Abstract
Macrophage-mediated inflammation is a major contributor to obesity-associated insulin resistance. The co-repressor NCoR interacts with inflammatory pathway genes in macrophages, suggesting that its removal would result in increased activity of inflammatory responses. Surprisingly, we find that macrophage-specific deletion of NCoR instead results in an anti-inflammatory phenotype along with robust systemic insulin sensitization in obese mice. We present evidence that de-repression of LXRs contributes to this paradoxical anti-inflammatory phenotype by causing increased expression of genes that direct biosynthesis of palmitoleic acid and ω3 fatty acids. Remarkably, the increased ω3 fatty acid levels primarily inhibit NF-κB-dependent inflammatory responses by uncoupling NF-κB binding and enhancer/promoter histone acetylation from subsequent steps required for pro-inflammatory gene activation. This provides a mechanism for the in vivo anti-inflammatory insulin sensitive phenotype observed in mice with macrophage-specific deletion of NCoR. Therapeutic methods to harness this mechanism could lead to a new approach to insulin sensitizing therapies.
Keywords: nuclear co-repressor, insulin resistance, obesity, macrophage, inflammation
INTRODUCTION
Insulin resistance is a characteristic pathophysiologic defect in the great majority of patients with type 2 diabetes mellitus (Haffner and Taegtmeyer, 2003; Olefsky and Glass, 2010; Reaven, 2005). Obesity is the most common cause of insulin resistance, and the current obesity epidemic in westernized countries is driving the parallel type 2 diabetes epidemic (Shoelson et al., 2007). Obesity-associated chronic tissue inflammation is a key mechanism for decreased insulin sensitivity (Olefsky and Glass, 2010) and in obesity, excessive numbers of proinflammatory, M1-like, macrophages accumulate in adipose tissue (Xu et al., 2003) and liver (Lanthier et al., 2009), where they locally release a variety of cytokines which act on insulin target cells to impair insulin signaling. While other immune cell types, such as lymphocytes, esosinophils, and neutrophils can also contribute to the tissue inflammatory state in obesity (Kintscher et al., 2008; Wu et al., 2011; Yang et al., 2010), their function in this context is largely to regulate macrophage migration and activation..
Inflammatory pathways in macrophages are under stringent control by a variety of transcription factors and co-regulatory molecules. Among these are NF-κB, AP1, the PPAR family, LXR, and their associated co-activators and co-repressors. As a general rule, co-activators are recruited to nuclear receptors (NR) by receptor agonists, whereas, co-repressors are resident at promoter regions in the absence of ligands (Glass and Ogawa, 2006). The major co-repressors nuclear receptor corepressor (NCoR) and silencing mediator of retinoid and thyroid hormone receptors (SMRT) are important in the regulation of metabolic processes. In the basal state, NCoR is resident on a number of inflammatory pathway genes, keeping them in the repressed state (Medzhitov and Horng, 2009). Upon signal-dependent inflammatory pathway activation, NCoR dissociates from the promoter complex, allowing proinflammatory transcription factors, such as NF-κB and AP1, to induce increased expression of these genes (Glass and Saijo, 2010; Ogawa et al., 2004).
Although NCoR can be detected on the promoter regions of genes which are targets of a variety of transcription factors, our previous studies (Li et al., 2011), as well as those of others (Yamamoto et al., 2011) have shown that deletion of NCoR does not lead to broad de-repression of a large number of transcriptional programs. For example, in adipocytes, the dominant effect of NCoR KO is PPARγ derepression (Li et al., 2011), whereas, in skeletal muscle PPARδ and MEF activation are the major effects of NCoR gene deletion (Yamamoto et al., 2011). This leads to the concept that NCoR has a restricted set of functions within the context of a particular cell type. Based on these ideas, we used lys M Cre-mediated excision (Clausen et al., 1999) of NCoR to generate macrophage/neutrophil NCoR KO mice. Although NCoR can cause basal repression of inflammatory pathways (Glass and Saijo, 2010), we unexpectedly found that macrophage NCoR KO mice displayed an anti-inflammatory, insulin-sensitive phenotype. Mechanistic studies traced this surprising phenotype to LXR de-repression. Thus, a major effect of NCoR KO in macrophages is to de-repress LXR, which leads to the induction of lipogenic pathway genes. This causes increased biosynthesis of palmitoleic acid (POA) and ω3 fatty acids, within macrophages, which exert robust local anti-inflammatory effects, and are associated with the in vivo insulin sensitive phenotype.
RESULTS
Creation of macrophage/neutrophil specific NCoR knock out mice
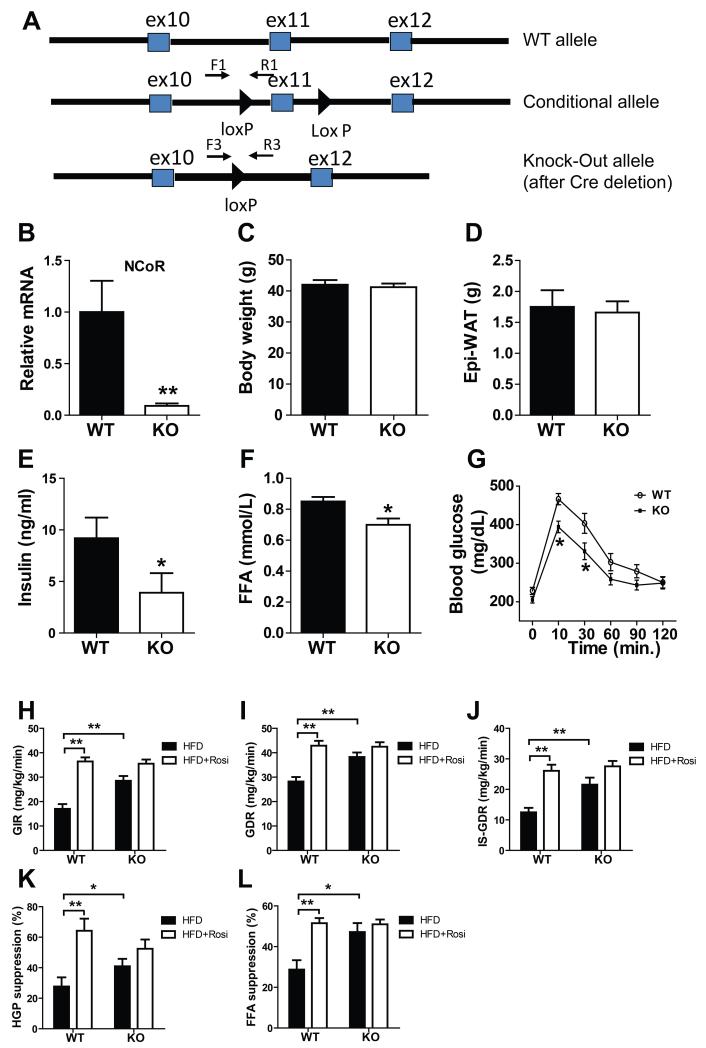
Mice carrying “floxed” NCoR alleles, with lox p sites flanking exon11 of the NCoR gene, were crossbred with mice transgenic for Lys M driven Cre expression. Both of these mouse lines have been extensively bred (> 10 generations) to the C57B/6J background. Since lys M is highly expressed in macrophage/neutrophil lineages, the resulting NCoRF/F•lys M cre mice exhibit NCoR deletion in these cell types. Consistent with our previous studies (Hevener et al., 2007), this system is highly efficient for generation of tissue specific NCoR KO mice and resulted in 80-90% deletion of NCoR from macrophages (Figs. 1A-B). For convenience, these animals are hereafter referred to as MNKO mice.
Figure 1.
Improved glucose metabolism and insulin sensitivity of MNKO mice. (A) Shown (top to bottom) are wild-type, floxed, and deleted NCoR gene loci. (B) Relative messenger RNA levels of NCoR in macrophages. Values are fold induction of gene expression normalized to the housekeeping gene 36B4 and expressed as mean ± s.e.m. (C) Body weight of WT and KO mice on 60% HFD. (D) Epi-WAT mass. (E) Fasting blood insulin levels. (F) Fasting FFA levels. (G) Glucose tolerance tests. (H) Glucose infusion rate (GIR) during hyperinsulinmic euglycemic clamp. (I) Glucose disposal rate (GDR). (J) Insulin-stimulated glucose disposal rate (IS-GDR). (K) Percent suppression of HGP by insulin (HGP suppression). (L) Percent suppression of free fatty acid levels (FFA suppression). Values are expressed as mean ± s.e.m. * P<0.05, ** P<0.01 for KO versus WT, or for comparisons as indicated.
Metabolic Studies in WT and MNKO Mice
MNKO mice exhibited normal fertility and their offspring followed the predicted Mendelian pattern with no developmental abnormalities noted. On normal chow diet, WT and MNKO mice reached the same body weight while the MNKO group was modestly more insulin sensitive during ITT testing (Data not shown). When placed on 60% HFD for 12 weeks, WT and MNKO mice exhibited similar increases in body weight (Fig. 1C) and fat mass (Fig. 1D). Given the known function of NCoR to repress basal expression of macrophage inflammatory pathway genes, we predicted increased inflammation and insulin resistance in the HFD MNKO mice. Surprisingly, insulin and FFA levels were significantly lower in HFD KO mice compared to WT littermates (Figs. 1E-F) indicating an improved overall metabolic phenotype. This phenotype was more robust following oral glucose administration, which revealed enhanced glucose tolerance (Fig. 1G) in MNKO animals. Together, with the reduced basal insulin levels, these results indicate that deletion of NCoR from macrophages confers a systemic insulin sensitive phenotype.
To quantitate this effect and to identify tissue specific responses, we performed hyperinsulinemic-euglycemic clamp studies (Li et al., 2011), which quantitatively assess insulin action in target tissues in vivo. The HFD MNKO mice were markedly more insulin sensitive compared to HFD WT mice. This was manifested by an increased overall glucose infusion rate (GIR) (Fig. 1H) and glucose disposal rate (GDR) (Fig. 1I). In addition, HFD MNKO mice exhibited enhanced insulin stimulated glucose disposal rate (IS-GDR, a measure of skeletal muscle insulin sensitivity) (Fig. 1J) and an increased ability of insulin to suppress hepatic glucose production (HGP, Fig. 1K) and circulating FFA levels (Fig. 1L). Thus, the MNKO mice displayed greater in vivo insulin sensitivity in all 3 major insulin target tissues, muscle, liver, and fat. Interestingly, treatment of HFD WT mice with rosiglitazone (3 weeks), led to the expected increase in systemic insulin sensitivity; however, the untreated MNKO mice were already insulin sensitive and rosiglitazone treatment had little, if any, additive beneficial effect (Figs. 1H-L)
Inflammatory Signaling and Macrophage Function
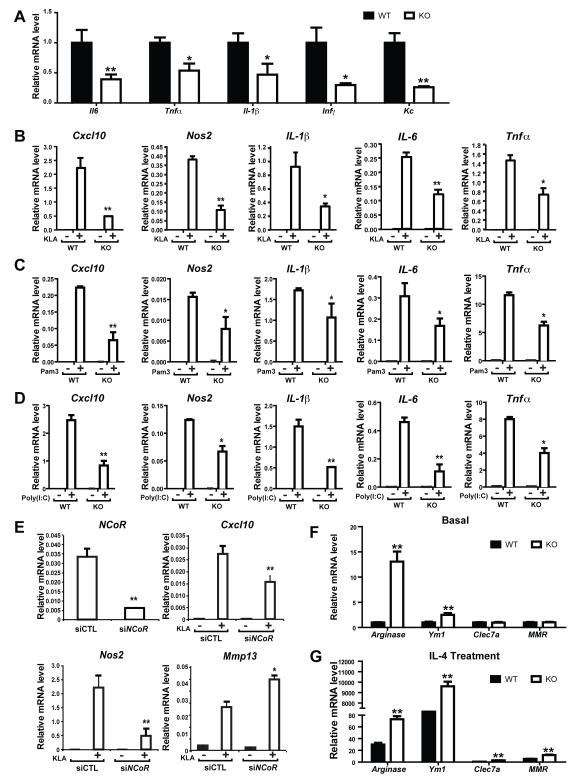
Given the unexpected insulin sensitive phenotype in the MNKO mice, we characterized inflammatory pathway signaling in WT and NCoR depleted macrophages. We measured mRNA expression levels of a variety of inflammatory genes in the basal state and after TLR4 (KLA), TLR3 (Poly(I:C)) and TLR2 (Pam3CSK4) stimulation in thioglycollate-elicited intraperitoneal macrophages (IP Macs) obtained from WT and MNKO mice (Fig. 2). Consistent with the in vivo results for glucose homeostasis and insulin sensitivity, basal gene expression was decreased in the MNKO macrophages (Fig. 2A), and, more importantly, NCoR deletion led to broadly impaired inflammatory responses to TLR4, TLR2, and TLR3 agonists (Figs. 2B-D). Complementing the results in macrophages from MNKO mice, RNAi-mediated depletion of NCoR in WT macrophages results in the same attenuation of inflammatory gene responses (Fig. 2E). Interestingly, a subset of genes characteristic of alternatively activated, M2-like, macrophages exhibited increased expression in NCoR KO cells in the basal state and after IL-4 stimulation (Figs. 2F-G).
Figure 2.
Reduced inflammation in IP-macrophages of MNKO mice. (A) Relative mRNA levels of inflammatory cytokines in IP-macrophages without treatment, or (B) with TLR4 agonist treatment. (C) Relative mRNA levels of inflammatory cytokines in IP-macrophages after TLR2 agonist treatment, or (D) TLR3 agonist treatment. (E) Relative mRNA levels of NCoR and indicated inflammatory cytokines in control (siCTL) versus siNCoR treated IP-macrophages after TLR4 agonist treatment. (F) Relative mRNA levels of M2-like cytokines in IP-macrophages without IL-4 treatment, or (G) with IL-4 treatment. Values are relative to GAPDH and are expressed as mean ± s.e.m. * P<0.05, ** P<0.01 for KO versus WT, or siCTL versus si NCoR in 2E.
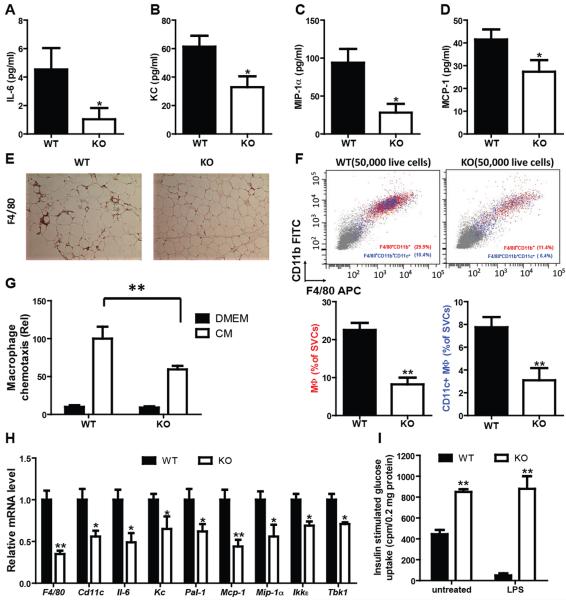
In light of the reduced inflammatory state in the MNKO mice, we went on to measure systemic aspects of HFD-mediated inflammation (Fig. 3). As can be seen in figures 3A-D, circulating levels of multiple cytokines, typically released by macrophages, were lower in the MNKO mice compared to WT, fully consistent with the gene expression data in Fig. 2. In addition, the circulating TNFα levels were lower in MNKO mice as compared to WT mice following LPS administration in vivo (Fig. S1A). Immunohistochemical analyses of adipose tissue sections was conducted by staining of adipose tissue sections for the macrophage marker F4/80 (Fig. 3E) which revealed that adipose tissue from MNKO mice contained fewer macrophages compared to WT mice. We extended this finding by performing FACS studies of adipose tissue stromal vascular cells (SVCs) (Fig. 3F), which demonstrated reduced macrophage content, as well as decreased CD11c+ proinflammatory macrophages in the MNKO mice.
Figure 3.
Hypoinflammatory phenotype of MNKO mice. Inflammatory cytokine expression in serum, including IL-6 (A), KC (B), MIP-1α (C), and MCP-1 (D). (E) F4/80 staining in Epi-WAT of WT and MNKO mice. (F) FACS analysis of macrophages and CD11c positive macrophage content in Epi-WAT. (G) Chemotaxis assay on the IP-macrophages from WT and MNKO mice. (H) Proinflammatory cytokine expression in Epi-WAT from WT and MNKO. (I) Effect of conditioned medium from IP-macrophages treated with LPS on glucose uptake in L6 cells. Values are expressed as mean ± s.e.m. * P<0.05, ** P<0.01. See also Fig. S1.
Macrophages and adipocytes are the cellular sources of chemokines in adipose tissue of obese mice and the decreased expression of MCP-1 and KC (Keratinocyte-derived chemokine) (Fig. 3A) is one mechanism for decreased adipose tissue macrophage (ATM) accumulation. However, we considered whether the intrinsic chemotactic properties of NCoR depleted macrophages were impaired. To this end we conducted in vitro chemotaxis experiments. We found that the ability of NCoR KO IP Macs to migrate towards adipocyte-conditioned medium (CM) was substantially impaired compared to WT cells (Fig. 3G), providing an additional mechanism for reduced ATMs and decreased inflammatory status in the MNKO mice.
Consistent with the reduced intrinsic inflammatory state of NCoR depleted macrophages and the decreased ATM accumulation in HFD MNKO mice, we also found decreased epididymal white adipose tissue (eWAT) expression of a number of proinflammatory genes typically associated with the inflammatory/insulin resistant state (Fig. 3H). A similar decrease in inflammatory gene expression was observed in perirenal, subcutaneous and BAT from HFD MNKO mice (Fig. S1B). Interestingly, although the overall degree of inflammation and ATM accumulation was less in subcutaneous WAT and BAT relative to eWAT and perirenal fat, the decreased inflammation in the MNKO fat depots was still evident. Similar to adipose tissue, hepatic inflammation also occurs in obesity with increased numbers of recruited hepatic macrophages (RHMs) which are distinct from Kupffer cells, and increased inflammatory gene expression. As seen in figure S1C, macrophage markers (F4/80 and CD11c) and inflammatory gene expression are reduced in liver tissue from HFD MNKO mice compared to WT controls.
The mechanism of inflammation-induced insulin resistance largely involves local release of inflammatory mediators, which exert paracrine effects to directly diminish insulin signaling in insulin target cells (Jager et al., 2007; Rotter et al., 2003). To assess this mechanism in MNKO and WT mice, conditioned media (CM) was harvested from basal and LPS treated WT and MNKO macrophages. L6 myocytes were then treated with the CM, followed by measurements of insulin stimulated glucose transport (Fig. 3I). After treatment with basal CM, myocyte glucose transport was substantially higher with MNKO CM administration. LPS stimulation leads to a marked increase in cytokine release by macrophages, and consequently CM harvested from the LPS treated WT cells robustly decreased the ability of insulin to stimulate transport in myocytes (Fig. 3I). In contrast, CM from LPS treated MNKO cells was without effect to inhibit insulin action. These findings indicate that CM from WT macrophages contains factors (most likely cytokines), which attenuate insulin signaling and that this is more pronounced after LPS stimulation. CM from the MNKO cells contains less insulin resistance-promoting cytokines, consistent with the results in figures 1-3 and S1. It is also possible that CM from MNKO cells contains factors that potentiate insulin-stimulated glucose transport.
Genome-wide Consequences of NCoR Deletion on Inflammatory Response Genes
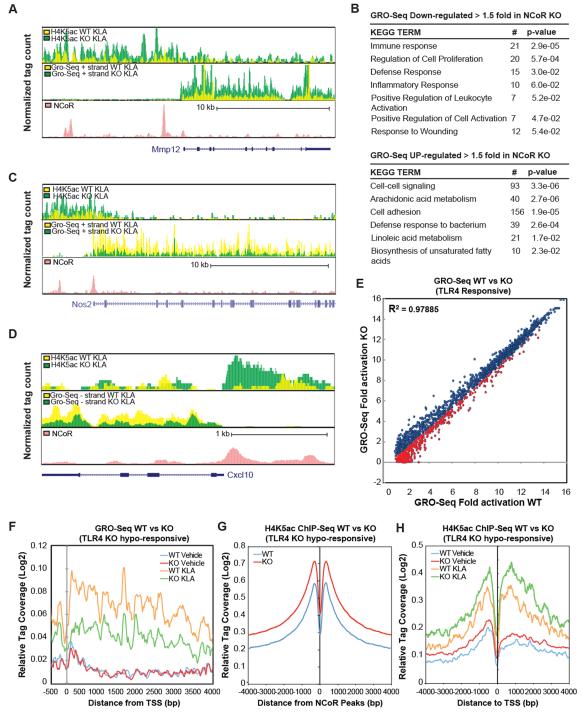
The unexpected insulin-sensitive phenotype of MNKO mice and the hypo-responsiveness of the corresponding macrophages to TLR agonists prompted us to evaluate the consequences of NCoR deletion in macrophages on a genome-wide scale. We next used global run-on sequencing (GRO-Seq) analysis to quantify nascent RNAs which are actively being transcribed in macrophages from WT and MNKO mice under basal conditions or upon activation with the TLR4 agonist KLA. GRO-Seq thereby provides a more direct measure of transcription rates than total mRNA levels. Using this approach, we identified 3397 expressed transcripts with RefSeq annotations that were > 1.5-fold increased in MNKO vs WT macrophages under basal conditions, exemplified by Mmp12 (Fig. 4A). Conversely, 1746 expressed transcripts were reduced > 1.5-fold in MNKO compared to WT macrophages. Unexpectedly, the overall pattern of differential gene regulation under basal conditions is consistent with roles of NCoR in both positive and negative regulation of innate immune functions. In particular, down-regulated genes were significantly enriched for functional annotations linked to immune response, defense response, and inflammatory response (Fig. 4B). Furthermore, a number of the RefSeq transcripts induced more than 2-fold in WT macrophages following 1h of KLA treatment exhibited <50% of maximal activation in MNKO macrophages, as exemplified by Nos2 and Cxcl10 (Figs. 4C-D). Indeed, a direct comparison of the KLA-stimulated induction of these induced genes in WT macrophages showed that ~26% of these genes achieved levels of induction in MNKO macrophages that were 66% or less than in WT macrophages (indicated by red data points in Figure 4E, and box plot in Fig. S2). We define this subset of KLA-induced genes as ‘hypo-responsive’, and collectively, they specify a genome-wide signature of the hypo-responsive MNKO phenotype. Visualization of GRO-Seq tag density for the hypo-responsive set of genes indicated no changes in levels of transcripts from paused Pol II at the transcriptional start site, but uniform reduction in tag densities throughout the gene bodies under KLA-stimulated conditions (Fig. 4F).
Figure 4.
Genome-wide impact of NCoR deletion. (A) UCSC genome browser image illustrating normalized tag counts for H4K5Ac ChIP-Seq, GRO-Seq, and NCoR ChIP-Seq (Barish et al., 2012)in WT and MNKO macrophages at the Mmp12 locus. (B) Functional KEGG annotations associated with genes demonstrating increased or reduced expression in MNKO macrophages by more than 1.5-fold. (C-D) UCSC genome browser images corresponding to panel A for the Nos2 and Cxcl10 genomic loci. (E) Scatter plot of fold change of genes induced > 2-fold by 1h KLA treatment in WT macrophages vs fold change of the same genes in MNKO macrophages. Fold activation is plotted as normalized GRO-Seq tag counts for genes comparing log2 values of KLA versus vehicle treated. MNKO hypo-responsive genes are highlighted in red. The Pearson’s correlation coefficient is reported in the figure (p<0.0001). (F) Normalized distribution of GRO-Seq tag counts in the vicinity of the transcriptional start sites of TLR4-responsive genes exhibiting compromised activation in MNKO macrophages. (G) Normalized distribution of H4K5Ac ChIP-Seq tag counts in the vicinity of NCoR binding sites under basal conditions. (H) Normalized distribution of H4K5Ac ChIP-Seq tag counts in the vicinity of transcriptional start sites (TSS) under basal conditions. For Figures 4G-H, H4K5Ac tag counts are presented as averages at indicated positions of KLA-activated genes demonstrating hypo-responsiveness in MNKO macrophages. TLR4 MNKO hypo-responsive loci are chosen according to those genes demonstrating 1.5-fold decrease in KLA response of MNKO relative to WT, as measured by comparison of normalized GRO-Seq tag counts of MNKO versus WT macrophages. See also Fig. S2.
To investigate consequences of NCoR deletion on deposition of a histone acetylation mark associated with transcriptional activation (Hargreaves et al., 2009), we performed ChIP-Seq analysis of histone H4 lysine 5 acetylation (H4K5ac) and of NCoR in WT and MNKO macrophages. As expected, H4K5ac was increased in the vicinity of NCoR binding sites in MNKO macrophages (Fig. 4G), consistent with a role of NCoR in recruiting histone deacetylase 3 (HDAC3) to these locations (Fig. S2B). Evaluation of H4K5ac at specific loci illustrated the expected increase at genes exhibiting up-regulation in MNKO cells, exemplified by the Mmp12 locus (Fig. 4A). However, H4K5ac was also increased at genes exhibiting hypo-responsiveness to KLA stimulation, exemplified by the Nos2 and Cxcl10 genes (Fig. 4 C-D). As shown in figure 4H, this pattern was observed for the entire set of hypo-responsive genes in MNKO cells. ChIP analysis confirmed that, in MNKO macrophages, HDAC3 binding is significantly reduced at NCoR sites on inflammatory loci demonstrating both hyper-acetylation and TLR4 hypo-responsiveness, as exemplified by Nos2, IL-1β and Cxcl10 (Fig. S2C).
NCoR Deficiency Induces an LXR Program Leading to Altered Fatty Acid Metabolism
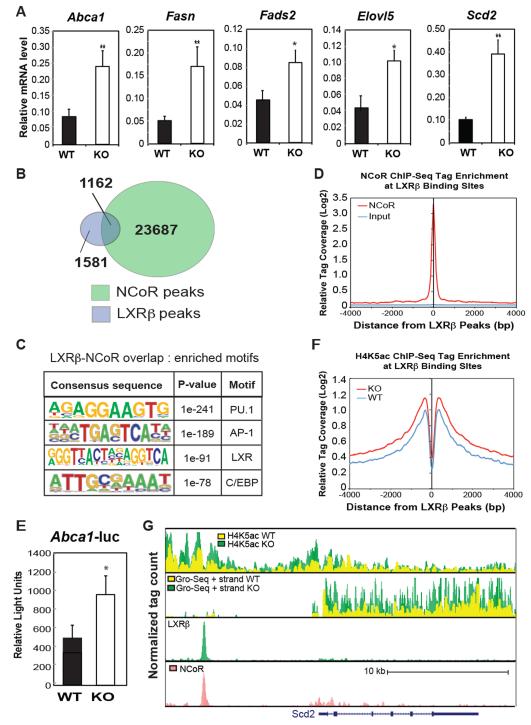
Based on these results, we evaluated genes that were de-repressed in MNKO macrophages for functions that might negatively regulate the TLR4 signaling pathway. Interestingly, de-repressed genes were significantly enriched for functional annotations linked to fatty acid metabolism, including the terms linoleic acid metabolism, biosynthesis of unsaturated fatty acids, and alpha linoleic acid metabolism (Fig. 4B). Further, several of the de-repressed genes encode enzymes involved in the synthesis of monounsaturated (MUFA) and polyunsaturated fatty acids (PUFA) that have been documented to have anti-inflammatory and/or insulin-sensitizing effects (Oh et al., 2010; Spann et al., 2012). These included Elovl5, Fads1 and Fads2, involved in the elongation and desaturation of long chain fatty acids to generate anti-inflammatory PUFAs, and Scd1/Scd2 which catalyze the synthesis of 16:1 POA, suggested to be an insulin sensitizing adipokine (Cao et al., 2008). Quantitative PCR assays confirmed significant up-regulation of Fads2, Elovl5 and Scd2 (Fig. 5A).
Figure 5.
Impact of NCoR KO on LXR target gene expression in macrophages. (A) Q-PCR analysis of the indicated LXR target genes in WT and MNKO macrophages. (B) Venn diagram of overlap between LXRβ and NCoR peaks in macrophages. (C) Sequence motifs associated with LXRβ and NCoR cobound sites. (D) Distribution of NCoR ChIP-Seq tag density in the vicinity of LXRβ binding sites. (E) LXR-dependent Abca1-luciferase reporter assays in WT and MNKO macrophages. (F) Distribution of H4K5ac ChIP-Seq tag density in the vicinity of LXRβ binding sites. (G) UCSC genome browser image illustrating normalized tag counts for H4K5Ac ChIP-Seq, GRO-Seq, LXRβ ChIP-Seq and NCoR ChIP-Seq (Barish et al., 2012) at the Scd2 locus. Values are expressed as mean ± s.e.m. * P<0.05, ** P<0.01 for KO versus WT.
Prior ChIP-Seq studies of the LXRβ cistrome in macrophages indicate that several of these genes are direct targets of liver X receptors (LXRs) (Heinz et al., 2010). Studies in LXR-deficient mice demonstrated that, in the unliganded state, LXRs mediate active repression at select LXR target genes dependent on interaction with NCoR (Wagner et al., MCB 2003). Consistent with these prior findings,, a significant fraction of known LXR target genes are derepressed in MNKO cells, including Abca1 and Fasn (Fig. 5A), suggesting that up-regulation of LXR target genes involved in fatty acid metabolism in MNKO macrophages could be a consequence of locus-specific de-repression of LXR. In agreement with this possibility, overlap of ChIP-Seq data revealed a high degree of co-localization of NCoR with LXRβ binding sites in the macrophage (Fig. 5B). Given that motif analysis verifies these sites as likely bona fide LXR binding sites (Fig. 5C), and that NCoR tag densities demonstrating most significant enrichment precisely centered on LXRβ peaks (Fig. 5D), it is likely that loss of NCoR would have a significant role at many of these loci containing LXR-NCoR complexes normally poised for active repression under basal conditions. Consistent with this, direct measure of LXR complex activity by use of luciferase reporter constructs, revealed that MNKO macrophages demonstrated significantly higher LXR reporter activity from a construct driven by LXR-binding sites from the Abca1 locus (Fig. 5E). Further, LXR binding sites were associated with increased H4K5ac in MNKO macrophages (Fig. 5F) and increased expression of nearby genes, exemplified by Scd2 (Fig. 5G, 2nd track). Accordingly, the Scd2 locus enhancer region occupied by LXR and NCoR (Fig. 5G, tracks 3&4) indicates a marked increase in H4K5ac (Fig. 5G, upper track). Collectively, these data indicate that one significant consequence of NCoR deletion in the macrophage is de-repression of LXR target genes, including those that control fatty acid metabolism.
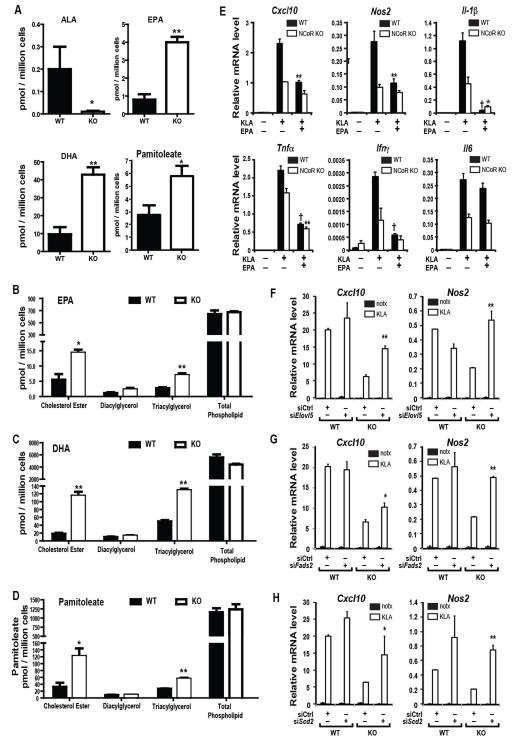
These findings suggested that NCoR KO macrophages might exhibit reprogramming of fatty acid metabolism, generating lipid products with anti-inflammatory effects. To assess this, we performed a lipidomic analysis on WT and NCoR KO IP Macs (Fig. 6A). 18:3n3 acid (ALA) is a precursor for the biosynthetic pathway, containing the gene products of Fads1, Fads2, and Elovl5, whose enzymatic activities enable production of the long chain ω3 FAs 20:5n3(EPA) and 22:6n3(DHA). Importantly, ALA concentrations are markedly decreased in the NCoR KO macrophages, whereas, levels of EPA and DHA were 4-6-fold increased (Fig. 6A). The potent anti-inflammatory effects of ω3 FAs are well known (Mori and Beilin, 2004; Oh et al., 2010), providing a potential mechanistic explanation for the decreased overall inflammatory tone of the MNKO mice. Furthermore, Cao et al. (Cao et al., 2008) identified palmitoleic acid (16:1n7, POA) as a “lipokine” which can exert insulin-like actions. In addition, recent studies have demonstrated that POA exhibits potent anti-inflammatory activity in macrophages (Spann et al., 2012). In line with increased expression of Scd1 and Scd2, the levels of POA were also 2-3-fold increased in the MNKO macrophages (Fig. 6A). We also measured EPA, DHA, and POA concentrations within each of the major lipid classes, finding increases in the cholesterol ester and triacylglycerol fractions, but not total phospholipids (Figs. 6B-D).
Figure 6.
Increased ω3 fatty acid in IP-macrophages from MNKO mice. (A) ALA, EPA, DHA and Pamitoleic acid content in vehicle or KLA-treated IP-macrophages from WT and MNKO mice. (B) EPA level, DHA level (C), and pamitoleate level (D) in different lipid fractions in IP-macrophages from WT and MNKO mice. (E) Effects of ω3 fatty acid treatment on inflammatory gene expression in KLA-treated WT and MNKO IP-macrophages. (F) KLA-induced inflammatory gene expression in IP-macrophages after siRNA knockdown of Elovl5, Fads2 (G), or Scd2 (H). Values are expressed as mean ± s.e.m. * P<0.05, ** P<0.01, † P<0.001 for KO versus WT, or EPA-KLA versus KLA in 6E, or KLA treated target specific siRNA (Scd2, Elovl5, or Fads2) versus siCTL in 6F-H. See also Fig. S3.
No appreciable differences in circulating EPA, DHA, or POA levels were observed in MNKO mice compared to WT mice (data not shown), suggesting that the biological consequences of increased production of these lipid species would most likely result from autocrine or paracrine mechanisms. To investigate this possibility, we first evaluated the impact of EPA on TLR responses in macrophages derived from WT and MNKO animals. As expected, EPA treatment strongly suppressed the KLA response of Cxcl10, Nos2, Il1b, Tnf, and Ifng in WT macrophages (Fig. 6E). In MNKO macrophages, responses of Cxcl10, Nos2, Il1b, Tnf, and Ifng to TLR4 ligation were reduced, as previously noted. With the exception of IL6, the inhibitory effects of EPA were much less robust in MNKO versus WT macrophages, consistent with increased endogenous levels of ω3 FAs in the MNKO macrophages.
To further investigate roles of endogenous POA and ω3 FAs in determining the MNKO phenotype, we used an RNAi approach to knockdown Fads2, Elovl5, and Scd2 in both WT and MNKO macrophages. Knockdown of these enzymes responsible for the production of POA and ω3 FAs (Fig. S3), led to increased KLA stimulated inflammatory gene expression of MNKO hypo-responsive genes, exemplified by Cxcl10 and Nos2, (Figs. 6F-H). The knockdowns demonstrated direct target specificity, as known LXR target genes such as Abca1 and Abcg1 were not reduced by the siRNAs, relative to the specific target genes Fads2, Elovl5, and Scd2 (Fig. S3A-C). Further, the siRNA-mediated derepression of TLR target genes was mainly selective for the hypo-responsive genes in the MNKO cells. Thus, many of the KLA-stimulatd genes, either hyper-responsive or unaffected by loss of NCoR, failed to exhibit the same increases with Fads or Scd2 knockdown (Fig. S3B-C). Taken together, these data are consistent with the hypothesis that NCoR depletion de-represses LXR, leading to increased expression of select lipid metabolic target genes in the POA and ω3 FA biosynthetic pathways.
ω3 FAs suppress transcriptional outputs of NF-κB regulatory elements
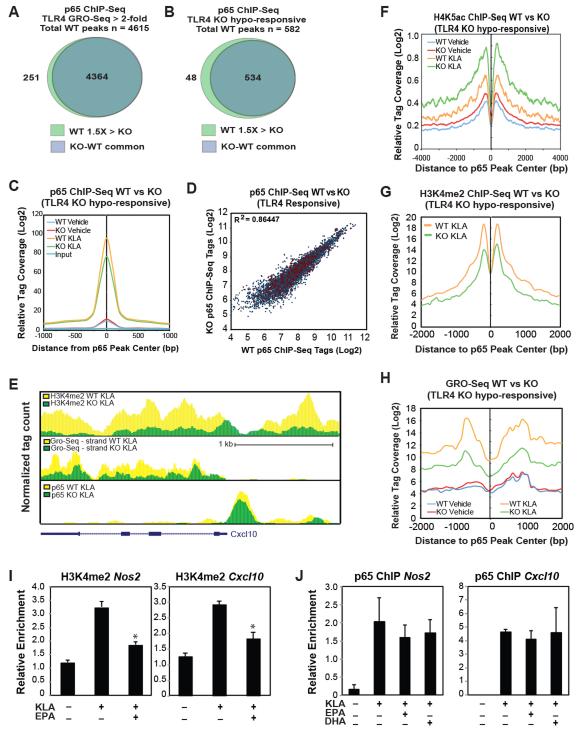
To specifically address the relationship of the hyporesponsive phenotype of MNKO macrophages to NF-κB-dependent gene expression, we assessed NF-κB-driven reporter gene activity in MNKO and WT macrophages. Reporter activity of constructs, driven by either NF-κB-bound enhancer regions from MNKO hypo-responsive genes (Nos2 and Cxcl10) or a consensus NF-κB response element, exhibited significantly reduced activity in MNKO versus WT macrophages (Fig. S4A). Next we performed ChIP-Seq measurements of the p65 component of NF-κB to determine whether reduced NF-κB activity was due to a reduction in DNA binding. Unexpectedly, ChIP-Seq analysis revealed a very similar pattern of p65 binding following KLA stimulation in both WT and MNKO macrophages. Motif analysis confirmed these as bona fide p65 binding sites, with highest enrichment for NF-κB motifs, as well as motifs for macrophage lineage-determining factors PU.1, AP-1, and C/EBP (Fig. S4B). The similarity in p65 binding was observed not only for the entire set of p65 binding sites associated with TLR4 responsive genes induced >2-fold in WT macrophages (Fig. 7A), but also for the subset of p65 binding sites associated with MNKO hypo-responsive loci (Fig. 7B-D), exemplified by the Cxcl10 locus (Fig. 7E). These sites were also associated with increased H4K5ac following TLR4 stimulation (Fig. 7F), indicating that binding of NF-κB resulted in recruitment of histone acetyltransferases. These findings indicate that much of the reduction in expression of NF-κB target genes in MNKO macrophages is independent of changes in NF-κB binding and occurs at a step subsequent to histone acetylation. Recent studies have implicated histone H3 lysine 4 mono- and dimethylation (H3K4me1 and H3K4me2) as additional indicators of active local transcription (Ng et al., 2003; Ostuni et al., 2013). In an effort to resolve the paradox of increased histone acetylation and undiminished p65 binding at hypo-responsive genes in the MNKO cells, we performed ChIP-Seq analysis of H3K4me2 in WT and MNKO macrophages. These studies revealed that MNKO macrophages demonstrated significantly reduced levels of TLR4-dependent H3K4me2 deposition at hypo-responsive genes, as shown for the Cxcl10 locus (Fig. 7E). This pattern of reduced H3K4me2 deposition, at both p65-bound enhancer elements and within the associated gene bodies was observed for the entire set of hypo-responsive genes (Fig. 7G and S4C). Conversely, genes that are de-repressed in MNKO macrophages exhibit increased H3K4me2 levels, exemplified by the Mmp12 locus (Fig. S4D).
Figure 7.
Increased ω3 FA production leads to decreased NF-κB activity in IP-macrophages of MNKO mice. (A) Venn diagram of overlap between p65 peaks within 50kb of TLR4 responsive genes (WT fold change >2, RPKM> 0.5, FDR<0.01) in KLA-treated WT and MNKO macrophages. (B) Venn diagram of overlap between p65 peaks within 50kb of MNKO hypo-responsive genes in KLA-treated WT and MNKO macrophages. (C) Normalized distribution of for p65 ChIP-Seq tag counts at enhancer-like regions associated with MNKO hypo-responsive genes (defined in 7B) in vehicle or KLA-treated WT and MNKO macrophages. (D) Scatter plot of normalized tag counts for p65 at enhancer-like regions associated with TLR4 responsive genes (defined in 7A) in KLA-treated MNKO and WT macrophages. The p65 peaks corresponding to hypo-responsive genes in MNKO macrophages (defined in 7B) are highlighted in red. The Pearson’s correlation coefficient is reported in the figure (p<0.0001). (E) UCSC genome browser image illustrating normalized tag counts for H3K4me2 ChIP-Seq, GRO-Seq, p65 ChIP-Seq, and NCoR ChIP-Seq (from Barish, et al, 2012) in WT and MNKO macrophages for the Cxcl10 genomic locus as indicated. (F) Normalized distribution of H4K5Ac ChIP-Seq tag density in the vicinity of p65 binding sites at enhancers associated with MNKO hypo-responsive genes (defined in 7B) in vehicle or KLA-treated WT and MNKO macrophages. (G) Normalized distribution of H3K4me2 ChIP-Seq tag density in the vicinity of p65 binding sites at enhancers associated with MNKO hypo-responsive genes (defined in 7B) in KLA-treated cells. (H) Normalized distribution of GRO-Seq tag counts in the vicinity of p65 peaks associated with MNKO hypo-responsive genes (defined in 7B) in vehicle or KLA-treated cells. (I) ChIP for H3K4me2 at Nos2 and Cxcl10 loci in WT macrophages treated with KLA in the presence or absence of EPA. (J) ChIP for p65 at Nos2 and Cxcl10 loci in WT macrophages treated with KLA in the presence or absence of EPA. Values are expressed as mean ± s.e.m. * P<0.05, ** P<0.01 for KLA versus EPA-KLA. See also Fig. S4.
In addition to H3K4 methylation, recent studies have demonstrated that enhancers direct the expression of eRNAs that are linked to enhancer activity (Lai et al., 2013). Because GRO-Seq analysis not only identifies nascent mRNA transcripts, but also enables quantification of eRNAs, we also evaluated eRNA expression at p65 binding sites associated with hypo-responsive genes. This analysis indicated significantly reduced eRNA production in MNKO-derived macrophages following TLR4 activation (Fig. 7H). Interestingly, in contrast to these defects in the NF-κB transcriptional pathway, we found no differences in JNK activity between WT and MNKO macrophages (Fig. S4E), consistent with both the transcriptomic results and the view that the primary effects of ω3 FAs in the MNKO cells are intracellular. Together these findings provide evidence that loss of NCoR leads to uncoupling of p65 binding and local TLR-dependent of histone acetylation from histone methylation and eRNA production, resulting in reduced signal-responsive gene expression.
To directly link these observations to effects of ω3 FAs on NF-κB activity, we evaluated effects of EPA and DHA on NF-κB-driven reporter genes in primary WT macrophages. These experiments demonstrated the expected decrease in reporter gene activity (Fig. S4F). Importantly, ChIP assays indicated that ω3-treatment of WT macrophages resulted in decreased TLR-dependent deposition of H3K4me2 (Fig. 7I) with no changes in p65 binding (Fig. 7J) at p65 binding sites in enhancers associated with the Nos2 and Cxcl10 genes. These observations suggest that ω3 fatty acids suppress the transcriptional functions of p65 at a step downstream of NF-κB binding.
DISCUSSION
Here we present evidence that deletion of NCoR from macrophages results in an anti-inflammatory phenotype with protection from systemic insulin resistance in the setting of HFD-induced obesity. This surprising phenotype in the MNKO mice was manifested by decreased macrophage accumulation in adipose tissue, decreased expression of inflammatory pathway genes and reduced cytokine protein levels. In addition, we observed improved glucose tolerance, as well as systemic insulin sensitivity, as measured by the hyperinsulinemic euglycemic clamp method, in these animals. Interestingly, treatment with rosiglitazone had little further effect to augment insulin sensitivity in the MNKO mice, possibly because they were already insulin sensitive in the untreated state, or because the anti-inflammatory effects attributable to rosiglitazone (Bouhlel et al., 2007; Odegaard et al., 2007) require NCoR (Pascual et al., 2005). This improvement in inflammatory status and insulin sensitivity reinforces the connection between macrophage-mediated tissue inflammation and insulin resistance. In vitro studies further demonstrated that while some inflammatory response genes were up-regulated in MNKO cells, a substantially larger number exhibited hypo-responsiveness to TLR4 ligation.
NCoR and the related protein SMRT were initially discovered based on their functions as co-repressors of retinoic acid and thyroid hormone receptors (Chen and Evans, 1995; Horlein et al., 1995). Subsequent studies extended these co-repressor functions to additional nuclear receptors, including LXRs (Hu et al., 2003), as well as a number of signal-dependent transcription factors, such as AP-1, NF-κB, and Ets (Mottis et al., 2013) The co-repressor activities of NCoR and SMRT are based on their ability to serve as platforms for the assembly of large co-repressor complexes, with a core NCoR/SMRT complex containing Tbl1, TblR1, HDAC3, and GPS2 (Rosenfeld et al., 2006). HDAC3 contributes directly to the co-repressor activity of NCoR/SMRT complexes by catalyzing the removal of histone acetylation marks that are involved in transcriptional activation (Codina et al., 2005).
Genome wide analysis confirmed increased H4K5 acetylation in MNKO cells at genome sites where NCoR is bound in WT cells, consistent with loss of recruitment of HDAC3 due to the NCoR deletion. At these sites, acetylation of histone H4 at lysine residues is linked to recruitment of pTEFb, which is associated with increased transcriptional elongation (Yang et al., 2005). However, increased H4K5ac was observed not only at genes exhibiting increased expression in MNKO cells, but also at the large subset of TLR4-responsive genes exhibiting impaired KLA stimulation (hypo-responsive gene set, Fig 4E). In addition, our p65 ChIP-Seq results showed unimpaired ongoing p65 occupancy of the promoters/enhancers for these hypo-responsive genes. Thus, even when NF-κB binds to its target genes at hyperacetylated sites, its transcriptional functions are compromised in NCoR deficient macrophages. These observations indicate a paradoxical uncoupling of histone acetylation and p65 DNA binding from transcriptional activation.
Investigation of possible causes of the hypo-responsive phenotype of MNKO macrophages led to the finding of de-repression of LXR target genes involved in generation of anti-inflammatory fatty acids, including long chain ω3-FAs and POA. Thus, NCoR can bind to LXR, mediating basal transcription repression at NCoR/LXR binding sites. In the absence of NCoR, LXR is de-repressed, leading to increased transcription of these targets, which include the lipogenic genes Elovl5, Fads1, Fads2, Scd1 and Scd2. These lipogenic genes drive the biosynthesis of ω3 FAs and POA within macrophages, effectively reprogramming macrophage lipid metabolism. De-repression of these genes was not observed in fetal liver-derived macrophages from systemic NCoR KO mice (Ghisletti et al., 2009), which may account for the relatively activated phenotype reported for those cells. Administration of these ω3 FAs was less effective at inhibiting TLR4 responses in MNKO cells, consistent with the elevated endogenous levels. Conversely, knockdown of the lipogenic enzymes responsible for their production partially restored inflammatory responses in NCoR KO cells. These results support the conclusion that increased expression of anti-inflammatory FAs contribute to the hypo-responsive phenotype of MNKO cells. The relative importance of anti-inflammatory FAs appears to vary in a gene-specific manner, indicating that additional mechanisms contribute.
Genome-wide and locus-specific analyses indicate that ω3 FAs provide at least two levels of suppression of inflammatory response genes. ω3 FAs exert anti-inflammatory effects by serving as agonists for GPR120 on macrophages (Oh et al., 2010). GPR120 ligation inhibits proinflammatory signaling by blocking activation of TAK1 with reduced downstream signaling to IKKβ/NF-κB (Oh et al., 2010). This, in turn would result in reduced translocation of NF-κB to the nucleus. Consistent with this prediction, reduced occupancy of p65 was observed on a subset of inflammatory gene promoters and enhancers in the NCoR deficient macrophages and in WT macrophages treated with ω3 FAs. However, this effect was quantitative and locus-specific, with the great majority of TLR4 hypo-responsive genes remaining largely or fully occupied by p65. Consistent with this, we found no decrease in JNK activity in the MNKO macrophages compared to WT. This indicates that in MNKO cells, the major effects of endogenously produced ω3 FAs are intracellular and largely independent of GPR120. Furthermore, these sites of p65 binding exhibited increased H4K5ac, which requires the active recruitment of histone acetyltransferases such as CBP and p300 by active NF-κB. In contrast, H3K4 dimethylation and enhancer RNA production were consistently inhibited at genomic regulatory elements associated with the hypo-responsive genes. Previous studies have demonstrated local deposition of H3K4me2 as a valid measure of signal-responsive transcription factor activity (He et al., 2010). Further, signal dependent inductions of H3K4me2 and transcriptional output from local transcription factor activated enhancers, reflects the increased induction of associated genes (He et al., 2010; Ostuni et al., 2013). Reduced H3K4me2 levels were further correlated with reduced eRNA production at p65 enhancers associated with hypo-responsive genes, further indicative of a loss of transcriptional output of NF-κB at these locations. Thus, while histone acetylation and p65 DNA binding are required for activation, they are not sufficient to promote transcriptional activation of the hypo-responsive inflammatory gene set in the context of NCoR deletion or ω3 FA treatment.
Together, the present studies provide evidence in support of two main conclusions. First, these observations suggest that macrophages can be a source of sufficient ω3 FAs and POA to exert autocrine and/or paracrine effects that result in improved insulin sensitivity. This concept is also exemplified by our findings that the normal effects of CM harvested from WT macrophages to inhibit insulin-stimulated glucose transport do not occur using CM from MNKO macrophages. That these ω3 FAs effects are local is supported by our data demonstrating increased levels of ω3 FAs in macrophages, without changes in systemic concentrations. This concept raises the possibility that treatments, which could reprogram macrophage lipid metabolism into the more metabolically favorable state characteristic of NCoR macrophage KO mice, could have important therapeutic effects to reduce tissue inflammation and promote insulin sensitivity. Second, these findings reveal a new molecular function of unsaturated ω3 FAs by which they reduce H3K4 methylation, enhancer transcription, and mRNA expression at regulatory elements that control inflammatory response genes and thereby establish a hypo-responsive state. These effects are due to reduced NF-κB complex activity independent of effects on NF-κB localization itself. Identification of the molecular targets and mechanisms responsible for these effects could also lead to development of new approaches for diseases characterized by pathogenic forms of inflammation.
METHODS
Animal care and use
Animals were maintained on a 12 h/12 h light/dark cycle with free access to food and water. All animal procedures were in accordance with University of California San Diego research guidelines for the care and use of laboratory animals.
Creation of control and monocyte/neutrophil specific NCoR KO mice
NCoRfl/fl mice were generated as described previously (Li et al., 2011). To generate macrophage/neutrophil specific NCoR null mice, NCoRfl/fl were bred with transgenic mice harboring Cre recombinase driven by myeloid-specific lysozyme M promoter (Clausen et al., 1999) to create the following genotypes: NCoRfl/fl (control), NCoRfl/fl-LysMCre (MNKO). These mice were backcrossed to the C57BL/6J strain for more than 10 generations.
Lipid Measurements
Thioglycollate-elicited peritoneal macrophages were plated at 2.0 × 106 cells per well of a 6-well plate in growth medium containing RPMI 1640 (with L-glutamine) (Invitrogen) plus 10% heat-inactivated FBS (Hyclone). After 24 hr cells were washed twice with PBS and switched to culture medium containing RPMI 1640 (Invitrogen) without serum and supplemented with fatty-acid free BSA (sigma) for 6 hr. Media and cells were collected for detection and quantitation of lipids by mass spectrometric methods as described previously (Brown et al., 2007).
Lipid fraction analyses in macrophages
The lipids from thioglycollate-elicited macrophages were extracted in the presence of authentic internal standards by the method of Folch et al. (Folch et al., 1957). Using chloroform:methanol (2:1 v/v), individual lipid classes within each extract were separated by liquid chromatography (Agilent Technologies model 1100 series). Each lipid class was trans-esterified in 1% sulfuric acid in methanol in a sealed vial under a nitrogen atmosphere at 100°C for 45 min. The resulting fatty acid methyl esters were extracted from the mixture with hexane containing 0.05% butylated hydroxytoluene and prepared for gas chromatography by sealing the hexane extracts under nitrogen. Fatty acid methyl esters were separated and quantified by capillary gas chromatography (Agilent Technologies model 6890) equipped with a 30 m DB-88MS capillary column (Agilent Technologies) and a flame-ionization detector.
High Throughput Sequencing and Data Analysis
ChIP fragments were sequenced for 36 or 50 cycles on an Illumina Genome Analyzer or HiSeq 2000, respectively, according to the manufacturer’s instructions. GRO-Seq results were trimmed to remove A-stretches originating from the library preparation. Each sequence tag returned by the Illumina Pipeline was aligned to the mm9 assembly using ELAND allowing up to 2 mismatches. Only tags that mapped uniquely to the genome were considered for further analysis.
Data analysis was performed using HOMER and the detailed instructions for analysis can be found at http://biowhat.ucsd.edu/homer/ or as previously described in (Heinz et al., 2010). Each sequencing experiment was normalized to a total of 107 uniquely mapped tags by adjusting the number of tags at each position in the genome to the correct fractional amount given the total tags mapped. Sequencing experiments were visualized by preparing custom tracks for the UCSC Genome browser. GRO-Seq density within gene bodies was determined by adding the number of tags within the gene body defined by RefSeq and normalizing by the length of the gene. Differentially expressed genes were identified as described previously (Escoubet-Lozach et al., 2011), using log values of gene body tag densities to calculate indicated fold change thresholds. For gene ontology analysis DAVID Bioinformatics Resources 6.7 was used.
Other methods
Statistical analyses
Statistical analyses were performed using Excel or GraphPad Prism 5. The images were prepared using Photoshop CS5.1 or GraphPad Prism 5. Data are presented as the mean ± SEM For experiments involving two factors, data were analyzed by two-way ANOVA followed by Bonferroni post tests. Individual pair-wise comparisons were performed using student t test. The p value < 0.05 was considered significant.
Supplementary Material
HIGHLIGHTS.
Macrophage NCoR KO mice are protected from HFD-induced insulin resistance.
NCoR deletion results in a paradoxical hypo-responsiveness to TLR4 signaling.
De-repression of LXRs results in increased anti-inflammatory ω3 fatty acids.
ω3 fatty acids inhibit enhancer/promoter activation at hypo-responsive genes.
ACKNOWLEDGEMENTS
We thank the Flow Cytometry Resource (Dennis Young) for FACS analysis at the Rebecca & John Moores Cancer Center and thank Jachelle M. Ofrecio for genotyping at UCSD. This study was funded in part by grants to J M O (DK033651, DK074868, DK-063491, DK-09062), to C K G (DK074868, GM069338), and to J A (the Ecole Polytechnique Federale de Lausanne, the Swiss National Science Foundation, and the EU Ideas program (ERC-AdG-23118)), and the Eunice Kennedy Shriver NICHD/NIH through a Cooperative Centers Program in Reproduction and Infertility Research (JMO).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barish GD, Yu RT, Karunasiri MS, Becerra D, Kim J, Tseng TW, Tai LJ, Leblanc M, Diehl C, Cerchietti L, et al. The Bcl6-SMRT/NCoR cistrome represses inflammation to attenuate atherosclerosis. Cell Metab. 2012;15:554–562. doi: 10.1016/j.cmet.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhlel MA, Derudas B, Rigamonti E, Dievart R, Brozek J, Haulon S, Zawadzki C, Jude B, Torpier G, Marx N, et al. PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 2007;6:137–143. doi: 10.1016/j.cmet.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Brown HA, Henage LG, Preininger AM, Xiang Y, Exton JH. Biochemical analysis of phospholipase D. Methods in enzymology. 2007;434:49–87. doi: 10.1016/S0076-6879(07)34004-4. [DOI] [PubMed] [Google Scholar]
- Cao H, Gerhold K, Mayers JR, Wiest MM, Watkins SM, Hotamisligil GS. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell. 2008;134:933–944. doi: 10.1016/j.cell.2008.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- Codina A, Love JD, Li Y, Lazar MA, Neuhaus D, Schwabe JW. Structural insights into the interaction and activation of histone deacetylase 3 by nuclear receptor corepressors. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:6009–6014. doi: 10.1073/pnas.0500299102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoubet-Lozach L, Benner C, Kaikkonen MU, Lozach J, Heinz S, Spann NJ, Crotti A, Stender J, Ghisletti S, Reichart D, et al. Mechanisms establishing TLR4-responsive activation states of inflammatory response genes. PLoS genetics. 2011;7:e1002401. doi: 10.1371/journal.pgen.1002401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG, Glass CK. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes & development. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Ogawa S. Combinatorial roles of nuclear receptors in inflammation and immunity. Nat Rev Immunol. 2006;6:44–55. doi: 10.1038/nri1748. [DOI] [PubMed] [Google Scholar]
- Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol. 2010;10:365–376. doi: 10.1038/nri2748. [DOI] [PubMed] [Google Scholar]
- Haffner S, Taegtmeyer H. Epidemic obesity and the metabolic syndrome. Circulation. 2003;108:1541–1545. doi: 10.1161/01.CIR.0000088845.17586.EC. [DOI] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevener AL, Olefsky JM, Reichart D, Nguyen MT, Bandyopadyhay G, Leung HY, Watt MJ, Benner C, Febbraio MA, Nguyen AK, et al. Macrophage PPAR gamma is required for normal skeletal muscle and hepatic insulin sensitivity and full antidiabetic effects of thiazolidinediones. J Clin Invest. 2007;117:1658–1669. doi: 10.1172/JCI31561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Hu X, Li S, Wu J, Xia C, Lala DS. Liver X receptors interact with corepressors to regulate gene expression. Mol Endocrinol. 2003;17:1019–1026. doi: 10.1210/me.2002-0399. [DOI] [PubMed] [Google Scholar]
- Jager J, Gremeaux T, Cormont M, Le Marchand-Brustel Y, Tanti JF. Interleukin-1beta-induced insulin resistance in adipocytes through down-regulation of insulin receptor substrate-1 expression. Endocrinology. 2007;148:241–251. doi: 10.1210/en.2006-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintscher U, Hartge M, Hess K, Foryst-Ludwig A, Clemenz M, Wabitsch M, Fischer-Posovszky P, Barth TF, Dragun D, Skurk T, et al. T-lymphocyte infiltration in visceral adipose tissue: a primary event in adipose tissue inflammation and the development of obesity-mediated insulin resistance. Arterioscler Thromb Vasc Biol. 2008;28:1304–1310. doi: 10.1161/ATVBAHA.108.165100. [DOI] [PubMed] [Google Scholar]
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanthier N, Molendi-Coste O, Horsmans Y, van Rooijen N, Cani PD, Leclercq IA. Kupffer cell activation is a causal factor for hepatic insulin resistance. Am J Physiol Gastrointest Liver Physiol. 2009;298:G107–116. doi: 10.1152/ajpgi.00391.2009. [DOI] [PubMed] [Google Scholar]
- Li P, Fan W, Xu J, Lu M, Yamamoto H, Auwerx J, Sears DD, Talukdar S, Oh D, Chen A, et al. Adipocyte NCoR knockout decreases PPARgamma phosphorylation and enhances PPARgamma activity and insulin sensitivity. Cell. 2011;147:815–826. doi: 10.1016/j.cell.2011.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- Mori TA, Beilin LJ. Omega-3 fatty acids and inflammation. Curr Atheroscler Rep. 2004;6:461–467. doi: 10.1007/s11883-004-0087-5. [DOI] [PubMed] [Google Scholar]
- Mottis A, Mouchiroud L, Auwerx J. Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Genes Dev. 2013;27:819–835. doi: 10.1101/gad.214023.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Molecular Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, Red Eagle A, Vats D, Brombacher F, Ferrante AW, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, Jepsen K, Sawka-Verhelle D, Perissi V, Sasik R, Rose DW, Johnson RS, Rosenfeld MG, Glass CK. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proc Natl Acad Sci U S A. 2004;101:14461–14466. doi: 10.1073/pnas.0405786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh DY, Talukdar S, Bae EJ, Imamura T, Morinaga H, Fan W, Li P, Lu WJ, Watkins SM, Olefsky JM. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell. 2010;142:687–698. doi: 10.1016/j.cell.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaven GM. The insulin resistance syndrome: definition and dietary approaches to treatment. Annu Rev Nutr. 2005;25:391–406. doi: 10.1146/annurev.nutr.24.012003.132155. [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Rotter V, Nagaev I, Smith U. Interleukin-6 (IL-6) induces insulin resistance in 3T3-L1 adipocytes and is, like IL-8 and tumor necrosis factor-alpha, overexpressed in human fat cells from insulin-resistant subjects. J Biol Chem. 2003;278:45777–45784. doi: 10.1074/jbc.M301977200. [DOI] [PubMed] [Google Scholar]
- Shoelson SE, Herrero L, Naaz A. Obesity, inflammation, and insulin resistance. Gastroenterology. 2007;132:2169–2180. doi: 10.1053/j.gastro.2007.03.059. [DOI] [PubMed] [Google Scholar]
- Spann NJ, Garmire LX, McDonald JG, Myers DS, Milne SB, Shibata N, Reichart D, Fox JN, Shaked I, Heudobler D, et al. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell. 2012;151:138–152. doi: 10.1016/j.cell.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Molofsky AB, Liang HE, Ricardo-Gonzalez RR, Jouihan HA, Bando JK, Chawla A, Locksley RM. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science. 2011;332:243–247. doi: 10.1126/science.1201475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Williams EG, Mouchiroud L, Canto C, Fan W, Downes M, Heligon C, Barish GD, Desvergne B, Evans RM, et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell. 2011;147:827–839. doi: 10.1016/j.cell.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Youm YH, Vandanmagsar B, Ravussin A, Gimble JM, Greenway F, Stephens JM, Mynatt RL, Dixit VD. Obesity increases the production of proinflammatory mediators from adipose tissue T cells and compromises TCR repertoire diversity: implications for systemic inflammation and insulin resistance. J Immunol. 2010;185:1836–1845. doi: 10.4049/jimmunol.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.