Abstract
The suprachiasmatic nucleus (SCN) of the hypothalamus is the principal circadian pacemaker in mammals, coordinating daily metabolic and physiological rhythms with the cycle of sleep and wakefulness. SCN neurons define circadian time via an auto-regulatory feedback loop in which the activation of Period (Per) and Cryptochrome genes is periodically suppressed by their own protein products. Casein kinase 1 (CK1) enzymes have a critical role in circadian pacemaking because they phosphorylate PER proteins and thereby direct their proteasomal degradation. In human pedigrees, individual mutations in either hCK1 or hPER2 lead to advanced sleep phase disorders, whereas in rodents, the Tau mutation of CK1 epsilon (CK1ϵTau) accelerates rest-activity cycles and shortens the period of the SCN molecular pacemaker. Biochemical analyses of recombinant PER proteins in cultured cells and endogenous proteins in peripheral tissues have identified PER1 and PER2, but not PER3, as direct substrates of CK1ϵ. The purpose of this study, therefore, was to determine the relative contributions of endogenous PER proteins to the period-accelerating effects of CK1ϵTau, both in vivo and in vitro. CK1ϵTau mice were mated onto Per1-, Per2-, and Per1-Per2 (Per1/2) double-null backgrounds, in all cases carrying the Per1-luciferase bioluminescent circadian reporter gene. Mice lacking both PER1 and PER2 were behaviorally arrhythmic, confirming the inadequacy of PER3 as a circadian factor. Individual loss of either PER1 or PER2 had no significant effect on the circadian period or quality of wheel-running behavior, and CK1ϵTau accelerated behavioral rhythms in both Per1- and Per2-null mice. CK1ϵTau also accelerated in vitro molecular pacemaking in SCN lacking either PER1 or PER2, with a greater effect in PER2-dependent (i.e., Per1-null) SCN than in PER1-dependent slices. In double-null slices, some SCN were arrhythmic, whereas others exhibited transient rhythms, which trended nonsignificantly toward a shorter period. Both short-period and long-period rhythms could be identified in individual SCN neurons imaged by charge-coupled device camera. CK1ϵTau had no effect, however, on SCN-level or individual neuronal rhythms in the absence of PER1 and PER2. Thus, the CK1ϵTau allele has divergent actions, acting via both endogenous PER1 and PER2, but not PER3 protein, to mediate its circadian actions in vivo. Moreover, PER-independent cellular oscillations may contribute to pacemaking, but they are unstable and imprecise, and are not affected by the Tau mutation.
Keywords: Sleep, bioluminescence, organotypic, suprachiasmatic nucleus, circadian, wheel-running, gene expression
The suprachiasmatic nucleus (SCN) of the hypothalamus is the principal circadian pacemaker in mammals, coordinating daily metabolic and physiological rhythms with the cycle of sleep and wakefulness (Reppert and Weaver, 2002). In common with other tissues, SCN neurons define circadian time via an auto-regulatory feedback loop in which the Period (Per) and Cryptochrome (Cry) genes are first activated by CLOCK-BMAL1 heterodimers acting at E-box enhancer sequences, then subsequently suppressed by their own protein products (Mohawk and Takahashi, 2011; Koike et al., 2012). Casein kinase 1 (CK1) enzymes have a critical role in circadian pacemaking because they phosphorylate PER proteins and thereby direct their proteasomal degradation (Lee et al., 2004; Gallego and Virshup, 2007). Individual mutations in either hCK1 or hPER2 lead to advanced sleep phase disorders (Jones et al., 1999; Toh et al., 2001; Xu et al., 2005), whereas in rodents the Tau mutation of CK1 epsilon (CK1ϵTau) accelerates rest-activity cycles and shortens the period of the SCN molecular pacemaker (Ralph and Menaker, 1988; Lowrey et al., 2000; Meng et al., 2008). Biochemical analyses of recombinant PER proteins in cultured cells and endogenous proteins in peripheral tissues ex vivo have identified PER1 and PER2, but not PER3, as direct substrates of CK1ϵ (Lee et al., 2004). The purpose of the current study, therefore, was to complement and extend this earlier work by examining the real-time dynamics of circadian pacemaking and determining the relative contributions of endogenous PER proteins to the period-accelerating effects of CK1ϵTau. Specifically, the aim was to examine the effects of CK1ϵTau at 2 levels: the rest-activity cycle of the whole animal and the molecular feedback loop of the SCN in culture, as revealed by rhythms of bioluminescence. A related study uncovered distinct and separable contributions of endogenous CRY1 and CRY2 to pacemaking in vivo and in vitro (Anand et al., 2013). The question here was whether similarly distinct roles for PER1 and PER2 might be revealed, and whether more dynamic measures of pacemaking at the level of SCN slices and individual neurons might identify a contribution of PER3 to the effects of CK1ϵTau. Such information would add to the growing understanding of how different elements within gene families (e.g., NPAS and CLOCK [Debruyne et al., 2006], and REV-ERBa and REV-ERBb [Cho et al., 2012]) sculpt circadian behavior and physiology.
Materials and Methods
All experiments with animals were licensed under the UK Animals (Scientific Procedures) Act of 1986 and subject to local ethical review. Mice lacking Per1 or Per2 (Bae et al., 2001) were obtained from Dr. D. Weaver (University of Massachusetts Medical School, USA). The Per1-luciferase (Per1-luc) bioluminescent reporter line (Yamaguchi et al., 2003) was provided by Dr. H. Okamura (University of Kyoto, Japan), and the CK1ϵTau line (Meng et al., 2008) was obtained from our own colony. The various lines, all on a C57Bl6 background and after at least 8 back-crosses prior to the current studies, were intercrossed to generate the required genotypes, all of which carried the Per1-luc reporter. Genotyping by PCR followed the methods published in the respective original descriptions of these mouse lines (Yamaguchi et al., 2003; Meng et al., 2008; Bae et al., 2001).
Wheel-running activity of single-housed adult male mice (aged between 8 and 20 weeks) was recorded and analyzed with Clocklab (Actimetrics, Evanston, IL, USA). Organotypic SCN slice cultures were prepared as described (Hastings et al., 2005) from pups aged between 5 and 13 days. Slices were held in culture for at least 1 week (and up to 3 weeks) prior to recording bioluminescence rhythms, either by photomultiplier tube (PMT) for whole-slice emission or by charge-coupled device (CCD) camera to monitor circadian gene expression by individual cells in the intact SCN (Maywood et al., 2006). The parameters of the bioluminescence rhythms (period, amplitude, and relative amplitude error [RAE], a measure of coherence of the rhythm, were determined using Biological Rhythms Analysis Software System (BRASS) software running on BioDARE, courtesy of Prof. A. Millar, University of Edinburgh, UK; see www.biodare.ed.ac.uk for details). Circular data were analyzed using Oriana Software (Kovach Computing Services, UK). The data from the initial 24 h of recording were not included in the analysis to avoid potential artifacts arising from transfer of the slice to fresh medium and a recording chamber. Rhythm phase was calculated relative to the start of this data set.
Results
Behavioral Rhythms
Wild-type (WT) mice presented very clear and stable rhythms of wheel-running behavior (Fig. 1A). The loss of either PER1 or PER2 did not compromise circadian behavior; all mice of both mutant lines exhibited rhythms with a quality indistinguishable from that of WT mice. In contrast, all double-mutant mice lacking both PER1 and PER2, with otherwise WT or Tau backgrounds (n = 4, 4), were unable to express circadian behavior. Rather, sporadic activity bouts were distributed among all time points, and they lacked the consolidation and coherence typical of WT and single-null mice. Thus, periodogram analysis did not generate significant peaks in the circadian domain, indicating that PER1 and PER2 are necessary but mutually redundant clock components.
Figure 1.
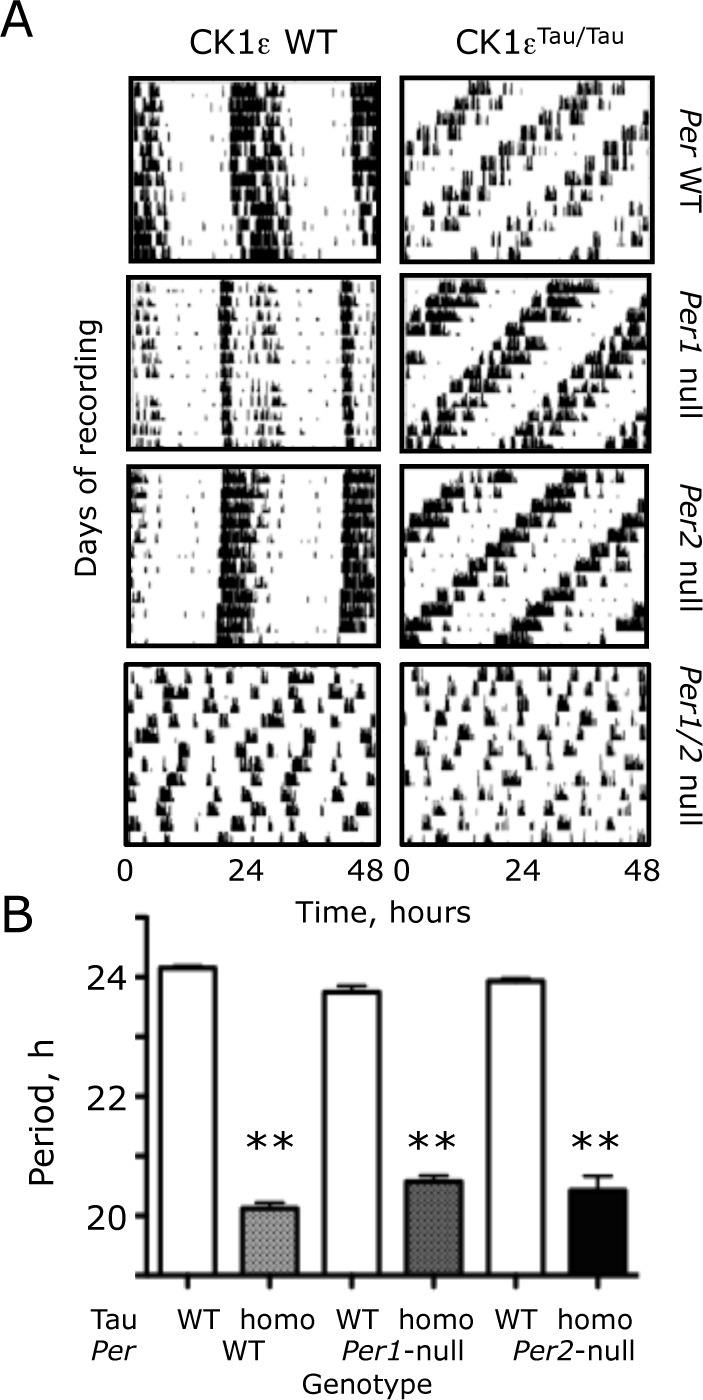
The Tau mutation of CK1 epsilon (CK1ϵTau) shortens circadian locomotor activity rhythms in Period-1 (Per1-) and Per2-null mice. (A) Representative actograms, recorded under continuous dim red light, of wild-type (WT) and CK1ϵTau/Tau mutant mice with homozygous Per1-null, Per2-null, and Per1-Per2 (Per1/2)-null backgrounds. (B) Free-running circadian period of WT, CK1ϵTau/Tau mutant, and Per-deficient mice, as in (A) (group data, mean + standard error of the mean). Note that no circadian behavior was detected in Per1/2 double-null mice. **p < 0.01 versus corresponding Tau WT, ANOVA, and Tukey post hoc.
The free-running circadian period of wheel-running behavior was shortened by 4 h in mice homozygous for the Tau mutation with an otherwise WT background (Tau WT period = 24.1 + 0.1 h [mean + standard error of the mean {SEM}]; Tau homo = 20.1 + 0.1 [n = 23, 14]) (Fig. 1B). Comparison by 2-way ANOVA of circadian periods in PER1 and PER2 single-mutant mice, with and without Tau (Per1-null n = 14, 13; Per2-null n = 10, 12), revealed a highly significant effect of Tau (F1,80 = 1909, p < 0.001) but not of Per genotype (F2,80 = 0.099, p = 0.91). There was, however, a significant interaction between Tau and Per genotypes (F2,80 = 11, p < 0.001). This interaction arose because the period-shortening effects of Tau were different among Per backgrounds. In Per1-null mice, Tau accelerated behavior by 3.2 h (23.8 + 0.1 vs. 20.6 + 0.1 h), whereas in Per2-null mice, the acceleration was 3.5 h (23.9 + 0.1 vs. 20.4 + 0.3 h). The impact of Tau was therefore attenuated by loss of either PER1 or PER2, but there was no significant difference of circadian period between Per1- and Per2-null mice carrying Tau. Thus, both PER1 and PER2 are comparable in vivo targets of Tau.
Circadian Pacemaking in PER1- and PER2-Deficient SCN Slices
The circadian rhythms of bioluminescence reported by the Per1-luc transgene in WT SCN slices were very clear and precise (Fig. 2A). Equally robust rhythms were evident in Tau WT slices lacking either PER1 or PER2. In SCN homozygous for the Tau mutation with an otherwise WT background, the circadian period of SCN was shortened by 3.7 h (WT period = 24.6 + 0.1 h; Tau homo = 20.9 + 0.2 [n = 36, 28]) (Fig. 2B). Comparison by 2-way ANOVA of circadian periods in Per1 and Per2 single-mutant mice, with and without Tau (Per1-null n = 17, 9; Per2-null n = 17, 28), revealed a highly significant effect of Tau (F1,129 = 582, p < 0.001) but not of Per genotype (F2,129 = 1.90, p = 0.15), although there was a significant interaction between Tau and Per genotypes (F2,129 = 7.60, p < 0.001). This interaction arose because the period-shortening effect of Tau was different according to Per background. In Per1-null SCN, Tau accelerated the clock by 4.5 h (24.6 + 0.1 vs. 20.1 + 0.2 h), whereas in Per2-null SCN, the acceleration was 2.9 h (24.1 + 0.1 vs. 21.2 + 0.1 h). The impact of Tau was therefore affected by loss of either PER1 or PER2, and in a PER2-dependent clock (i.e., Per1-null), Tau caused a greater acceleration than in a PER1-dependent (Per2-null) clock. Thus, the period of Per1-null–Tau homozygous SCN was significantly (p < 0.01, Bonferroni post hoc) shorter than that of the corresponding Per2-null SCN, even though there were no significant differences in the periods of Tau WT slices among the Per backgrounds. At the level of the SCN, therefore, both PER1 and PER2 are mediators of the period-shortening effect of Tau, with a greater contribution from PER2.
Figure 2.
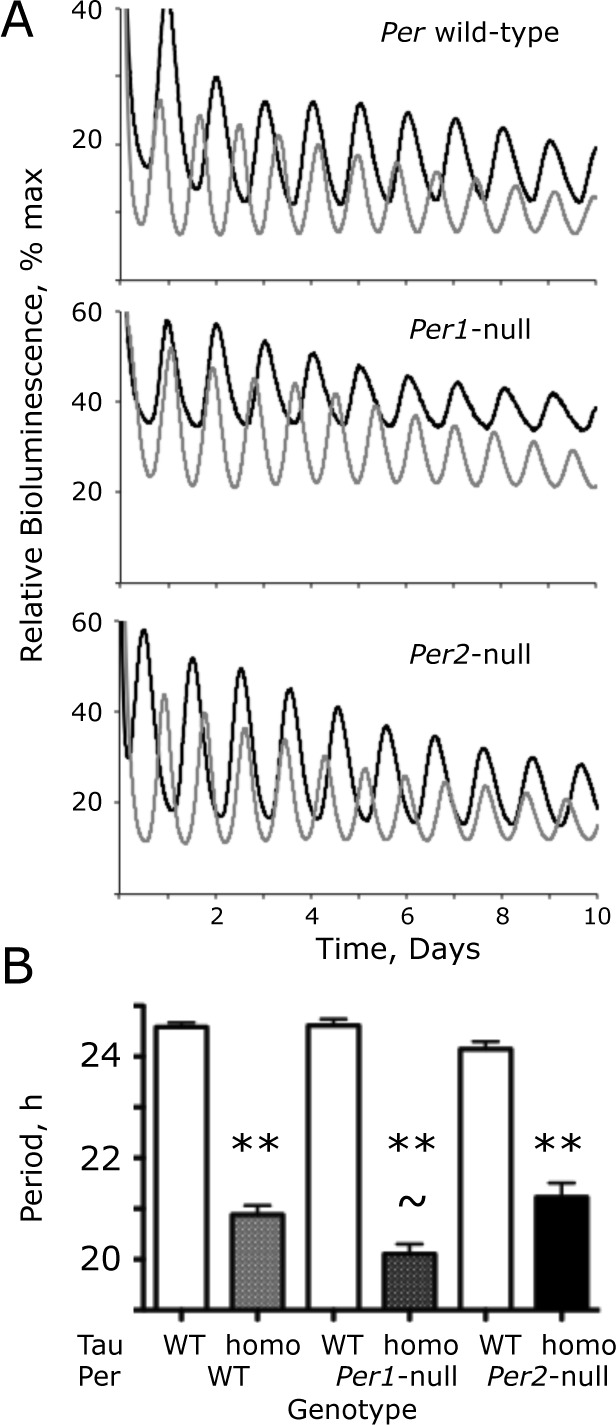
The Tau mutation of CK1 epsilon (CK1ϵTau) shortens circadian Period1-luciferase (Per1-luc) bioluminescence rhythms in Per1- and Per2-null suprachiasmatic nucleus (SCN) slices. (A) Representative recordings by photomultiplier tube (PMT) of bioluminescence rhythms from wild-type (WT) (black) and CK1ϵTau/Tau mutant (gray) SCN with Per WT, Per1-null, or Per2-null backgrounds. (B) Free-running circadian period of bioluminescence rhythms from WT, CK1ϵTau/Tau mutant, and Per-deficient SCN, as in (A) (group data, mean + standard error of the mean). **p < 0.01 versus corresponding Tau WT, ~p < 0.05 versus Per2-null, Tau homozygote, ANOVA, and Tukey post hoc.
Circadian Pacemaking in SCN Slices Deficient in Both PER1 and PER2
Mice lacking PER1 and PER2 were behaviorally arrhythmic. In generating such mice for SCN studies, slices were created that carried a single copy of either Per1 or Per2. A single Per allele was nevertheless sufficient to sustain a functional SCN clock, with a mean period of 23.8 + 0.4 h (n = 8) and amplitude of 775 + 310, neither significantly different from WT slices with 4 Per alleles (period of 24.6 + 0.1 h; amplitude = 839.5 + 219; n = 9). Their coherence was, however, significantly reduced in SCN with a single copy of Per (WT RAE = 0.036 + 0.004; single Per RAE = 0.068 + 0.011; p < 0.05 t-test). The presence of only a single Per allele did not make the SCN more sensitive to the homozygous Tau mutation (Period: Per WT = 20.6 + 0.4; single Per = 19.8 + 0.3 h, n = 7, 6). Because the single Per alleles were distributed unevenly among Tau backgrounds (the Tau WT group contained one Per2 heterozygote and 7 Per1 heterozygotes; the Tau homozygote group contained one Per1 heterozygote and 5 Per2 heterozygotes), it is not possible to make an assessment of their differential contributions. Nevertheless, the available data suggest that in the interaction between Tau and PER1/2 (for these purposes, they are considered equivalent), there was no obvious dose dependence or sensitivity to the ratio of their relative expression.
In contrast to the clear rhythms in slices carrying one Per allele, SCN slices completely lacking both PER1 and PER2 exhibited very poorly organized rhythms of Per1-luc bioluminescence, consistent with the behavioral arrhythmicity of Per1-Per2 (Per1/2) double-null mice (Fig. 3A). Closer examination did, however, reveal transient oscillations during the first few days of recording that immediately followed transfer of the slices to fresh recording medium (Fig. 3B). These tended to have a shorter period than wild-type slices but not significantly so (Per1/2-null = 22.0 + 1.6 h, n = 7, assessed during hours 24-96) (Fig. 3C). There was, however, a significantly greater incoherence (RAE = 0.147 + 0.099, p < 0.01 t-test) in the Per-null oscillations, and they progressively damped out. Although the overall level of Per1-luc bioluminescence was not significantly lower, the amplitude of oscillation was < 10% of the WT slices (50.8 + 18, p < 0.05 t-test). This reduction is too great to be accounted for by unequal segregation of hemizygous and homozygous reporter animals among genotypes: rather, it reflects a major loss of circadian control.
Figure 3.
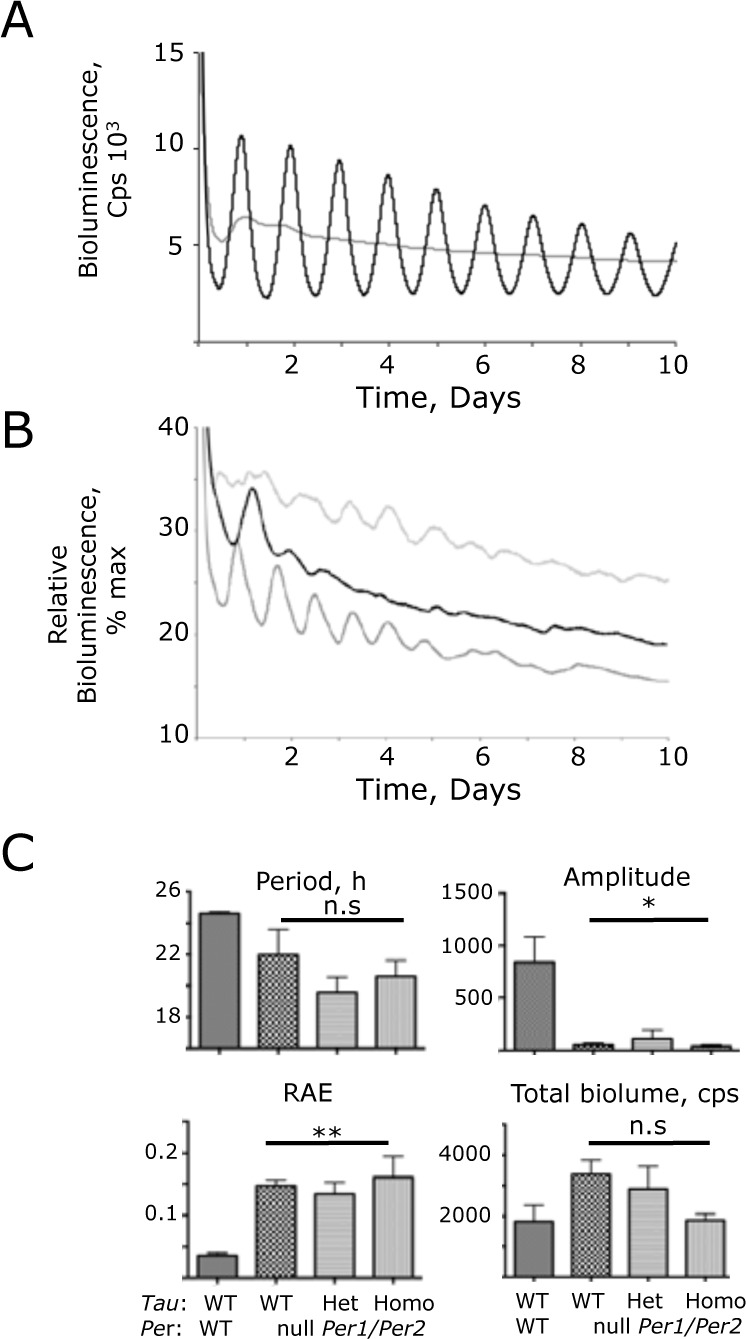
Compromised ensemble molecular pacemaking in Period 1 (Per1)- and Per2-null (Per1/2-null) suprachiasmatic nucleus (SCN), revealed by Per1-luciferase (Per1-luc) bioluminescence recording. (A) Representative bioluminescence recording by photomultiplier tube (PMT) of wild-type (WT) (black) and Per1/2-null (gray) SCN. (B) Three representative recordings from Per1/2-null SCN rescaled to reveal transient, short-period ensemble oscillations. The maximum value of the individual recording was scaled to 100, and all other counts were rescaled accordingly (i.e., % maximum). (C) Circadian parameters, determined by Biological Rhythms Analysis Software System fast Fourier transform (BRASS-FFT) of Per1/2 WT control SCN, and SCN deficient in PER1 and PER2, with WT or CK1ϵTau alleles (group data, mean + standard error of the mean). n.s., not significantly different; *p < 0.05; **p < 0.01 versus control WT SCN (ANOVA and Tukey post hoc).
Importantly, the period of the transient oscillation in Per1/2-null SCN was not significantly affected by the presence of one or two copies of the Tau allele (Tau Het = 19.3 + 0.8, Homo = 20.6 + 1.0 h, n = 7, 9) (Fig. 3C). Thus, the Tau mutation requires the presence of at least one copy of Per1 or Per2 to affect the period of the SCN clockwork. In addition, whatever the nature of the mechanism underlying the transient oscillations present in Per1/2-null SCN, it is not sensitive to Tau.
Cellular Circadian Pacemaking in SCN Slices Deficient in Both PER1 and PER2
Deletion of Bmal1 or both Cry genes renders mice behaviorally arrhythmic and compromises bioluminescence rhythms in the SCN, similar to the effect of Per1/2 deletion seen here. Nevertheless, at the level of individual cells imaged in SCN slices by CCD camera, ongoing oscillations in the circadian range can be detected in these mutants, albeit with poor synchrony throughout the SCN circuit. Oscillatory behavior was therefore tested in single cells of Per1/2 double-null SCN. As noted in this article with PMT recordings, the ensemble rhythm of Per1/2-null SCN was poorly defined relative to WT controls (RAE WT = 0.082 + 0.022, Per1/2 double null = 0.588 + 0.044, n = 3, 6), and the period tended to be shorter than that of WT slices (WT 24.3 + 0.5 h, double-null 21.2 + 3.1 h). Poorly defined ensemble rhythms were also detected by bioluminescent imaging in Per1/2-null SCN with a single or two copies of Tau (RAE Het = 0.414 + 0.076, Homo = 0.446 + 0.079, n = 2 [one slice failed fast Fourier transform, or FFT], 3). The period of these residual rhythms was not significantly shortened by one or two copies of the Tau allele (Tau het 28.3 + 1.3 h, Tau homo 25.4 + 4.6 h, n = 2, 3). In Per1/2 WT slices, Per1-luc bioluminescence emission was identifiable from a large number of individual cells (>150 per slice, n = 3 slices) (Fig. 4A and 4B). In Per1/2-null SCN with a wild-type Tau background (n = 6 slices), a similar number of bioluminescent cells could be identified, but whereas BRASS analysis was able to fit a rhythm of a broadly circadian period (15-35 h) in circa 96.0 + 2.6% of cells in control slices, this fell significantly to 34.1 + 5.6% in the Per1/2-null slices (Table 1). The mean period of the Per1/2-null cells was not significantly different from that of cells in control slices (Table 1), but the coefficient of variance of period (a measure of how variable the period is during the recording, determined by BRASS) within Per1/2-null slices was significantly higher (Table 1). Furthermore, the phase distribution of the rhythmic cells, assessed by Rayleigh plots, was significantly greater in the null mutant SCN (Fig. 4C and Table 1), which is indicative of poor synchrony throughout the circuit. Thus, molecular circadian pacemaking can arise spontaneously in individual neurons of Per1/2-null SCN, although it is poorly organized within cells, has comparatively low amplitude, and lacks synchrony throughout the slice.
Figure 4.
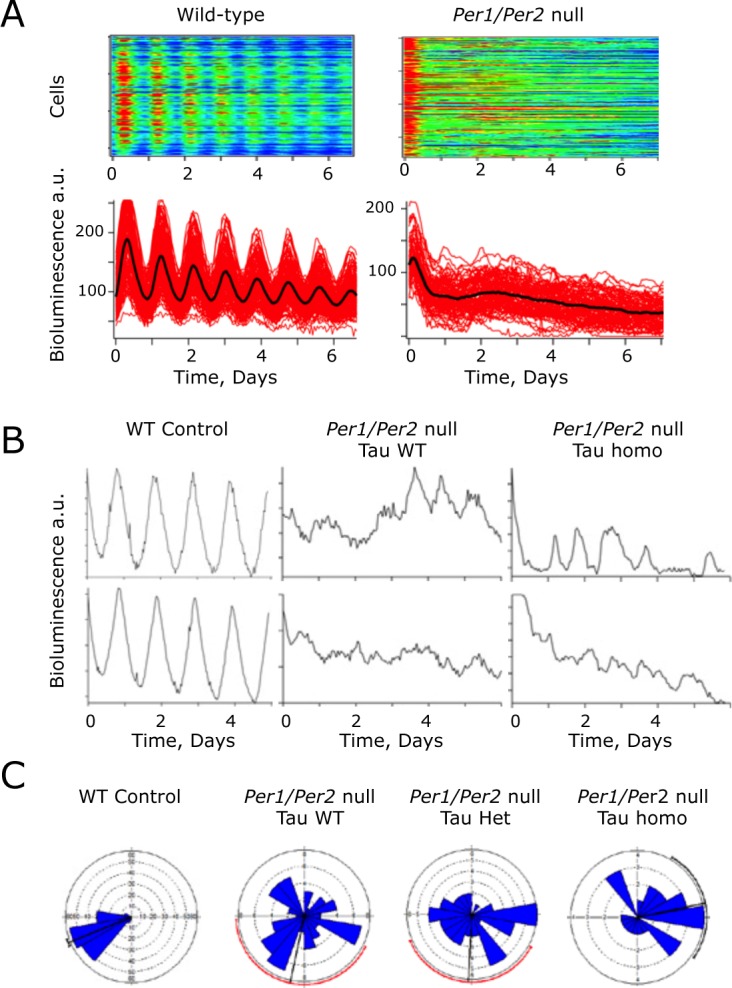
Cellular molecular pacemaking in Period1 and Period2 (Per1/2)-null suprachiasmatic nucleus (SCN), as revealed by Per1-luciferase (Per1-luc) bioluminescence rhythms, is not sensitive to the Tau mutation of casein kinase 1 epsilon (CK1ϵTau). (A) Representative raster (above) and graphical (below) plots of bioluminescence recordings of individual cells from Per1/2 wild-type (WT) control and Per1/2-null SCN. Note the disorganization in the Per1/2-null slice. (B) Representative recordings from individual SCN cells from control, and Per1/2-null SCN without (WT) or with (homo) the CK1ϵTau allele. (C) Representative Rayleigh plots of cellular bioluminescence rhythms recorded from WT control and Per1/2-null SCN, without or with CK1ϵTau (het, homo) (n = 156, 61, 45, and 25, respectively).
Table 1.
Circadian analysis of Period1-luciferase (Per1-luc) bioluminescence rhythms of individual cells in Per1 and Per2 wild-type (WT) control and Per1/2-null suprachiasmatic nucleus (SCN) slices, the latter with WT, heterozygous, or homozygous Tau backgrounds. The mean value is the mean of the individual slice values (n). For each individual slice, a large number (ca. >150) of cells was analyzed. *p < 0.05; **p < 0.01 versus control slices; Tukey multiple-comparison test.
| WT controls (n = 3) | Per-null Tau WT (n = 6) | Per-null Tau het (n = 3) | Per-null Tau homo (n = 3) | p value (ANOVA) | |
|---|---|---|---|---|---|
| Fast Fourier transform pass rate, % + standard error of the mean (SEM) | 96.0 + 2.6 | 34.1 + 5.6** | 32.1 + 9.6** | 45.0 + 18.0* | <0.01 |
| Period, h + SEM | 24.7 + 0.2 | 27.5 + 1.0 | 25.5 + 1.4 | 25.5 + 1.3 | 0.30; n.s. |
| Period variance + SEM | 2.0 + 0.8 | 23.5 + 4.1* | 20.5 + 1.1 | 34.1 + 14.9* | 0.03 |
| Relative amplitude error | 0.18 + 0.07 | 0.67 + 0.04** | 0.70 + 0.03** | 0.63 + 0.05** | <0.01 |
| Amplitude | 13.4 + 6.1 | 2.3 + 0.9* | 2.5 + 0.6 | 4.3 + 1.3 | 0.04 |
| Rayleigh mean vector + SEM | 0.92 + 0.02 | 0.16 + 0.05** | 0.31 + 0.15* | 0.20 + 0.07** | <0.01 |
| Rayleigh p value (mean) | 1*E−12 | 0.4108 | 0.1636 | 0.2497 |
The effect of the Tau allele on cellular circadian bioluminescence rhythms was then examined in Per1/2-null SCN. As with the Tau wild-type slices, bioluminescent cells were identified in Tau heterozygous and homozygous SCN, although only a minority were identified as rhythmic by BRASS analysis (32.1 + 9.6% and 45.0 + 18%, respectively; n = 3 for both). Again, these rhythmic cells were highly asynchronous with a wide phase dispersal (Fig. 4C), and they also exhibited a high coefficient of variance for their period (Table 1). Importantly, the cellular periods were not significantly affected by the presence of the Tau allele (heterozygote 25.5 + 1.4 h, homozygotes 24.6 + 1.1 h) when compared to Tau WT Per1/2-null SCN (1-way ANOVA p > 0.05), but as with the Per-null Tau WT, the Per-null Tau heterozygote and homozygote slices had significantly higher period variance when compared to the PER-proficient control SCN (Table 1).
Discussion
The aim of this study was to explore the relative contributions of endogenous PER1, PER2, and PER3 to circadian pacemaking and as targets of the CK1ϵTau mutation. It shows that both PER1 and PER2 are individually competent to sustain circadian behavior and molecular rhythms in the SCN. In contrast, PER3 cannot support clock function in the absence of PER1 and PER2. Both PER1 and PER2 are subject to the period-accelerating effects of Tau. Indeed, a single allele of either Per1 or Per2 can maintain clock function and show a shortened period in a Tau background. The effectiveness of the Tau allele was not affected by the number of Per alleles (i.e., there was no effect of the relative doses of PER and Tau). At the level of individual neurons, weak oscillatory behavior in the circadian range was detected in PER1/PER2-deficient SCN, but it was poorly defined, not synchronized among cells within a slice, and not sensitive to the Tau mutation. Thus, at all levels of analysis (whole animal, SCN slice, and individual SCN neurons), endogenous PER1 and PER2 are necessary and mutually redundant components of circadian pacemaking and mediators of the period-accelerating effect of the CK1ϵTau mutation. Moreover, a weak PER- and Tau-independent oscillator within the circadian domain can be detected in SCN slices and cells.
In the current study, there was a close correspondence, among all genotypes, between the circadian rhythms measured in vivo and in vitro on Per1-null and Per2-null genetic backgrounds. This contrasts with the report that endogenous rhythms in Per1-null mutant SCN do not represent circadian behavior in vivo (Pendergast et al., 2009): specifically, SCN from Per1-null mice, carrying a Per1-luc reporter, were poor oscillators at tissue and cellular levels, even though such mice were behaviorally rhythmic. Ruan et al. (Ruan et al., 2012) also reported poor circadian rhythms in Per1-null SCN, but rhythms could be established by a change of medium. These results contrast, however, with another report that is consistent with the present study, in which PER1-deficient SCN comprised perfectly effective circadian pacemakers (Liu et al., 2007). The conflicting results may arise because of the age at which SCN slices were made: Pendergast et al. worked with postnatal mice “aged 25-204 d (mean ± SD: 65.2 ± 38.4 d),” whereas in the present study, SCN were made from neonatal pups (all younger than 14 days). Liu et al., however, stated that “SCN slices prepared from early postnatal (2-7 days old) or adult animals (1-4 months old) produced highly comparable rhythms for all genotypes.”
PER1/PER2 double-null mice were behaviorally arrhythmic, and their SCN were either totally arrhythmic or exhibited transient short-period oscillations, likely initiated by medium change. Given that PER3 continues to be expressed in the absence of PER1 and PER2, albeit arrhythmically (Lee et al., 2004), this confirms our earlier conclusion (Bae et al., 2001) that endogenous PER3 is not able to sustain circadian behavior, and that PER1 and PER2 are necessary but mutually redundant clock components in the SCN. We cannot exclude the possibility, however, that PER3 contributes to circadian control in other tissues. Indeed, Pendergast et al. (2012) provided compelling data that loss of PER3, in the presence of both PER1 and PER2, can shorten the circadian period in vitro and advance the circadian phase in vivo of some peripheral tissues.
The mutual redundancy between PER1 and PER2 in supporting circadian pacemaking contrasts markedly with the differential contributions of CRY1 and CRY2 to circadian rhythms in vivo and in vitro. Specifically, loss of CRY1 accelerates the clock, whereas loss of CRY2 lengthens the period (van der Horst et al., 1999). Stabilization of either CRY in the Fbxl3 Afterhours mutant further lengthens the circadian period (Anand et al., 2013). These effects were interpreted from the viewpoint that both proteins are negative transcriptional regulators, but CRY2 is the weaker partner and is also able to attenuate the stronger actions of CRY1 when both proteins are present. No comparable effects on period are seen, however, with individual loss of the PER proteins. Insight into a possible molecular basis for the differential roles of CRY1 and CRY2 came from the demonstration that their access to E-box sequences occurred at different phases of the circadian molecular cycle (Koike et al., 2012). Specifically, CRY2 binds earlier in the phase of negative feedback than does CRY1 (a difference of approximately 8 h in respective peak phases), and its presence may well delay access of CRY1 to the E-boxes, thus attenuating the molecular effects of CRY1. Consistent with their mutual redundancy, however, Koike et al. (2012) showed that the peaks of access to E-boxes for PER1 and PER2 were almost overlapping (phase difference of ca. 1.4 h). These observations thereby provide indirect evidence that whereas CRY1 and CRY2 perform significantly different tasks in the circadian molecular cycle, the actions of PER1 and PER2 are closely aligned in time and function.
The presence of weak circadian oscillations in Per1/2-null SCN neurons was comparable to reports of similar oscillations in Cry1/2-null (Maywood et al., 2011; Ono et al., 2013) and Bmal1-null SCN (Ko et al., 2010). Although transient at the level of the SCN slice and poorly synchronized between cells within a slice, they reveal the presence of an underlying oscillatory or resonant structure that is independent of both the positive and negative limbs of the conventional E-box–dependent circadian transcriptional feedback loop (Koike et al., 2012). The presence of calcium- and cyclic AMP−responsive elements (CREs) in the promoters of the Per1-luc and PER2::luc reporters used to reveal these oscillations is consistent with the view that calcium- and cAMP-dependent cytosolic signaling pathways that converge onto these elements have intrinsic periodicity, independent of the transcriptional loop (Hastings et al., 2008; Ko et al., 2010). A critical role of interneuronal signaling is to engage these cytosolic pathways and thereby mutually synchronize the transcriptional feedback loops of all cells throughout the SCN (O’Neill et al., 2008; Liu et al., 2007; Brancaccio et al., 2013). The loss of transcriptional regulators intrinsic to the loop will destabilize the loop directly. It will also destabilize it indirectly by compromising transcriptional regulation of these interneuronal signals and the cytosolic inputs that they, in turn, drive. Hence, the Per1-luc rhythms observed in Per1/2-null SCN neurons are poorly defined and desynchronized, although they are subject to transient re-initiation by medium change, which will activate the intrinsically rhythmic cytosolic signaling pathways. This likely involves calcium- and cAMP-dependent cascades, but there remains the possibility that acutely induced cellular rhythms of adenosine triphosphate availability arising from PER-independent circadian changes in the metabolic state would also lead to rhythms in bioluminescence separately from any changes in luciferase abundance (Edgar et al., 2012).
The central role of CK enzymes and PER proteins in setting the speed of the circadian pacemaker has been established by genetic, biochemical, and pharmacological approaches in experimental models. Moreover, the causation of familial advanced sleep phase syndromes by mutations in hCK1 and hPER2 emphasizes their relevance to human health (Jones et al., 1999; Toh et al., 2001; Xu et al., 2005). The current results obtained by a genetic approach, accompanied by dynamic measures of circadian pacemaking in SCN slices and cells, complement and extend earlier biochemical studies that focused on PER-CK1 interactions between recombinant proteins in cell lines and endogenous proteins in ex vivo peripheral tissues. Specifically, both endogenous PER1 and PER2, but not PER3, are effective in vivo mediators of circadian period shortening by CK1ϵTau. There was, however, a trend in vitro for a PER2-dependent (i.e., Per1-null) SCN to be more sensitive to the mutation than a PER1-dependent (Per2-null) SCN (4.5 vs. 2.9 h, respectively). It may therefore be the case that other factors (e.g., retinal and brain stem afferents to the SCN) modify the function of the SCN in vivo, with their absence unmasking this PER1-versus-PER2 difference in vitro. In this regard, it would be of interest to determine the effects of Tau on the stability of PER1 and PER2 in the SCN (currently reported solely for PER2; Meng et al., 2008). Aside from this limited effect, there was no significant contrast between the PER paralogs in their contribution to pacesetting. As for PER3, its particular role in circadian pacemaking remains difficult to define. It is clear that polymorphisms in hPer3 can influence diurnal preference, but this is likely related to an effect on sleep homeostasis rather than circadian function per se (Viola et al., 2007). Consistent with the biochemical evidence that PER3 is unable directly to interact with CK1ϵ (Lee et al., 2004), our in vivo and in vitro results exclude endogenous PER3 both as an essential circadian factor in the SCN and as a mediator of CK1ϵTau actions.
Finally, our observation that the Tau mutation can accelerate the clock of mice via actions on PER2 is consistent with the report that a mutation in hPER2 that affects the binding with, and phosphorylation by, WT CK1ϵ shortens the circadian period and leads to a sleep phase disorder (Toh et al., 2001). In a previous study, inhibitors of CK1δ/ϵ were shown to slow the circadian clock of both WT and Tau mutant mice, and in the latter case they can correct the period of the homozygous mutant SCN to WT (i.e., cause a deceleration of 4 h) (Meng et al., 2010). Thus, the development of compounds that affect CK1δ/ϵ function directly, or alter the ability of either PER1 or PER2 to interact with CK1δ/ϵ, may provide a suitable means to control aberrant circadian period or disrupted entrainment arising from genetic or environmental (e.g., rotational shiftwork) perturbations.
Acknowledgments
This work was supported by the Medical Research Council, UK. The authors are grateful to the staff of the Biomedical Facilities at the MRC Laboratory of Molecular Biology for excellent technical support.
Footnotes
Conflict of Interest Statement: The authors declare no competing financial interests.
References
- Anand SN, Maywood ES, Chesham JE, Joynson G, Banks GT, Hastings MH, Nolan PM. (2013) Distinct and separable roles for endogenous CRY1 and CRY2 within the circadian molecular clockwork of the suprachiasmatic nucleus, as revealed by the Fbxl3(Afh) mutation. J Neurosci 33:7145-7153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. (2001) Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30:525-536 [DOI] [PubMed] [Google Scholar]
- Brancaccio M, Maywood ES, Chesham JE, Loudon AS, Hastings MH. (2013) A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron 78:714-728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. (2012) Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature 485:123-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM. (2006) A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron 50:465-477 [DOI] [PubMed] [Google Scholar]
- Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, et al. (2012) Peroxiredoxins are conserved markers of circadian rhythms. Nature 485:459-464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. (2007) Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol 8:139-148 [DOI] [PubMed] [Google Scholar]
- Hastings MH, Maywood ES, O’Neill JS. (2008) Cellular circadian pacemaking and the role of cytosolic rhythms. Curr Biol 18:R805-R815 [DOI] [PubMed] [Google Scholar]
- Hastings MH, Reddy AB, McMahon DG, Maywood ES. (2005) Analysis of circadian mechanisms in the suprachiasmatic nucleus by transgenesis and biolistic transfection. Methods Enzymol 393:579-592 [DOI] [PubMed] [Google Scholar]
- Jones CR, Campbell SS, Zone SE, Cooper F, DeSano A, Murphy PJ, Jones B, Czajkowski L, Ptacek LJ. (1999) Familial advanced sleep-phase syndrome: a short-period circadian rhythm variant in humans. Nat Med 5:1062-1065 [DOI] [PubMed] [Google Scholar]
- Ko CH, Yamada YR, Welsh DK, Buhr ED, Liu AC, Zhang EE, Ralph MR, Kay SA, Forger DB, Takahashi JS. (2010) Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol 8:e1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. (2012) Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338:349-354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Weaver DR, Reppert SM. (2004) Direct association between mouse PERIOD and CKIepsilon is critical for a functioning circadian clock. Mol Cell Biol 24:584-594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, et al. (2007) Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129:605-616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS. (2000) Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288:483-492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Chesham JE, O’Brien JA, Hastings MH. (2011) A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc Natl Acad Sci U S A 108:14306-14311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Reddy AB, Wong GK, O’Neill JS, O’Brien JA, McMahon DG, Harmar AJ, Okamura H, Hastings MH. (2006) Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol 16:599-605 [DOI] [PubMed] [Google Scholar]
- Meng QJ, Logunova L, Maywood ES, Gallego M, Lebiecki J, Brown TM, Sladek M, Semikhodskii AS, Glossop NR, Piggins HD, et al. (2008) Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron 58:78-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng QJ, Maywood ES, Bechtold DA, Lu WQ, Li J, Gibbs JE, Dupre SM, Chesham JE, Rajamohan F, Knafels J, et al. (2010) Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc Natl Acad Sci U S A 107:15240-15245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk JA, Takahashi JS. (2011) Cell autonomy and synchrony of suprachiasmatic nucleus circadian oscillators. Trends Neurosci 34:349-358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. (2008) cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 320:949-953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono D, Honma S, Honma K. (2013) Cryptochromes are critical for the development of coherent circadian rhythms in the mouse suprachiasmatic nucleus. Nature Commun 4:1666 [DOI] [PubMed] [Google Scholar]
- Pendergast JS, Friday RC, Yamazaki S. (2009) Endogenous rhythms in Period1 mutant suprachiasmatic nuclei in vitro do not represent circadian behavior. J Neurosci 29:14681-14686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendergast JS, Niswender KD, Yamazaki S. (2012) Tissue-specific function of Period3 in circadian rhythmicity. PLoS ONE 7:e30254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph MR, Menaker M. (1988) A mutation of the circadian system in golden hamsters. Science 241:1225-1227 [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. (2002) Coordination of circadian timing in mammals. Nature 418:935-941 [DOI] [PubMed] [Google Scholar]
- Ruan GX, Gamble KL, Risner ML, Young LA, McMahon DG. (2012) Divergent roles of clock genes in retinal and suprachiasmatic nucleus circadian oscillators. PLoS ONE 7:e38985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptacek LJ, Fu YH. (2001) An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science 291:1040-1043 [DOI] [PubMed] [Google Scholar]
- van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, et al. (1999) Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398:627-630 [DOI] [PubMed] [Google Scholar]
- Viola AU, Archer SN, James LM, Groeger JA, Lo JC, Skene DJ, von Schantz M, Dijk DJ. (2007) PER3 polymorphism predicts sleep structure and waking performance. Curr Biol 17:613-618 [DOI] [PubMed] [Google Scholar]
- Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptacek LJ, Fu YH. (2005) Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature 434:640-644 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. (2003) Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302:1408-1412 [DOI] [PubMed] [Google Scholar]


