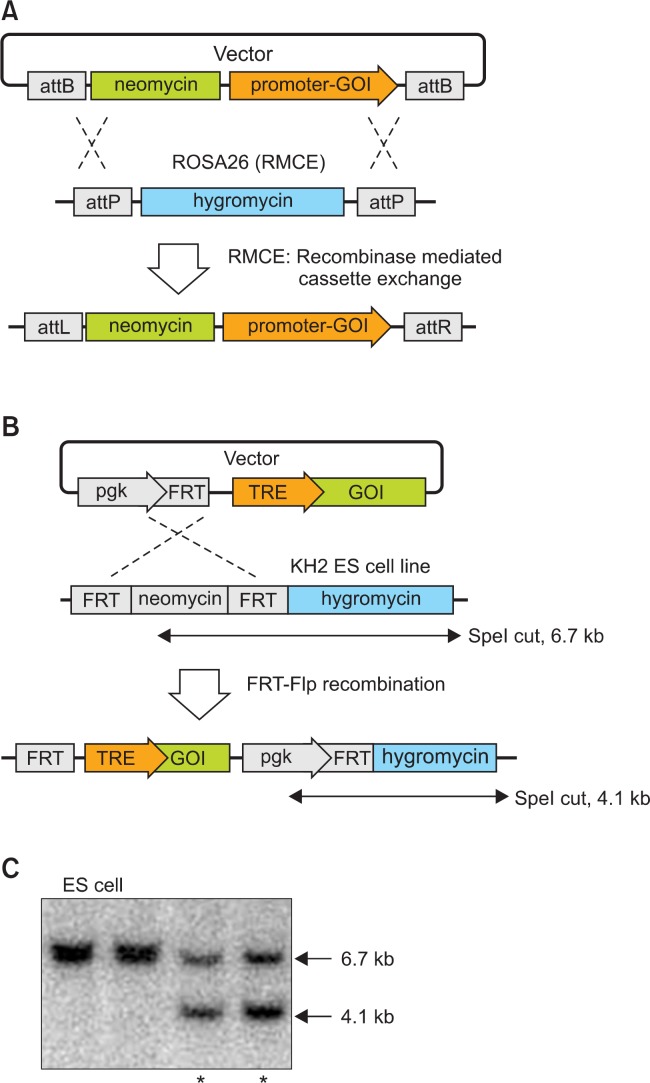
The recombinase mediated cassette exchange (RMCE) method was developed to enhance the efficiency of targeted transgenesis at the ROSA26 locus in ES cells (Seibler et al., 2005) (Fig. 1A). This strategy facilitates the transgene insertion at the ROSA26 locus with high efficiency and consistently achieves greater than 90% correct ES cell clones. Because of the direct cloning of the transgenes into the RMCE vector and the highly efficient integration of the vectors at the ROSA26 locus in ES cells, the RMCE approach readily enables the generation of targeted ES cell clones and transgenic mice. For these reasons, RMCE reduces the cost and time of GEMMs generation.
Fig. 1.
Targeted transgenesis. (A) Targeted transgenesis at the ROSA26 locus by recombinase mediated cassette exchange (RMCE) results in genomic integration of the target gene into the Rosa26.10 allele of mouse ES cells (Kleinhammer et al., 2011b). The hygromycin resistance gene was exchanged by C31 Integrase-mediated recombination of both pairs of attB and attP sites. Recombined ES cell clones were selected by the neomycin resistance gene and identified by PCR or Southern blot analysis. GOI: Gene of interest. (B) Flpe-mediated recombination in the KH2 ES cell line (Beard et al., 2006). The ES cell line contained an FRT-hygromycin-polyA homing cassette downstream of the COL1A1 locus. Upon co-electroporation of the targeting vector and an Flpe transient expression vector, recombination resulted in the loss of the neomycin cassette and the insertion of the gene of interest. This FRT-Flpe recombination also restored and conferred hygromycin resistance. (C) Southern blot analysis to identify recombined ES cell clones. For SpeI digestion, the bands representing alleles before and after recombination were 6.7 and 4.1 kb, respectively, as described in (B). Asterisks indicate recombined ES cell clones.
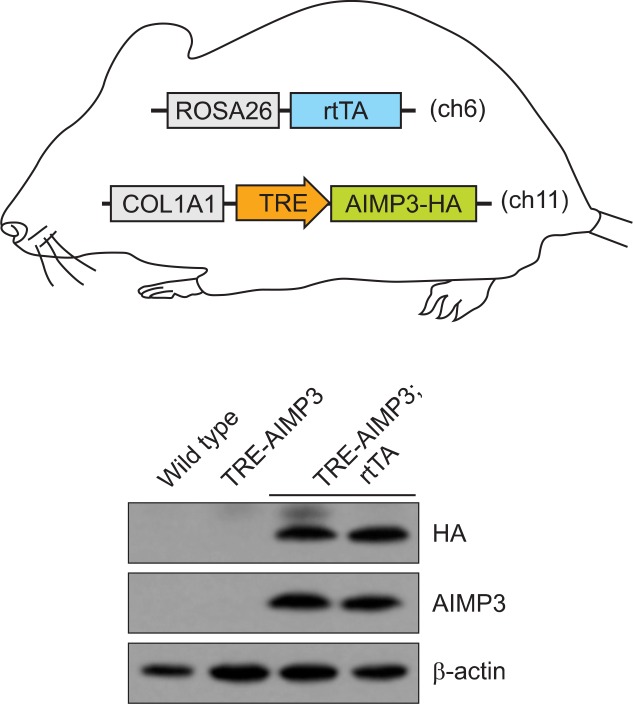
The COL1A1 3’-untranslated region (UTR) is also used for targeted transgenesis (Beard et al., 2006). The COL1A1 locus has been shown to support high transgene expression even in cell types that do not normally express the type 1 collagen gene (McCreath et al., 2000). Beard and colleagues developed a new system of modification of the COL1A1 locus for FRT-mediated targeting. They positioned the FRT target site in a region that lies approximately 500 base pairs downstream of the 3’-UTR and generated the KH2 ES cell line. Using this ES cell line and Flpe recombinase, the target transgene can be integrated into the FRT homing site, which supports the stable expression of the transgene (Fig. 1B, 1C, 2).
Fig. 2.
In vivo expression of a tetracycline-inducible gene in mice. Mice were derived from KH2 ES cells that carry both the R26-rtTA allele and the Flp-in TRE-AIMP3-HA allele (Beard et al., 2006). Doxycyline was administered to the mice in drinking water for 5 days (2 mg/ml, supplemented with sucrose 10 mg/ml). Thymocytes were harvested and analyzed by immunoblot with anti-HA or anti-AIMP3 antibodies. β-actin was used as a loading control. AIMP3: ARS-interacting multifunctional protein 3.
There are several advantages to generating a collection of these ES cell lines using RMCE method or FRT-mediated targeting (Beard et al., 2006; Kleinhammer et al., 2011b). First, once the FRT or attP sites have been targeted using recombination, transgenes can be introduced without building additional targeting vectors with homologous recombination arms. Second, the same targeting vector can introduce cDNA into several selected sites in the genome. Third, a series of cDNAs can be introduced into the same integration site to evaluate gene function without the variation in level and pattern of gene expression due to position effects. Fourth, targeting vectors for tissue-specific expression of cDNA can be constructed and efficiently integrated into different loci to evaluate their patterns of gene expression. Transgenic mice generated using these ES cell clones will also be able to show how the transgenes of interest function in vivo. Taken together, these methods provide a novel approach to efficiently modify gene expression in ES cells and in mice in an easier, faster and more predictable manner than that achieved by conventional techniques.
COMBINED USE OF TARGETED TRANSGENESIS AND TETRAPLOID BLASTOCYST TO ACCELERATE THE GENERATION OF GEMMs
The combined use of targeted insertion of a transgene or shRNA into the ROSA26 locus using RMCE or into the COL1A1 locus using FRT-mediated targeting and subsequent ES cell injection using tetraploid blastocysts significantly accelerates the generation of transgenic or knockdown mice (Seibler et al., 2003; Mackay and West, 2005; Ohta et al., 2008). Only 2 weeks are needed for cloning the DNA vector, 3 weeks are needed for generating targeted ES cell clones, and 4 weeks are needed for producing newborn mice. Therefore, this system enables the rapid performance of in vivo gene function studies and provides a generally applicable tool for reverse mouse genetics research. Moreover, the investigation of the effects of gene expression or knockdown after the onset of a chronic or acute disease is simplified. Taken together, these technologies provide a novel and readily applicable approach for the inducible and reversible analysis of gene function in mice that overcomes the limitations of homologous recombination-based gene targeting and random transgenesis.
THE PROS AND CONS OF GEMMS GENERATION USING ENGINEERED NUCLEASE
Recently, engineered nucleases provide us alternative methods of generating GEMMs. To date, three kinds of engineered nuclease have been developed: zinc-finger nuclease (ZFNs), transcription activator-like effector (TALEN) nuclease, and clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) system (Gaj et al., 2013). These systems allow the direct generation of li ve knockout or knock-in mice without producing chimeras, therefore saving the generation time and cost compared to the conventional methods using ES cell and homologous recombination.
However, despite these advantages, there are several issues which must be carefully considered before generating and applying these mice by using engineered nuclease. First, one of the major concerns is the presence of substantial off-target effects, which are due to innate properties of these methods. Now, many methods to reduce the off-target effects have been tried and developed. Second, as mutations of target gene generated by these methods are heterogeneous, separation and identification of these mutations are required. Third, these methods make bi-allelic mutation in the target site. If these mutations have harmful effect on embryo development or survival of neonates, mutant mice cannot be generated by these methods. Taken together, in spite of great technical advances and many advantages in GEM generation using these methods, application of GEMMs generated by these methods are restricted in specific cases because of the several problems as described above.
THE GENERATION OF INDUCIBLE TRANSGENIC MICE
Inducible gene expression has several advantages in the study of gene function and genetic regulatory mechanisms. This regulation of gene expression is a powerful tool for studying the biological role of genes whose constitutive expression might be lethal, harmful, or detrimental to cells. Furthermore, temporal, spatial and titratable control of gene expression is important in the study of genes involved in disease development and progression.
The tetracycline-dependent system is one of several inducible systems that has been widely used for several decades (Lewandoski, 2001; Jonkers and Berns, 2002; Sun et al., 2007). This system works effectively and efficiently in vitro and in vivo to control gene expression. It was first developed for in vivo use in 1995 (Gossen et al., 1995), and this system has been used in many fields to analyze the outcomes of gene expression after the onset of disease in mice.
The tetracycline-dependent system has two systems, Tetoff and Tet-on, which have two components that direct the expression of the gene of interest, the tetracycline-controlled transactivator (tTA) or a reverse tTA (rtTA) and the tetracycline-responsive promoter element (TRE) (Sun et al., 2007). In both systems, target gene expression can be regulated both reversibly and quantitatively in cells or in mice with varying concentrations of tetracycline or tetracycline derivatives such as doxycycline. In the Tet-off system, transcription of the target gene is inactive in the presence of tetracycline or doxycycline; in the Tet-on system, transcription is active in the presence of tetracycline or doxycycline. The Tet-off and Tet-on systems are complementary, so the decision to choose one over the other depends on the specific experimental strategy and plan.
In the Tet-off system, the tTA is composed of the Tet repressor DNA binding protein (TetR) originating from Escherichia coli transposon Tn10 fused to the transactivating domain of VP16 of human cytomegalovirus (Sun et al., 2007). The target gene is under transcriptional control of a TRE. The TRE is composed of a Tet operator (tetO) sequence fused to a minimal promoter. The commonly used minimal promoter sequence is derived from the human cyctomegalovirus immediate-early promoter. In the absence of tetracycline or doxycycline, the tTA binds to the TRE and activates transcription of the target gene. In the presence of tetracycline or doxycycline, the tTA cannot bind to the TRE and expression of the target gene remains inactive.
The Tet-on system uses the rtTA, which is a fusion protein of the TetR repressor and the VP16 transactivation domain. However, a change in four amino acids in the TetR DNA binding moiety alters the rtTA’s binding characteristics and the protein can only recognize the tetO sequences in the TRE in the presence of tetracycline or doxycycline. Therefore, in the Tet-on system, transcription of the TRE-regulated target gene is stimulated by the rtTA only in the presence of tetracycline or doxycycline (Fig. 2).
Several tTA and rtTA transgenic mice have been developed in which tTA and rtTA, respectively, are expressed only in a specific cell type (Jaisser, 2000). Therefore, it is possible to express the TRE-regulated target transgene in a tissue-specific manner. Furthermore, strict regulation of the level of tTA or rtTA activity allows both quantitative and temporal regulation of the activation of the target gene. Additionally, targeted transgenesis is useful for generating these transgenic mice because the expression of a TRE-regulated transgene may be influenced by its chromosomal insertion site.
APPLICATION OF INDUCIBLE TRANSGENIC MICE
Tetracycline-inducible expression systems have many applications and are of crucial importance in rigorous target validation and preclinical trials for several reasons (Jonkers and Berns, 2002). First, the ability to control expression via the tetracycline-dependent system has been used to establish an oncogene’s tumor initiation and maintenance requirements. Using a regulated reversible system, the sustained requirement for oncogenic activity in a fully established tumor can be directly demonstrated by extinguishing the expression of the transgene and monitoring the phenotypic consequences in the tumor. For example, this approach showed that persistent RAS activation is required for melanoma maintenance (Wong and Chin, 2000), persistent MYC-activation is required for maintenance of hematopoietic tumors and a subset of breast adenocarcinoma (Boxer et al., 2004), mutant isocitrate dehydrogenase-2 (IDH2) has a proto-oncogenic role in leukemia initiation and maintenance (Kats et al., 2014), and oncogenic BRAF is required for tumor growth and maintenance in melanoma models (Hoeflich et al., 2006). Also, the overexpression of Mad2 in transgenic mice induces a variety of neoplasias and the acceleration of myc-induced lymphomagenesis; however, continued overexpression of Mad2 is not required for tumor maintenance (Sotillo et al., 2007; Sotillo et al., 2010). Second, the capacity to introduce or retrieve additional cooperating oncogenes or tumor-suppressor genes allows for the determination of whether specific mutations influence the tumor maintenance capacity of a specific oncogene. Third, these inducible systems permit the study of several additional important aspects of tumor biology beyond target validation. For example, identifying target-specific biomarkers to assess drug action is useful for discovering resistance mechanisms and understanding tumor dormancy. Lastly, the inducible Cre system is also useful in target validation. This system can mediate the deletion of somatic genes in nearly all cells or in only specific cells of the adult mouse (Jaisser, 2000; Belteki et al., 2005; Sun et al., 2007; Reinert et al., 2012). This gene deletion capability can provide information on inhibitor efficacy or potential mechanism-based toxicities that might result from pharmacological inactivation of the gene.
RNAi MICE AS AN ALTERNATIVE TO KNOCKOUT MICE
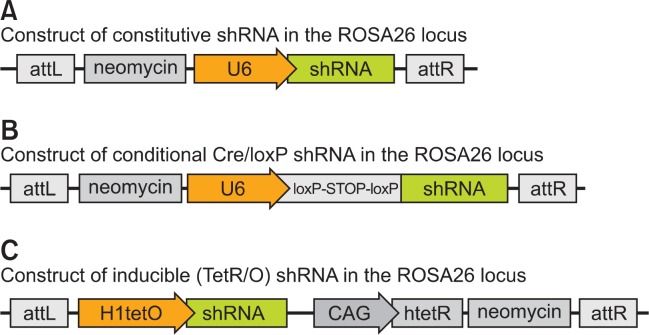
RNA interference (RNAi) has recently emerged as a practical approach for gene function studies, and it promises to be more rapid than gene knockout methods (Svoboda et al., 2001; Paddison et al., 2002; Rubinson et al., 2003; Prawitt et al., 2004; Coumoul and Deng, 2006). RNAi results in a knockdown rather than a complete loss of gene function. Therefore, RNAi mice could be an alternative to knockout mice. For reliable expression of the shRNA, the targeting vector of interest can be inserted into the ROSA26 locus or the COL1A1 3’-UTR by recombination of a site-specific recombinase in ES cells. Recombination efficiency is high at these sites and transgenesis in ES cells is a highly efficient process (Seibler et al., 2005; Beard et al., 2006; Seibler et al., 2007; Premsrirut et al., 2011). These ES cell clones can then be used to generate RNAi transgenic mice. This technology enables the production of adult knockdown mice within 10 months for in vivo validation of drug targets. This shRNA-based approach enables constitutive whole-body knockdown, inducible knockdown dependent on TRE and doxycycline, and Cre/loxP-dependent knockdown of target genes in mice (Kleinhammer et al., 2011b, 2013) (Fig. 3).
Fig. 3.
Construct of RNAi mice generated by recombinase mediated cassette exchange (RMCE). For stable integration of the shRNA expression vector into the ES cell genome, RMCE was used as described in Figure 1A. Constructs of commonly used RNAi mice are described as constitutive (A), conditional (B) and inducible (C).
GENERATION OF INDUCIBLE RNAi MICE
In mice, constant high-level expression of shRNAs may cause unexpected problems, including death, which may arise from the saturation of cellular miRNA processing pathways. Inducible RNAi technologies were devised and implemented to reduce the risk of such side effects (Dickins et al., 2007; Seibler et al., 2007; McJunkin et al., 2011; Zuber et al., 2011a). Temporal control of gene silencing can prevent impaired development until the induction of target gene expression.
Inducible expression of shRNA can be achieved by a tetracycline-dependent system as described previously. Figure 3C shows the construct for temporally-controlled gene silencing based on the TetR/tetO system. Binding of TetR to tetO blocks transcription of shRNA and de-repression can be achieved by adding the inducer doxycycline, which results in the release of TetR from tetO. Doxycycline-inducible gene silencing within 10 days of doxycycline administration achieves 80% to 90% repression in most tissues. The knockdown is reversible upon withdrawal of the inducer. Therefore, this inducible and reversible system permits the investigation of the effects of gene knockdown in a suitable mouse model, such as after the onset of a chronic or acute disease. This feature is especially interesting for pharmaceutical drug target validation because inducible gene silencing in mouse models of human disease is a surrogate for the treatment of patients with antagonistic drugs.
GENERATION OF CONDITIONAL RNAi MICE
Cre/loxP recombination is a controllable, but irreversible, me ans of achieving cell type-specific or temporally regulated RNAi that is comparable to that achieved with gene targeting by homologous recombination (Ventura et al., 2004; Coumoul et al., 2005; Coumoul and Deng, 2006; Kleinhammer et al., 2011a). Figure 3B shows the construct for gene silencing based on Cre/loxP recombination. A loxP-STOP-loxP cassette located between the U6 promoter and the shRNA sequence interrupts transcription from the U6 promoter. Cre/loxP recombination enables transcription to start producing shRNA. A number of Cre transgenic mice have been developed and Cre/loxP-dependent shRNA can be expressed temporally or spatially.
APPLICATIONS OF RNAi MICE MODELS
RNAi mice models have many applications in many fields. First, RNAi mice are useful in target validation (Zuber et al., 2011a; Zuber et al., 2011b). These RNAi mice models are an optimal surrogate for therapeutic drug action. Like antagonistic drugs, these models lower target gene activity in most tissues throughout the body without completely abolishing it. By observing the effects of a gene knockdown, researchers are able to assess the viability of a given gene product as a target for antagonist drug intervention. Second, RNAi mice can be used to determine target specificity. The reversibility of gene knockdown enables researchers to examine target-specific effects of compounds in the same mice. Third, RNAi mice can be used in safety studies. After mimicking transient drug action by inducing shRNA expression, target gene expression can be reverted to endogenous levels by withdrawing the inducer. This simulation allows the study of both short-term and long-lasting target-dependent side effects of drug action. Fourth, RNAi mice can be used to model human diseases, such as cancer, neurodegenerative diseases, type II diabetes, and genetic disorders, in an inducible and reversible manner. Fifth, RNAi mice allow for the assessment of gene function in vivo (Premsrirut et al., 2011). The inducible and reversible features of the system are beneficial for studying the effect of switching off a gene and reverting to the normal state.
CONCLUSIONS
We have reviewed the recent progress, challenges, and applications associated with the generation and use of GEMMs for drug development and preclinical trials. Since the 1980’s, several powerful and reproducible technologies have been developed that allow the precise manipulation of the murine genome and the generation of complicated mouse models of human diseases (van der Weyden et al., 2011). These methods have facilitated the evaluation of gene function by classical approaches of gain and loss of function mutagenesis in mice. Subsequently, these improvements in technologies guided application of GEMMs in drug discovery. Many obstacles in the process of using mouse models are being and will be overcome. No single model type is appropriate for every situation and understanding the needs of a drug discovery project will determine the appropriate GEMMs to develop (Abate-Shen, 2006; Sharpless and Depinho, 2006; Politi and Pao, 2011). Validating the efficacy of novel therapeutics in humans is becoming more difficult, so the development of improved pre-clinical methods to validate and prioritize novel compounds and therapies is important (Begley and Ellis, 2012). Finally, we predict that GEMMs will become a necessary tool, along with cell culture-based and xenograft model systems, in the preclinical evaluation of disease targets and the development of their respective compounds and therapies.
Acknowledgments
This work was supported by National Research Foundation grants (NRF-2013M3A6A4045755 and 2013044427) funded by the Korean government (MSIP). The authors have no conflict of interest to declare.
REFERENCES
- Abate-Shen C. A new generation of mouse models of cancer for translational research. Clin Cancer Res. 2006;12:5274–5276. doi: 10.1158/1078-0432.CCR-06-0500. [DOI] [PubMed] [Google Scholar]
- Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483:531–533. doi: 10.1038/483531a. [DOI] [PubMed] [Google Scholar]
- Belteki G, Haigh J, Kabacs N, Haigh K, Sison K, Costantini F, Whitsett J, Quaggin SE, Nagy A. Conditional and inducible transgene expression in mice through the combinatorial use of Cre-mediated recombination and tetracycline induction. Nucleic Acids Res. 2005;33:e51. doi: 10.1093/nar/gni051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolon B. Genetically engineered animals in drug discovery and development: a maturing resource for toxicologic research. Basic Clin Pharmacol Toxicol. 2004;95:154–161. doi: 10.1111/j.1742-7843.2004.pto950402.x. [DOI] [PubMed] [Google Scholar]
- Boxer RB, Jang JW, Sintasath L, Chodosh LA. Lack of sustained regression of c-MYC-induced mammary adenocarcinomas following brief or prolonged MYC inactivation. Cancer Cell. 2004;6:577–586. doi: 10.1016/j.ccr.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Zhang L, Conklin DS, Hannon GJ, Rosen-quist TA. Germline transmission of RNAi in mice. Nat Struct Biol. 2003;10:91–92. doi: 10.1038/nsb896. [DOI] [PubMed] [Google Scholar]
- Coumoul X, Deng CX. RNAi in mice: a promising approach to decipher gene functions in vivo. Biochimie. 2006;88:637–643. doi: 10.1016/j.biochi.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Coumoul X, Shukla V, Li C, Wang RH, Deng CX. Conditional knockdown of Fgfr2 in mice using Cre-LoxP induced RNA interference. Nucleic Acids Res. 2005;33:e102. doi: 10.1093/nar/gni100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickins RA, McJunkin K, Hernando E, Premsrirut PK, Krizhanovsky V, Burgess DJ, Kim SY, Cordon-Cardo C, Zender L, Hannon GJ. Tissue-specific and reversible RNA interference in transgenic mice. Nat Genet. 2007;39:914–921. doi: 10.1038/ng2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat. Rev. Cancer. 2007;7:645–658. doi: 10.1038/nrc2192. [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- Hansen K, Khanna C. Spontaneous and genetically engineered animal models: use in preclinical cancer drug development. Eur. J. Cancer. 2004;40:858–880. doi: 10.1016/j.ejca.2003.11.031. [DOI] [PubMed] [Google Scholar]
- Hoeflich KP, Gray DC, Eby MT, Tien JY, Wong L, Bower J, Gogineni A, Zha J, Cole MJ, Stern HM. Oncogenic BRAF is required for tumor growth and maintenance in melanoma models. Cancer Res. 2006;66:999–1006. doi: 10.1158/0008-5472.CAN-05-2720. [DOI] [PubMed] [Google Scholar]
- Jaisser F. Inducible gene expression and gene modification in transgenic mice. J Am Soc Nephrol. 2000;11:S95–S100. [PubMed] [Google Scholar]
- Jonkers J, Berns A. Conditional mouse models of sporadic cancer. Nat. Rev. Cancer. 2002;2:251–265. doi: 10.1038/nrc777. [DOI] [PubMed] [Google Scholar]
- Kats LM, Reschke M, Taulli R, Pozdnyakova O, Burgess K, Bha rgava P, Straley K, Karnik R, Meissner A, Small D. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell. 2014;14:329–341. doi: 10.1016/j.stem.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinhammer A, Deussing J, Wurst W, Kuhn R. Conditional RNAi in mice. Methods. 2011a;53:142–150. doi: 10.1016/j.ymeth.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Kleinhammer A, Wurst W, Kuhn R. Constitutive and conditional RNAi transgenesis in mice. Methods. 2011b;53:430–436. doi: 10.1016/j.ymeth.2010.12.015. [DOI] [PubMed] [Google Scholar]
- Kleinhammer A, Wurst W, Kuhn R. Target validation in mice by constitutive and conditional RNAi. Methods Mol Biol. 2013;986:307–323. doi: 10.1007/978-1-62703-311-4_19. [DOI] [PubMed] [Google Scholar]
- Kucherlapati R. Genetically modified mouse models for bio-marker discovery and preclinical drug testing. Clin Cancer Res. 2012;18:625–630. doi: 10.1158/1078-0432.CCR-11-2021. [DOI] [PubMed] [Google Scholar]
- Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- Mackay GE, West JD. Fate of tetraploid cells in 4n<->2n chimeric mouse blastocysts. Mech Dev. 2005;122:1266–1281. doi: 10.1016/j.mod.2005.09.001. [DOI] [PubMed] [Google Scholar]
- McCreath K, Howcroft J, Campbell K, Colman A, Schnieke A, Kind A. Production of gene-targeted sheep by nuclear transfer from cultured somatic cells. Nature. 2000;405:1066–1069. doi: 10.1038/35016604. [DOI] [PubMed] [Google Scholar]
- McJunkin K, Mazurek A, Premsrirut PK, Zuber J, Dow LE, Simon J, Stillman B, Lowe SW. Reversible suppression of an essential gene in adult mice using transgenic RNA interference. Proc Natl Acad Sci USA. 2011;108:7113–7118. doi: 10.1073/pnas.1104097108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H, Sakaide Y, Wakayama T. Generation of mice derived from embryonic stem cells using blastocysts of different developmental ages. Reproduction. 2008;136:581–587. doi: 10.1530/REP-08-0184. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Hannon GJ. Stable suppression of gene expression by RNAi in mammalian cells. Proc Natl Acad Sci USA. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palais G, Nguyen Dinh Cat A, Friedman H, Panek-Huet N, Millet A, Tronche F, Gellen B, Mercadier JJ, Peterson A, Jaisser F. Targeted transgenesis at the HPRT locus: an efficient strategy to achieve tightly controlled in vivo conditional expression with the tet system. Physiol. Genomics. 2009;37:140–146. doi: 10.1152/physiolgenomics.90328.2008. [DOI] [PubMed] [Google Scholar]
- Politi K, Pao W. How genetically engineered mouse tumor models provide insights into human cancers. J Clin Oncol. 2011;29:2273–2281. doi: 10.1200/JCO.2010.30.8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prawitt D, Brixel L, Spangenberg C, Eshkind L, Heck R, Oesch F, Zabel B, Bockamp E. RNAi knock-down mice: an emerging technology for post-genomic functional genetics. Cytogenet Genome Res. 2004;105:412–421. doi: 10.1159/000078214. [DOI] [PubMed] [Google Scholar]
- Premsrirut PK, Dow LE, Kim SY, Camiolo M, Malone CD, Miething C, Scuoppo C, Zuber J, Dickins RA, Kogan SC, Shroyer KR, Sordella R, Hannon GJ, Lowe SW. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145:145–158. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinert RB, Kantz J, Misfeldt AA, Poffenberger G, Gannon M, Brissova M, Powers AC. Tamoxifen-induced CreloxP recombination is prolonged in pancreatic islets of adult mice. PLoS One. 2012;7:e33529. doi: 10.1371/journal.pone.0033529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond A, Su Y. Mouse xenograft models vs GEM models for human cancer therapeutics. Dis Model Mech. 2008;1:78–82. doi: 10.1242/dmm.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles AI, Varticovski L. Harnessing genetically engineered mouse models for preclinical testing. Chem Biol Interact. 2008;171:159–164. doi: 10.1016/j.cbi.2007.01.014. [DOI] [PubMed] [Google Scholar]
- Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Seibler J, Küter-Luks B, Kern H, Streu S, Plum L, Mauer J, Kühn R, Brüning JC, Schwenk F. Single copy shRNA configuration for ubiquitous gene knockdown in mice. Nucleic Acids Res. 2005;33:e67. doi: 10.1093/nar/gni065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibler J, Kleinridders A, Kuter-Luks B, Niehaves S, Bruning JC, Schwenk F. Reversible gene knockdown in mice using a tight, inducible shRNA expression system. Nucleic Acids Res. 2007;35:e54. doi: 10.1093/nar/gkm122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibler J, Zevnik B, Küter-Luks B, Andreas S, Kern H, Hennek T, Rode A, Heimann C, Faust N, Kauselmann G. Rapid generation of inducible mouse mutants. Nucleic Acids Res. 2003;31:e12. doi: 10.1093/nar/gng012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless NE, Depinho RA. The mighty mouse: genetically engineered mouse models in cancer drug development. Nat Rev Drug Discov. 2006;5:741–754. doi: 10.1038/nrd2110. [DOI] [PubMed] [Google Scholar]
- Singh M, Johnson L. Using genetically engineered mouse models of cancer to aid drug development: an industry perspective. Clin Cancer Res. 2006;12:5312–5328. doi: 10.1158/1078-0432.CCR-06-0437. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, Benezra R. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Schvartzman JM, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–440. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathdee D, Ibbotson H, Grant SG. Expression of trans-genes targeted to the Gt(ROSA)26Sor locus is orientation dependent. PLoS One. 2006;1:e4. doi: 10.1371/journal.pone.0000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suggitt M, Bibby MC. 50 years of preclinical anticancer drug screening: empirical to target-driven approaches. Clin Cancer Res. 2005;11:971–981. [PubMed] [Google Scholar]
- Sun Y, Chen X, Xiao D. Tetracycline-inducible expression systems: new strategies and practices in the transgenic mouse modeling. Acta BiochimBiophys Sin. 2007;39:235–246. doi: 10.1111/j.1745-7270.2007.00258.x. [DOI] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Schultz RM. RNAi in mouse oocytes and preimplantation embryos: effectiveness of hairpin dsRNA. Biochem Biophys Res Commun. 2001;287:1099–1104. doi: 10.1006/bbrc.2001.5707. [DOI] [PubMed] [Google Scholar]
- van der Weyden L, White JK, Adams DJ, Logan DW. The mouse genetics toolkit: revealing function and mechanism. Genome Biol. 2011;12:224. doi: 10.1186/gb-2011-12-6-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc Natl Acad Sci USA. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AK, Chin L. An inducible melanoma model implicates a role for RAS in tumor maintenance and angiogenesis. Cancer Metastasis Rev. 2000;19:121–129. doi: 10.1023/a:1026537423753. [DOI] [PubMed] [Google Scholar]
- Zambrowicz BP, Sands AT. Knockouts model the 100 best-selling drugs--will they model the next 100? Nat Rev Drug Discov. 2003;2:38–51. doi: 10.1038/nrd987. [DOI] [PubMed] [Google Scholar]
- Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol. 2011a;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, Taylor MJ, Johns C, Chicas A, Mulloy JC, Kogan SC, Brown P, Valent P, Bradner JE, Lowe SW, Vakoc CR. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011b;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]