Abstract
In this study, 23 oleanane-type triterpenoid saponins were isolated from a methanol extract of the roots of Pulsatilla koreana. The NF-κB inhibitory activity of the isolated compounds was measured in TNFα-treated HepG2 cells using a luciferase reporter system. Compounds 19–23 inhibited TNFα-stimulated NF-κB activation in a dose-dependent manner, with IC50 values ranging from 0.75–8.30 μM. Compounds 19 and 20 also inhibited the TNFα-induced expression of iNOS and ICAM-1 mRNA. Moreover, effect of the isolated compounds on PPARs transcriptional activity was assessed. Compounds 7–11 and 19–23 activated PPARs the transcriptional activity significantly in a dose-dependent manner, with EC50 values ranging from 0.9–10.8 μM. These results suggest the presence of potent anti-inflammatory components in P. koreana, and will facilitate the development of novel anti-inflammatory agents.
Keywords: Pulsatilla koreana, NF-κB inhibitory activity, Tumor necrosis factor-α, PPAR
INTRODUCTION
The Pulsatilla genus, Pulsatilla koreana Nakai (Ranunculaceae) from Korea is an important herb in traditional medicine used to treat amoebic dysentery and malaria (Kim et al., 2004). Phytochemical studies on P. koreana roots have demonstrated the presence of protoanemonin, deoxypodophyllotoxin, oleanane, and lupane-type triterpenoid saponins (Martín et al., 1990; Kim et al., 2002; Kim et al., 2004; Bang et al., 2005; Lee et al., 2010; Suh et al., 2010). Root extracts of P. koreana possess antitumor, antibiotic, and anti-inflammatory activities (Martín et al., 1990; Cuong et al., 2009; Yang et al., 2010). Although, P. koreana contains a wide variety of metabolites and bioactivities, the active components that inhibit nuclear factor kappa B (NF-κB) and activate peroxisome proliferator-activated receptor (PPAR) have not been identified.
NF-κB is a family of Rel domain-containing proteins that includes RelA, RelB, c-Rel, NF-κB1, and NF-κB2. NF-κB activation has been linked to multiple pathophysiological conditions such as cancer, arthritis, asthma, inflammatory bowel disease, and other inflammatory conditions (Baldwin, 2001). NF-κB activation by various stimuli, including the inflammatory cytokines tumor necrosis factor alpha (TNF-α) and interleukin-1 (IL-1), T-cell activation signals, growth factors, and stress inducers cause transcription at κB sites, which are involved in a number of diseases, including inflammatory disorders and cancer (Baldwin, 2001; Pande and Ramos, 2005). In the present study, the effects of compounds 1–23 (oleanane-type triterpenoid saponins that were isolated from a methanol extract of the roots of Pulsatilla koreana) on TNFα-induced NF-κB transcriptional activity in human hepatocarcinoma (HepG2) cells were evaluated using an NF-κB-luciferase assay.
PPAR is a member of the nuclear receptor superfamily of ligand-dependent transcription factors. It is predominantly expressed in adipose tissue, adrenal glands, and the spleen (Moraes et al., 2006; Sharma and Staels, 2007). Three isoforms have been identified: PPARα, PPARγ, and PPARβ(δ). PPARs regulate the expression of genes involved in the regulation of glucose, lipid, and cholesterol metabolism by binding to specific peroxisome proliferator response elements (PPREs) in the enhancer sites of target genes (Berger and Moller, 2002; Balint and Nagy, 2006; Barish et al., 2006; Haluzik and Haluzik, 2006). Accordingly, compounds that modulate PPARs function are attractive for the treatment of type 2 diabetes, obesity, metabolic syndrome, inflammation, and cardiovascular disease (Kuroda et al., 2010). In this study, 23 oleanane-type triterpenoid saponins were isolated from a methanol extract of the roots of P. koreana. Their inhibition of NF-κB and activation of PPAR were measured in HepG2 cells using luciferase reporter systems.
MATERIALS AND METHODS
Chemical and sample preparation
Compounds 1–23 were isolated from the roots of P. koreana and identified in our previous reports. Sulfasalazine and benzafibrate (positive control) were the products of Sigma-Aldrich. All other chemicals and reagents were of analytical grade. The tested compounds and positive control were dissolved in DMSO.
Plant material
Dried roots of P. koreana were purchased from herbal market, Kumsan, Chungnam, Korea in March 2009 and identified by one of the authors (Prof. Young Ho Kim). A voucher specimen (CNU 09106) was deposited at the Herbarium of College of Pharmacy, Chungnam National University, Daejeon, Korea.
Extraction and isolation
Dried roots of P. koreana (2.0 kg) were extracted with MeOH under reflux for 10 h (7 L×3 times) to yield 500.0 g of extract. This extract was suspended in water and partitioned with ethyl acetate to yield 37.0 g ethyl acetate extract and 463.0 g water extract. The water extract was partitioned with n-BuOH to yield 130.0 g BuOH extract. The ethyl acetate extract was subjected to silica gel column chromatography with a gradient of CHCl3-MeOH (50:1, 20:1,10: 1 and 1:1; 2 L for each step) to give 6 fractions (Fr. E1–E6). The fraction E4 was separated using an YMC column with a MeOH-actone-H2O (0.25:0.3:1–1.3:1.3:1, 1.2 L for each step) elution solvent to give compound 23 (75.0 mg). The fraction E5 was separated using an YMC column with a MeOH-H2O (3.2:1 1.4 L) elution solvent to give compound 11 (19.0 mg).
The n-BuOH extract was subjected to silica gel column chromatography with a gradient of CHCl3-MeOH-H2O (5:1:0.1, 2:1:0.1 and 0:1:0; 3 L for each step) to give 6 fractions (Fr. B1-B6). The fraction B3 was separated using a silica gel column with CHCl3-MeOH-H2O (5:1:0.1, 4:1:0.1 and 3:1:0.1, 1 L for each step) to give 4 sub-fractions (Fr. B3.1-B3.4). Fraction B3.1 was separated using an YMC column with a MeOH-H2O (4.5:1, 1.1 L) elution solvent to give compound 20 (120.0 mg). Fraction B3.2 was separated using an YMC column with a MeOH-acetone-H2O (2.5:0.7:1, 2.5 L) elution solvent to give compounds 8 (78.0 mg) and 19 (30.0 mg). Fraction B3.3 was separated using an YMC column with a MeOH-acetone-H2O (1.5:0.7:1, 750 mL) elution solvent to give compound 22 (180.0 mg). Fraction B3.4 was separated using an YMC column with a MeOH-acetone-H2O (2:0.5:1, 2.5 L) elution solvent to give compound 9 (2.1 g). The fraction B4 was separated using a silica gel column with CHCl3-MeOH-H2O (3.5:1:0.1, 2:1:0.1 and 1:1:0.2, 1 L for each step) to give 4 sub-fractions (Fr. B4.1–B4.4). Fraction B4.3 was separated using an YMC column with a MeOH-acetone-H2O (10.5:1:1–0.85:2:1, each 550 mL) elution solvent to give compound 10 (25.0 mg).
Water extract was chromatographed on a column of highly porous polymer (Diaion HP-20) and eluted with H2O and MeOH, successively, to give 4 fractions (Fr. W1-W4). Fraction W3 was subjected to silica gel column chromatography with a gradient of CHCl3-MeOH-H2O (6:1:0.1, 4:1:0.1, 2:1:0.1 and 0:1:0; 4 L for each step) to give 6 fractions (Fr. W3.1-W3.6). Fraction W3.3 using an YMC column with a MeOH-acetone-H2O (1:0.3:1–1:0.4:1.4, 650 mL for each step) elution solvent to give compounds 2 (50.0 mg), 3 (44.0 mg), 13 (17.0 mg), 14 (77.0 mg), and 16 (38.0 mg). Fraction W3.4 was separated using an YMC column with a MeOH-H2O (1.3:1–2.5:1, 750 mL for each step) elution solvent to give compounds 1 (44.0 mg) and 12 (110.0 mg). Fraction W3.5 was separated using an YMC column with an acetone-MeOH-H2O (0.25:1:1–0.32:1:1, 600 mL for each step) elution solvent to give compounds 4 (78.0 mg) and 18 (460.0 mg). Fraction W3.6 was separated using a silica gel column with CHCl3-MeOH-H2O (1.2:1:0.15, 1.5 L) to give compound 15 (130.0 mg). Fraction W4 was subjected to silica gel column chromatography with a gradient of CHCl3-MeOH-H2O (2.5:1:0.1, 1.5:1:0.15 and 0:1:0; 3 L for each step) to give 3 fractions (Fr. W4.1-W4.3). Fraction W4.1 was further chromatographed on RP chromatography column with acetone-MeOH-H2O (0.7:1.5:1–1:2:1, 1 L for each step) to yield compounds 7 (12.0 mg) and 21 (196.0 mg). Fraction W4.2 was further chromatographed on RP chromatography column with acetone-MeOH-H2O (0.6:1:1–1:1.7:1, 750 mL for each step) to yield compounds 5 (300.0 mg) and 17 (12.0 mg). Compound 6 (36.0 mg) was isolated from W4.3 using RP chromatography column with acetone-MeOH-H2O (0.3:1.7:1).
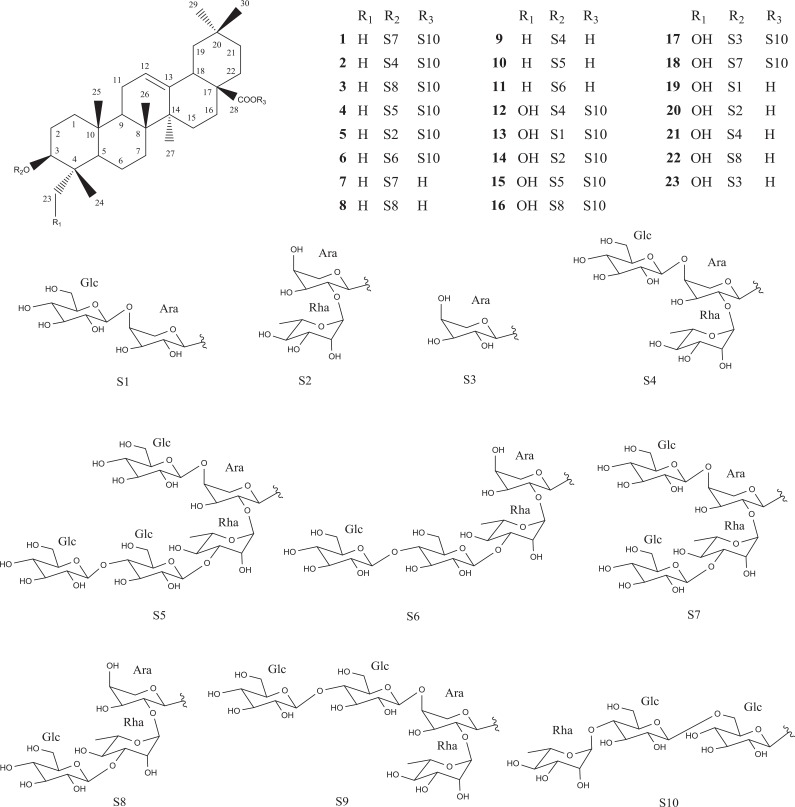
Their structures were elucidated as cernuoside A (1) (Zhang et al., 2000), hederacholchiside E (2) (Yang et al., 2010), beesioside Q (3) (Tommasi et al., 2000), 3-O-β-D-glucopyranosyl (1→4)-β-D-glucopyranosyl (1→3)-α-L-rha m no pyra nosyl (1→2) [β-D-glucopyranosyl(1→4)]-α-L-arabinopyranosyl ole a nolic acid 28-O-α-L-rhamnopyranosyl (1→4)-β-D-gluco py ranosyl (1→6)-β-D-glucopyranoside (4) (Liu et al., 2012), hederacoside B (5) (Majester-Savornin et al., 1991), raddeanoside R17 (6) (Sun et al., 2008), 3-O-β-D-glucopyranosyl (1→3)-α-L-rhamnopyranosyl (1→2) [β-D-glucopy ranosyl (1→4)]-α-L-arabinopyranosyl oleanolic acid (7) (Schenkel et al., 1991), 3-O-β-D-glucopyranosyl (1→3)-α-L-rhamno py ranosyl (1→ 2)-α-L-arabinopyranosyl oleanolic acid (8) (Bang et al., 2005), raddeanoside R13 (9) (Bang et al., 2005), 3-O-β-D-glucopyranosyl (1→4)-β-D-glucopyranosyl (1→3)-α- L-rhamnopyranosyl (1→2) [β-D-glucopyranosyl(1→4)]-α-L-ara binopyranosyl oleanolic acid (10) (Mimaki et al., 1999), 3-O-β-D-glucopyranosyl(1→4)-β-D-glucopyranosyl (1→3)-α-L-rhamnopyranosyl (1→2) [β-D-glucopyranosyl(1→4)]-α-L-arabinopyranosyl oleanolic acid (11) (Mimaki et al., 1999), hederacholchiside F (12) (Yang et al., 2010), fatsiaside G (13) (Li et al., 1990), Pulsatilla saponin F (14) (Tran et al., 2011), pulsatilloside F (15) (Li et al., 2013a), patrinia saponin H3 (16) (Kang et al., 1997), hederasaponin D (17) (Tran et al., 2011), cernuoside B (18) (Zhang et al., 2000), scabioside C (19) (Bang et al., 2005), hederoside C (20) (Li et al., 1990), Pulsatilla saponin D (21) (Shimizu et al., 1978), kalopanaxsaponin H (22) (Ye et al., 1996), and scabioside A (23) (Baykal et al., 1997) (Fig. 1). Their structures were elucidated on the basis of spectroscopic data and comparison of 1D- and 2D-NMR and mass spectral data with reported values.
Fig. 1.
Structures of compounds 1–23 from the roots of P. koreana.
Cell lines and culture
Human hepatocarcinoma HepG2 cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen, Carlsbad, CA, USA) containing 10% heat-inactivated fetal bovine serum, 100 units/mL penicillin, and 10 μg/mL streptomycin, at 37ºC and 5% CO2. Human TNF-α was purchased from ATgen (Seoul, Korea).
Cell toxicity assay
Cell-Counting Kit (CCK)-8 (Dojindo, Kumamoto, Japan) was used to analyze the effect of compounds on cell toxicity according to the manufacturer’s instructions. Cells were cultured overnight in 96-well plate (∼1×104 cells/well). Cell toxicity was assessed after the addition of compounds on dose-dependent manner. After 24 h of treatment, 10 μl of the CCK-8 solution was added to triplicate wells, and incubated for 1 h. Absorbance was measured at 450 nm to determine viable cell numbers in wells.
NF-κB-Luciferase assay
HepG2 cells were maintained in Dulbecco’s modified Eagles’ medium (DMEM) (Invitrogen, Carlsbad, CA) containing 10% heat-inactivated fetal bovine serum (FBS), 100 units/mL penicillin, and 10 μg/mL streptomycin at 37°C and 5% CO2. The luciferase vector was first transfected into HepG2 cells. After a limited amount of time, the cells were lysed, and luciferin, the substrate of luciferase, was introduced into the cellular extract along with Mg2+ and an excess of ATP. Under these conditions, luciferase enzymes expressed by the reporter vector could catalyze the oxidative carboxylation of luciferin. Cells were seeded at 2×105 cells per well in a 12-well plate and grown. After 24 h, cells were transfected with inducible NF-κB firefly luciferase reporter and constitutively expressing Renilla reporter. After 24 h of transfection, medium was changed to assay medium (Opti-MEM+0.5% FBS+0.1 mM NEAA+1 mM sodium pyruvate+100 units/ml penicillin+10 μg/ml streptomycin) and cells were pretreated for 1 h with either vehicle (1% DMSO in water) and compounds, followed by 1 h of treatment with 10 ng/mL TNFα for 23 hr. Unstimulated HepG2 cells were used as a negative control (−), PDTC was used as a positive control. Dual Luciferase assay was performed 48 h after transfection, and promoter activity values are expressed as arbitrary units using a Renilla reporter for internal normalization (Kim et al., 2010).
RNA preparation and reverse transcriptase polymerase chain reaction (RT-PCR)
Total RNA was extracted using Easy-blue reagent (Intron Biotechnology, Seoul, Korea). Approximately 2 μg total RNA was subjected to reverse transcription using Moloney murine leukemia virus (MMLV) reverse transcriptase and oligo-dT primers (Promega, Madison, WI, USA) for 1 h at 42°C. PCR for synthetic cDNA was performed using a Taq polymerase pre-mixture (TaKaRa, Japan). The PCR products were separated by electrophoresis on 1% agarose gels and stained with EtBr. PCR was conducted with the following primer pairs: iNOS sense 5’-TCATCCGCTATGCTGGCTAC-3’, iNOS antisense 5’-CTCAGGGTCACGGCCATTG-3’, ICAM-1 sense 5’-CTGCAGACAGTGACCATC-3’, ICAM-1 antisense 5’-GTCCAGTTTCCCGGACAA-3’, β-actin sense 5’-TCACCCACACT-GTGCCCATCTACG-3’, and β-actin antisense 5’-CAGCGGAACCGCTCATTGCCAATG-3’. The specificity of products generated by each set of primers was examined using gel electrophoresis and further confirmed by a melting curve analysis. HepG2 cells were pretreated in the absence and presence of compounds for 1 h, then exposed to 10 ng/mL TNFα for 6 h. Total mRNA was prepared from the cell pellets using Easy-blue. The levels of mRNA were assessed by RT-PCR.
PPRE-Luciferase assay
HepG2 cells were seeded at 1.5 × 105 cells per well in 12-well plates and grown for 24 h before transfection. An optimized amount of DNA plasmid (0.5 μg of PPRE-Luc and 0.2 μg of PPAR-inpCMV) was diluted in 100 μL of DMEM. All cells were transfected with the plasmid mixture using WelFect M Gold (WelGENE Inc.) as described by the manufacturer. After 30 min of incubation at room temperature, the DNA plasmid solution (100 μL) was introduced and mixed gently with cells. After 24 h of transfection, the medium was changed to TOM (Transfection Optimized Medium, Invitrogen) containing 0.1 mM NEAA, 0.5% charcoal-stripped FBS, and the individual compounds (test group), dimethyl sulfoxide (vehicle group), or benzafibrate (positive control group). The cells were then cultured for 20 h. Next, the cells were washed with PBS and harvested with 1× passive lysis buffer (200 μL). The intensity of emitted luminescence was determined using an LB 953 Autolumat (EG&G Berthold, Bad Wildbad, Germany).
Statistical analysis
Unless otherwise stated, all experiments were performed with triplicate samples and repeated at least three times. All results are expressed as the mean ± S.E.M. Data was analyzed by Dunnett’s multiple comparison test. Upon observation of a statistically significant effect, the Newman-Keuls test were performed to determine the difference between the groups. A p value *(<0.05) and **(<0.01) were considered to be significant.
RESULTS
Compounds 1–23 inhibit NF-κB activity in HepG2 cells
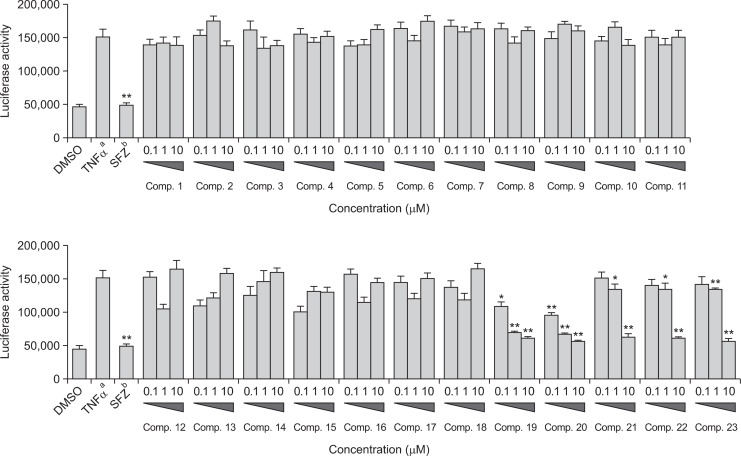
The NF-κB inhibitory activity of compounds 1–23 was evaluated using TNFα-induced NF-κB luciferase reporter assay. Cell viability was measured using Cell-Counting Kit (CCK)-8. The results showed that compounds 1–23 were not cytotoxic in HepG2 cells at the tested concentrations (data not shown). HepG2 cells were treated with 10 ng/mL TNFα, which increased NF-κB transcriptional activity compared with untreated control cells. Transfected HepG2 cells were pre-treated with various concentrations (0.1, 1, and 10 μM) of compounds 1–23, and then stimulated with TNFα. Sulfasalazine (SFZ) was used as a positive control (Fig. 2). Data revealed that compounds 19–23 significantly inhibited TNFα-induced NF-κB transcriptional activity in a dose-dependent manner, with IC50 values of 1.12 ± 0.36, 0.75 ± 0.15, 8.30 ± 3.61, 8.10 ± 2.55, and 7.50 ± 1.88 μM, respectively. Compound 20 was the most effective, and was more potent than the positive control, SFZ (IC50=0.9 μM). However, the other compounds (1–18) were inactive at the tested concentrations (IC50>10 μM, data not shown).
Fig. 2.
Effects of compounds 1–23 on the TNF α-induced NF-κB luciferase activity in HepG2 cells. aStimulated with TNFα. bStimulated with TNFα in the presence of 1–23 (0.1, 1, and 10 μM) and sulfasalazine. SFZ: sulfasalazine, positive control (10 μM). Statistical significance is indicated as *(p<0.05) and **(p<0.01) as determined by Dunnett’s multiple comparison test.
Effect of compounds 19 and 20 on the expression of NF-κB target genes
NF-κB target genes include iNOS and ICAM-1, which play important roles in the inflammatory response (Wong and Menendez, 1999; Ley et al., 2007). The ability of compounds 19 and 20 to inhibit the transcription of iNOS and ICAM-1 was assessed (Fig. 3). Both compounds inhibited the expression of iNOS and ICAM-1 mRNA significantly in a concentration-dependent manner, suggesting that they inhibited the transcription of these genes. The housekeeping, gene β-actin was unchanged by the same concentrations of compounds 19 and 20.
Fig. 3.
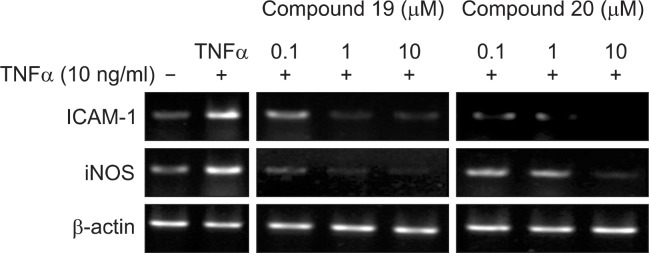
Effects of compounds 19 and 20 on iNOS and ICAM-1 mRNA expression in HepG2 cells. –: cells were treated without 10 μg/mL TNFα and compounds; +: cells were treated with 10 μg/mL TNFα only; + 0.1, 1, 10: cells were treated with 10 μg/mL TNFα and compounds.
PPAR transactivational activity of compounds 1–23
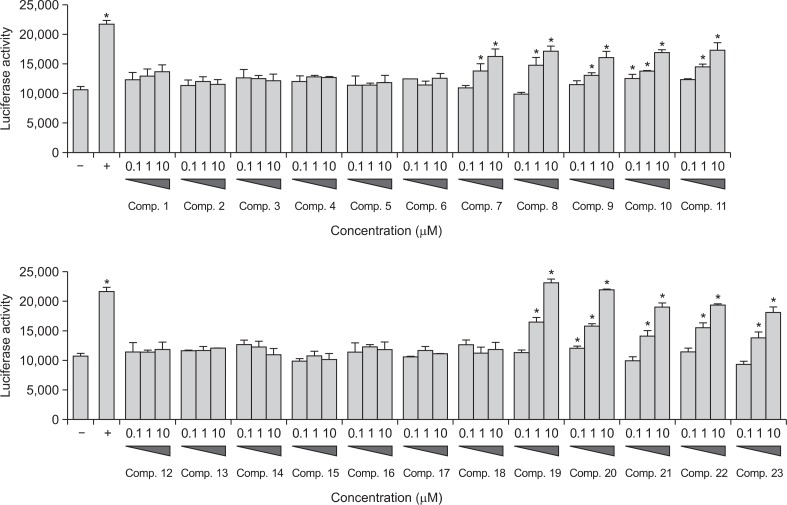
We evaluated the effects of compounds 1–23 on PPAR activity using a nuclear transcription PPRE cell-reporter system. The PPAR-responsive luciferase reporter construct, used carries a copy of the firefly luciferase gene under the control of a minimal CMV promoter, with tandem repeats of the PPRE sequence. Activated PPAR binds to the PPRE and activates transcription of the luciferase reporter gene. Benzafibrate was used as the positive control. HepG2 cells were co-transfected with the PPRE luciferase reporter and PPAR expression plasmids (Fig. 4). Compounds 7–11 and 19–23 activated the transcriptional activity of PPARs significantly in a dose-dependent manner, with EC50 values of 9.8 ± 1.7, 8.4 ± 2.0, 10.8 ± 4.2, 9.1 ± 1.3, 8.2 ± 1.8, 0.9 ± 0.2, 1.7 ± 0.5, 4.2 ± 1.2, 1.9 ± 0.3, and 6.9 ± 1.0 μM, respectively. Compound 19 was the most effective and was equivalent to the positive control, benzafibrate (IC50=0.9 μM). The remaining compounds were inactive at the tested concentrations (EC50>20 μM).
Fig. 4.
PPARs transactivational activity of compounds 1–23 in HepG2 cells. (−) Vehicle group; (+) positive control (1 μM): benzafibrate. Statistical significance is indicated as * (p < 0.05) as determined by Dunnett’s multiple comparison test.
DISCUSSION
The aim of this study was to identify novel inhibitors of NF-κB among 23 compounds isolated from the roots of P. koreana. In previously study, triterpenoid saponins from P. koreana showed anticancer, enhanced immunity, and inflammatory activities (Li et al., 2013a). However, this is the first report describing NF-κB inhibitory and PPAR activating effects of these compounds. The NF-κB inhibitory activities and structural properties of compounds 1–23 allowed us to infer information regarding the structure-function relationship. Compounds 19–23 had strong activity because the C-3 of the aglycone was linked to a sugar chain and C-28 was linked to a carboxyl group. Compounds (1–18), which have two sugar chains linked to C-3 and C-28, were active. Therefore, a sugar chain at C-3 and a carboxyl group at C-28 are likely to be key functional elements. The presence of a methyl group at C-23 of the aglycone (compounds 7–11), also resulted in no activity. This suggests that the hydroxyl group at C-23 plays an important role in the anti-inflammatory activity. These observations are consistent with previous reports (Mimaki et al., 2004; Zhang et al., 2011; Li et al., 2013b). Interestingly, compounds 19 and 20 exhibited stronger activity; they contained a disaccharide (Ara-Glc and Ara-Rha, respectively) linked to C-3. These data might be useful to evaluate the structure-function relationship of other triterpenoid saponins.
Compounds 7–11 and 19–23 significantly activated the transcriptional activity of PPARs. These results were similar to the structure-function relationships of cytotoxic activity. These results suggest that compounds 7–11 and 19–23 are promising PPAR agonists. PPARα/γ, PPARγ/β(δ) dual, and PPARα/γ/β(δ) agonist combinations can achieve a broad spec trum of metabolic effects and reduce undesired weight gain and mortality rates by improving insulin sensitity and decreasing obesity, dyslipidemia, and hypertension. They also exert beneficial effects on inflammatory markers (Shearer and Billin, 2007). Therefore, additional studies of individual PPAR subtypes are need to determine how the compounds influence the response to inflammatory stimuli.
In this study, 23 oleanane-type triterpenoid saponins were isolated from a methanol extract of P. koreana roots. To our knowledge, this is the first report describing NF-κB inhibitory and PPAR activating effects of oleanane-type triterpenoid saponins from P. koreana. Importantly, these results suggest that oleanane-type triterpenoid saponins are the major bioactive components from this plant that affect inflammation. These results might be useful to evaluate the structure-function relationships of other triterpenoid saponins. Our findings also suggest the presence of anti-inflammatory components in P. koreana, and will facilitate the development of novel anti-inflammatory agents.
Acknowledgments
This study was supported by the Priority Research Center Program (2009-0093815) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology, Republic of Korea.
REFERENCES
- Baldwin AS. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. J Clin Invest. 2001;107:241–246. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balint BL, Nagy L. Selective modulators of PPAR activity as new therapeutic tools in metabolic diseases. Endocr. Metab. Immune. Disord. : Drug Targets. 2006;6:33–43. doi: 10.2174/187153006776056620. [DOI] [PubMed] [Google Scholar]
- Bang SC, Kim Y, Lee JH, Ahn BZ. Triterpenoid saponins from the roots of Pulsatilla koreana. J Nat Prod. 2005;68:268–272. doi: 10.1021/np049813h. [DOI] [PubMed] [Google Scholar]
- Barish GD, Narkar VA, Evans RM. PPARδ: A dagger in the heart of the metabolic syndrome. J Clin Invest. 2006;116:590–597. doi: 10.1172/JCI27955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baykal T, Panayir T, Sticher O, Calis I. Scabioside A: A new triterpenoid saponoside from Scabiosa rotate. J Facul Pharm Gazi Univ. 1997;14:31–36. [Google Scholar]
- Berger J, Moller DE. The mechanisms of action of PPARs. Annu Rev Med. 2002;53:409–435. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- Cuong TD, Hung TM, Lee MK, Thao NTP, Jang HS, Min BS. Cytotoxic compounds from the roots of Pulsatilla koreana. Nat Prod Sci. 2009;15:250–255. [Google Scholar]
- Haluzik MM, Haluzik M. PPAR-α and insulin sensitivity. Physiol Res. 2006;55:115–122. doi: 10.33549/physiolres.930744. [DOI] [PubMed] [Google Scholar]
- Kang SS, Kim JS, Kim YH, Choi JS. A triterpenoid saponin from Patrinia scabiosaefolia. J Nat Prod. 1997;60:1060–1062. doi: 10.1021/np970175v. [DOI] [PubMed] [Google Scholar]
- Kim KK, Park KS, Song SB, Kim KE. Up regulation of GW112 gene by NF-κB promotes an antiapoptotic property in gastric cancer cells. Mol Carcinog. 2010;49:259–270. doi: 10.1002/mc.20596. [DOI] [PubMed] [Google Scholar]
- Kim Y, Bang SC, Lee JH, Ahn BZ. Pulsatilla saponin D: The antitumor principle from Pulsatilla koreana. . Arch Pharm Res. 2004;27:915–918. doi: 10.1007/BF02975843. [DOI] [PubMed] [Google Scholar]
- Kim Y, You YJ, Nam NH, Ahn BZ. Prodrugs of 4’-demethyl-4-deoxypodophyllotoxin: synthesis and evaluation of the antitumor activity. Bioorg Med Chem Lett. 2002;12:3435–3438. doi: 10.1016/s0960-894x(02)00758-8. [DOI] [PubMed] [Google Scholar]
- Kuroda M, Mimaki Y, Honda S, Tanaka H, Yokota S, Mae T. Phenolics from Glycyrrhiza glabra roots and their PPAR-γ ligand-binding activity. . Bioorg Med Chem. 2010;18:962–970. doi: 10.1016/j.bmc.2009.11.027. [DOI] [PubMed] [Google Scholar]
- Lee KY, Cho YW, Park JH, Lee DY, Kim SH, Kim YC, Sung SH. Quality control of Pulsatilla koreana based on the simultaneous determination of triterpenoidal saponins by HPLC-ELSD and principal component analysis. . Phytochem Anal. 2010;21:314–321. doi: 10.1002/pca.1201. [DOI] [PubMed] [Google Scholar]
- Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol. 2007;7:678–689. doi: 10.1038/nri2156. [DOI] [PubMed] [Google Scholar]
- Li W, Ding Y, Sun YN, Yan XT, Yang SY, Choi CW, Cha JY, Lee YM, Kim YH. Triterpenoid saponins of Pulsatilla koreana root have inhibition effects of tumor necrosis factor-α secretion in lipopolysaccharide-induced RAW264.7 Cells. . Chem Pharm Bull. 2013a;61:471–476. doi: 10.1248/cpb.c12-01034. [DOI] [PubMed] [Google Scholar]
- Li W, Ding Y, Sun YN, Yan XT, Yang SY, Choi CW, Kim EJ, Kang HK, Kim YH. Oleanane-type triterpenoid saponins from the roots of Pulsatilla koreana and their apoptosis-inducing effects on HL-60 human promyelocytic leukemia cells. Arch Pharm Res. 2013b;36:768–774. doi: 10.1007/s12272-013-0042-5. [DOI] [PubMed] [Google Scholar]
- Li XC, Wang DZ, Wu SG, Yang CR. Triterpenoid saponins from Pulsatilla campanella. Phytochemistry. 1990;29:595–599. [Google Scholar]
- Liu JY, Guan YL, Zou LB, Gong YX, Hua HM, Xu YN, Zhang H, Yu ZG, Fan WH. Saponins with neuro-protective effects from the roots of Pulsatilla cernua. Molecules. 2012;17:5520–5531. doi: 10.3390/molecules17055520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín ML, San Román L, Domínguez A. In vitro activity of protoanemonin, an antifungal agent. Planta Med. 1990;56:66–69. doi: 10.1055/s-2006-960886. [DOI] [PubMed] [Google Scholar]
- Majester-Savornin B, Elias R, Diaz-Lanza AM, Balansard G, Gasquet M, Delmas F. Saponins of the ivy plant, Hedera helix, and their leishmanicidic activity. Planta Med. 1991;57:260–262. doi: 10.1055/s-2006-960086. [DOI] [PubMed] [Google Scholar]
- Mimaki Y, Kuroda M, Asano T, Sashida Y. Triterpene saponins and lignans from the roots of Pulsatilla chinensis and their cytotoxic activity against HL-60 cells. J Nat Prod. 1999;62:1279–1283. doi: 10.1021/np9901837. [DOI] [PubMed] [Google Scholar]
- Mimaki Y, Yokosuka A, Hamanaka M, Sakuma C, Yamori T, Sashida Y. Triterpene saponins from the roots of Clematis chinensis. J Nat Prod. 2004;67:1511–1516. doi: 10.1021/np040088k. [DOI] [PubMed] [Google Scholar]
- Moraes LA, Piqueras L, Bishop-Bailey D. Peroxisome proliferator-activated receptors and inflammation. Pharmacol Ther. 2006;110:371–385. doi: 10.1016/j.pharmthera.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Pande V, Ramos MJ. NF-kappaB in human disease: current inhibitors and prospects for de novo structure based design of inhibitors. Curr Med Chem. 2005;12:357–374. doi: 10.2174/0929867053363180. [DOI] [PubMed] [Google Scholar]
- Sharma AM, Staels B. Peroxisome proliferator-activated receptor γ and adipose tissue-understanding obesity-related changes in regulation of lipid and glucose metabolism. J Clin Endocrinol Metab. 2007;92:386–395. doi: 10.1210/jc.2006-1268. [DOI] [PubMed] [Google Scholar]
- Schenkel EP, Werner W, Schulte KE. Saponins from Thinouia coriaceae. Planta Med. 1991;57:463–467. doi: 10.1055/s-2006-960152. [DOI] [PubMed] [Google Scholar]
- Shearer BG, Billin AN. The next generation of PPAR drugs: Do we have the tools to find them? Biochim. Biophys. Acta. 2007;1771:1082–1093. doi: 10.1016/j.bbalip.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Shingyouchi K, Morita N, Kizu H, Tomimori T. Triterpenoid saponins from Pulsatilla cenuiui spreng I. Chem Pharm Bull. 1978;26:1666–1671. [Google Scholar]
- Suh JH, Youm JR, Han SB. Simultaneous determination of triterpenoid saponins from Pulsatilla koreana using high performance liquid chromatography coupled with a charged aerosol detector (HPLC-CAD) Bull Korean Chem Soc. 2010;31:1159–1164. [Google Scholar]
- Sun Y, Li M, Liu J. Haemolytic activities and adjuvant effect of Anemone raddeana saponins (ARS) on the immune responses to ovalbumin in mice. Int Immunopharmacol. 2008;8:1095–1102. doi: 10.1016/j.intimp.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Tommasi N, Autore G, Bellino A, Pinto A, Pizza C, Sorrentino R, Venturella P. Antiproliferative triterpene saponins from Trevesia palmata. J Nat Prod. 2000;63:308–314. doi: 10.1021/np990231n. [DOI] [PubMed] [Google Scholar]
- Tran HQ, Nguyen TTN, Chau VM, Phan VK, Nguyen XN, Bui HT, Nguyen PT, Nguyen HT, Song SB, Kim YH. Anti-inflammatory triterpenoid saponins from the stem bark of Kalopanax pictus. J Nat Prod. 2011;74:1908–1915. doi: 10.1021/np200382s. [DOI] [PubMed] [Google Scholar]
- Wong HR, Menendez IY. Sesquiterpene lactones inhibit inducible nitric oxide synthase gene expression in cultured rat aortic smooth muscle cells. Biochem Biophys Res Commun. 1999;262:375–380. doi: 10.1006/bbrc.1999.1207. [DOI] [PubMed] [Google Scholar]
- Yang HJ, Cho YW, Kim SH, Kim YC, Sung SH. Triterpenoidal saponins of Pulsatilla koreana roots. Phytochemistry. 2010;71:1892–1899. doi: 10.1016/j.phytochem.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Ye WC, Ji NN, Zhao SX, Liu JH, Ye T, McKervey MA, Stevenson P. Triterpenoids from Pulsatilla chinensis. Phytochemistry. 1996;42:799–802. doi: 10.1016/0031-9422(96)00043-x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Samadi AK, Rao KV, Cohen MS, Timmermann BN. Cytotoxic oleanane-type saponins from Albizia inundata. J Nat Prod. 2011;74:477–482. doi: 10.1021/np100702p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Ye W, Yan X, Zhu G, Che CT, Zhao S. Cernuosides A and B, two sucrase inhibitors from Pulsatilla cernua. J Nat Prod. 2000;63:276–278. doi: 10.1021/np990207+. [DOI] [PubMed] [Google Scholar]