Abstract
Periodontal diseases reflect a tissue destructive process of the hard and soft tissues of the periodontium that are initiated by the accumulation of multispecies bacterial biofilms in the subgingival sulcus. This accumulation, in both quantity and quality of bacteria, results in a chronic immunoinflammatory response of the host to control this noxious challenge, leading to collateral damage of the tissues. As knowledge of the characteristics of the host-bacterial interactions in the oral cavity has expanded, new knowledge has become available on the complexity of the microbial challenge and the repertoire of host responses to this challenge. Recent results from the Human Microbiome Project continue to extend the array of taxa, genera, and species of bacteria that inhabit the multiple niches in the oral cavity; however, there is rather sparse information regarding variations in how host cells discriminate commensal from pathogenic species, as well as how the host response is affected by the 3-dimensional architecture and interbacterial interactions that occur in the oral biofilms. This review provides some insights into thes- processes by including existing literature on the biology of nonoral bacterial biofilms, and the more recent literature just beginning to document how the oral cavity responds to multispecies biofilms.
Keywords: oral bacteria, biofilm, epithelial cells, cytokines, chemokines
Periodontal diseases are one of the most frequent global infections in humans [1, 2]. While many medically important infections occur on a global scale, most of these are due to monoinfections with specific microbial pathogens, eg. Vibrio cholerae, Mycobacterium tuberculosis, influenza. Moreover, few of these are chronic infections of the host, although there are notable pathogens that represent long term disease processes, eg. Plasmodium falciparum, HIV. An even more select subset of pathogens initiate a disease process by creating organized biofilms that enhance their ability to adhere, replicate, accumulate, and express their virulence potential [3–6]. However, even in this scenario, most medically significant pathogens that form biofilms tend to be monospecies infections, eg. Pseudomonas aeruginosa, Staphylococcus aureus, Yersinia pestis.
Chronic periodontal infections provide a very different “face” to the host with regards to controlling these bacteria. First, it is estimated that over 700 species of bacteria can colonize the oral cavity of humans, and that in an individual the range of niches in the oral cavity host 100 or more species [7]. Second, these bacteria accrete and accumulate on host surfaces with tropisms for specific sites in the oral cavity, as well as specificity for interbacterial interactions that result in 3-dimensional structured and organized biofilms [8–10]. Third, the bacteria must survive within a milieu of host factors derived from saliva, serum, and gingival tissues [11]. Finally, this dynamic host-bacterial environment has evolved to help sustain a protective commensal microbial ecology, while minimizing the ability of opportunistic pathogens to emerge within these established biofilms [12]. The majority of data derived from studies of host-bacterial interactions in periodontitis has focused on the individual “trees” in isolation and have provided extensive information in controlled systems to delineate the capacity of single species to alter host cell functions. However, not available in these investigations was the ability to define how the microbial “forest” as a multicellular complex 3-dimensional structure could present a challenge quite different from simply the sum of the species in the forest. In order to extend our understanding of these interactions that are required to maintain homeostasis in the oral cavity, and the characteristics of a dysregulated response that leads to disease progression, it is necessary to begin to model the host cell-biofilm attributes at a molecular level.
Medically Important Pathogenic Biofilms
Acute medical infections are generally associated with planktonic bacteria and must be diagnosed rapidly and treated accurately to prevent tissue damage and/or death. In contrast, bacteria in biofilms demonstrate an infectious course usually accompanied by sustained host hyper-inflammation, with the biofilm species using plasma exudate as nutrition. This lifestyle is a particularly important bacterial adaptation to grow as part of a sessile community, which mimics an integrated multicellular organism with its own development cycle, cooperative behavior among the species, and coordinated management using quorum sensing signal molecules to communicate among the constituents. These microorganisms attach to both biotic and abiotic surfaces, followed by multiplication while becoming embedded in an extracellular matrix, leading to characteristic biofilms. The outcomes are 3-dimensional aggregates of bacteria that are now recognized as significant contributors to in many bacterial-associated infections (eg. endocarditis, dental caries, middle ear infections, osteomyelitis, medical device-related infections, chronic lung infections in cystic fibrosis patients, persistence of food-borne pathogens) [13–16]. Recent estimates suggest that 60–85% of all microbial infections involve biofilms developed on these natural tissues or artificial devices.
Historical studies of biofilms based upon medical infectious agents were described as sessile communities of microbes characterized by cells that are irreversibly attached to a substratum or co-aggregate with each other. Generally they are embedded in a matrix of extracellular polymeric substances. Additionally, the initial bacterial adherence and biofilm accretion are considered to proceed in two steps with attachment to a surface followed by cell-to-cell adhesion. This is followed by maturation of the biofilm that frequently is represented by stratification of bacteria within the biofilms and finally dispersal from the sessile community. For this process to occur, it would be expected that environmental conditions impact the type of bacteria and their ability to survive and replicate within a mutualistic complex. The success of this would require regulation of a range of genes specific for biofilm life versus those required for a successful planktonic existence. Biofilms have been shown to have a high persistence once the infections are established and thus responsible for many chronic infectious processes. This related directly to their phenotypic resistance to high concentrations of antimicrobials [13, 17], and ability to modulate the host immune systems [18, 19].
Numerous studies have been conducted on biofilms of Staphylococcus aureus due to its major role in infections of catheters and prosthetics. These studies have identified genetic regulation and molecular components involved in biofilm formation and maturation [20]. Similar types of studies were reported on novel biofilm gene regulation in Mycobacterium tuberculosis [21, 22]. An additional major feature of biofilms is the altered microenvironment that is created within and surrounding the development of the biofilms. Characteristics of these differences include both structural and physiological. The exracellular matrix accumulates and often encases the multicellular structures within the 3-D architecture of the biofilms. With Y. pestis the extracellular matrix of the biofilm contains a homopolymer of N-acetyl-d-glucosamine, which is a constituent of many bacterial biofilms and may adversely affect both host resistance mechanisms and extrinsic antibiotic administration. This extracellular polymeric substance has been reported to facilitate tolerance to environmental stresses, and contributes to physiological adaptation of individual bacterial cells in the heterogeneous microenvironments within the complex architecture of biofilms [23]. The physiological adaptation studies have identified available nutrients, temperature, and pH as factors that can contribute to the altered microbial ecologies in biofilms. Studies with P. aeruginosa, Klebsiella pneumoniae, and V. cholera have shown that increased pH leads to a higher biofilm production in vitro [24]. Similar studies examined the effect of pH on production of extracellular virulence factors of P. aeruginosa growing on catheter biofilms. Both alginate and proteinase production was higher at pH 8; in contrast, siderophores (pyochelin and pyoverdin) that were synthesized to a greater level at pH 5 [25]. It has also been suggested that the pH at the site of infection is one of a number of factors that may significantly influence the in vivo activity of an antibiotic prescribed for treatment of infection. Results by Moriarty et al., [26] showed that growth in an acidic environment might be expected to reduce the susceptibility of P. aeruginosa to certain antibiotics. A substantial decrease in the biofilm production was observed at 37°C compared with the amount of biofilm formed at 30°C with most strains of these species. Studies of S. aureus and Salmonella typhimurium biofilms demonstrated significant differences in gene expression in biofilms cultivated at different acidic, neutral or alakaline pH values [27]. A number of these genes have been directly related to virulence of these pathogens. As an additional environmental regulator of biofilms, data from cystic fibrosis would suggest that the P. aeruginosa biofilms likely form under anaerobic conditions [28]. This tenet can be extrapolated as both anaerobically grown planktonic bacteria and biofilm bacteria were significantly less susceptible to single and combination antibiotics compared to aerobic growth of planktonic bacteria. Additionally, antibiotic combinations that killed under anaerobic conditions frequently differed from those that were bactericidal against the same microorganisms grown as biofilms [29].
The vast majority of studies of medically significant pathogens have used bacteria in the planktonic state, although many express their virulence through the formation of biofilms. More recent in vivo and in vitro studies have documented innate and adaptive immune responses to biofilms. However, strategies used by bacteria in biofilms to resist/minimize various immune responses have also been demonstrated. Hence, with biofilm infections often present as persistent infections with simultaneous activation of both innate and adaptive immune responses can be detected. Unfortunately, in many cases, neither of these response arms can eliminate the biofilm pathogen, but frequently, as noted in periodontitis, causes collateral tissue damage.
It has been shown that S. aureus biofilms induce a distinct inflammatory response compared to their planktonic counterparts. The differential gene expression and production of inflammatory cytokines by the biofilm was predicted to have an adverse effect on the formation and persistence of chronic wounds [30]. Furthermore, S. aureus biofilms significantly reduced keratinocyte viability and significantly increased apoptosis compared with planktonic bacterial cells [31]. With S. aureus biofilms PMNs moved across the biofilm and took up bacteria as they moved, while with a related pathogen, S. epidermidis, the PMN were rather immobile, and phagocytosis was limited to bacteria closely associated with the surface of the PMNs. Consequently, it was interpreted that S. aureus biofilms are more sensitive towards PMN attack compared to S. epidermidis, and that these types of biofilms are not inherently protected against the attack by phagocytic cells, albeit the inherent sensitivity is related to bacterial species or strains [32]. A separate study showed that opsonization of S. aureus biofilms with human serum IgG binding resulted in complement activation. However, this immune process did not affect the adherence of PMN to the biofilms nor did it enhance degranulation or phagocytosis [33]. Finally, it was found that developing "young" biofilms of S. aureus were more sensitive towards attack by PMNs compared to mature biofilms [34] Thus, studies developing data regarding how host protective features and host cells interface with biofilms need to consider substantive differences between newly developing versus mature biofilms with respect to clearly elucidating how host cells try to manage biofilm accretion.
Biofilms of P. aeruginosa induced a higher production of TNF and IL-6 than planktonic bacteria [35]. In addition to the cytokine levels, reactive nitrogen species, and stimulated macrophage secretory products were generated in greater levels with biofilms of P. aeruginosa versus planktonic forms [36]. The switch between the two life-styles caused several reversible LPS structure modifications affecting the lipid A and polysaccharide moieties. In addition, LPS isolated from the biofilm bacteria induced slightly higher levels of inflammatory cytokines than LPS extracted from planktonic bacteria. Parallel reports attempting to better delineate effective host responses against various pathogens suggested that Th17 cells have a prominent role in host defense against M. tuberculosis [37]; however, chronic biofilms can redirect these immune cells down a pathway of tissue injury. Finally, with regards to host responses, neutrophils settle on P. aeruginosa biofilms and become phagocytically engorged. On this surface, they become partially degranulated, immobilized, and round-up. This results in an increase in oxygen consumption of the system, but with little increase in the soluble concentration of microbicidal factors, such as H2O2. Thus, host defense becomes compromised as biofilm bacteria escape, while neutrophils remain immobilized with a diminished oxidative potential [38]. Consequently, it remains undetermined as to whether particular/specific/unique immune responses to biofilm infections exist.
Host-Bacterial Interactions in Periodontal Disease
As the field of oral microbiology developed through that last few decades, substantial emphasis was placed on more accurately identifying the range of microbial species that occurred in the oral cavity, defining their various niches, and better discriminating the microbial components of healthy and disease site ecologies [39–42]. In parallel to these microbiologic studies a range of investigations used cell culture systems to explore the effect of oral bacteria and their products on responses of immune and nonimmune cells to microbial challenge. These reports routinely used individual planktonic bacteria or soluble/secreted molecules from the bacteria to stimulate host cells [43, 44]. These studies resulted in a wide array of observations comparing individual oral bacteria related to dose-dependency of profiles of host responses from many cell types, eg. epithelial cells, fibroblasts, lymphocytes, neutrophils, macrophages. These studies have identified a hierarchy of stimulatory capacity of individual species, demonstrating, in broad strokes, that the Gram-negative bacteria were generally more active for inducing host cellular responses compared to the Gram-positive oral bacteria. Selected bacteria, eg. Aggregatibacter actinomycetemcomitans, Fusobacterium nucleatum, were identified to be particularly active in stimulating immune and nonimmune cells. Oral bacteria such as A. actinomycetemcomitans had the capacity to kill neutrophils and immunosuppress lymphocytes [45], while the physiologic proteolytic activity of Porphyromonas gingivalis had detrimental effects on many cells types with increasing levels of challenge [46, 47]. Additionally, using isolated components from individual bacterial species (eg. LPS, fimbriae, outer membrane proteins) investigators deduced some information on the relationship of these bacterial biomolecules to individual members of the host repertoire of responses [48]. Examples of these findings included ompA from A. actinomycetemcomitans as a major stimulatory molecule [49], the S-layer protein and BpsA outer membrane protein of Tannerella forsythia highly active in stimulating cells [50], and the LPS from P. gingivalis in certain configurations actually being a rather poor stimulatory or endotoxic molecule [51, 52].
These results also contributed to an understanding of the function of pattern recognition receptors displayed by host cells (TLRs, NODs, etc.) in the capacity of innate immune responses to maintain homeostasis of oral tissues [53, 54]. These investigations helped to elucidate the potential types of host-bacterial interactions that occur in the oral cavity, which result in the identification of biomolecules in health and disease in gingival tissues, gingival crevicular fluid, and saliva. However, these approaches are limited by a reductionist approach to understanding the complexity of the interactions that likely occur in situ. Thus, these methodologies consciously removed the bacterial species from their native biofilms that exist in situ enabling them to interact with host cells floating in suspension, and/or dissociated these complex stimulatory agents into isolated components lacking the context of these molecules within the membrane milieu of the bacterium. Consequently, the field must advance by creating data that addresses the similarities or differences in host outcomes when challenged with a consortium of commensal and pathogenic species, and the types of communications that are signaled in the host cells, as well as the feedback on gene regulation in the microbes in the biofilms.
Biofilms in Periodontal Disease
Extensive literature over the last 3 decades has described findings of specific microbial consortia that appear supra- and subgingivally in the oral cavity of humans [10, 55]. These studies have identified a sequential nature of this colonization and the appearance and emergence of selected consortia associated with progressing disease [10, 56]. Furthermore, results from intervention studies demonstrated that successful therapy is generally accompanied by a reversal of the microbial ecology to one more consistent with health [57–59]. Kolenbrander and colleagues [60, 61] have laid the groundwork for understanding the characteristics of this sequential acquisition of the ecology, as well as factors affecting specific microbial consortia. They have demonstrated ligands and receptors for specific micoorganisms to adhere to salivary pellicle and identified cognate interactions among bacteria resulting in interbacterial co-aggregation. As examples research has shown the binding capabilities of pioneer species, such as the streptococci, for specific salivary proteins/peptides, and even unique domains on those molecules [62–67]. Binding of fimbria on Actinomyces spp. has been show to specific host molecules, selected streptococcal species, and even to members of the microbiota most associated with gingivitis and potential transition to more pathogenic biofilms [68, 69]. Finally, there is substantial physiologic evidence [8] and more recently deductions from genome sequencing of oral bacteria regarding the occurrence of “food webs” in these complex biofilms with interbacterial dependence on nutrition input and utilization of end-products of the bacterial metabolism [70–72].
These concepts have underpinned recent studies of the structure and organization of oral biofilms. Extending from observations of Costerton and co-workers [73–76] who provided paradigm shift of modeling biofilms as organized and structured 3-deminsional assemblies of bacteria species that develop into a multicellular unit. This model suggested the accretion of bacteria in specific scaffolds, passageways in the biofilms for fluid flow for nutrition and waste disposal, and the capacity of the bacteria to detach from the sessile biofilm forms and seed distant sites to create new structures. As importantly these in vitro studies have demonstrated an organization of multiple oral species in these biofilms [77], not simply an amorphous conglomeration of the bacteria, and reflect very well early TEM studies from Listgarten and colleagues [78–80] on dental plaque, ie. biofilms.
Models of Multispecies Biofilm Function
Over the last decades evidence has accumulated delineating a defined order of succession and kinetics, for both supra- and subgingival bacterial species, during development of biofilms following cleaning. These data support that the oral cavity facilitates the growth of a somewhat unique autochthonous microbiota in the range of ecological niches. Thus, the oral microbiome essentially exists within biofilms throughout the various niches in the oral cavity. Mature dental biofilms demonstrate an accumulation of bacterial species in microcolony towers [81, 82]. It is clear that the resident bacteria interact with one another through cognate binding, as well as having the capacity to communicate via specific signalling molecules, share DNA, and supply metabolites for communal nutrition. As an example, it has been shown that commensal Veillonellae spp. use lactic acid for growth in saliva, which enhances their ability to communicate metabolically with initial, early, middle, and late colonizers in establishing multispecies biofilm communities [83].
The bacterial complexes form an ecosystem that contributes directly to maintaining health when the system is in equilibrium. However, specific ecological shifts in this commensal microbiome allow pathogens to emerge, manifest their pathogenic potential, and cause disease. It is clear that this oral microbial ecology is influenced by temperature, pH, atmosphere, endogenous nutrients and a variety of surfaces for biofilm formation, and is affected by host defenses and genetics. Consequently, oral biofilms develop under a range of different conditions and different microenvironments, albeit, mutually beneficial interactions between the microbial cells are essential for the success of development of biofilms in the oral cavity. As such, the host environment dictates the composition and gene expression of the resident microbiota. Changes in oral environmental conditions disrupt the normal symbiotic/mutualistic relationship between the host and its resident microbes, with a resulting increase in the risk of disease. However, while the conditions under which oral biofilms develop are tightly linked to the overall health and biology of the host, it remains undetermined concerning the initiating factors that presage or drive these changes. For example, it has been shown that biofilm formation regulates the survival and invasiveness of F. nucleatum under aerobic conditions [84]. This was interpreted as the biofilm fundamentally enhancing the ability of the bacterium to survive in oxygen. The increased survival lead to heterogeneity within the biofilm and potential genetic variations in F. nucleatum that offered a strategy for the bacteria to resist host defenses and enable greater opportunity to invade the epithelial cells.
Much work is being conducted focusing on the functional genomics of oral bacteria when placed into multispecies biofilm growth environments. These investigations are using data derived from the progress in genome sequencing of a range of oral bacteria and allowing direct studies of variations in transcriptome of individual species within a complex biofilm [85–88]. These studies enable modification of the microbial constituents, the characteristics of nutritional availability, and the microenvironment of the biofilms, including pH and oxygen tension [89] to identify alterations in the transcriptome. Moreover, these changes in gene expression have been evaluated at the translational level to identify new molecules or increases in specific components that enhance the capacity of the individual bacterial species to compete effectively and thrive in these biofilms.
Thus, to better understand the parameters of the interactions between the complex oral biofilms and host cells and tissues, new types of models are required. The development of these in vitro models are necessary to delineate the molecular interactions that take place at the interface between the structured biofilms and host cells during periodontal health and disease. The literature has demonstrated the capability to cultivate single- and multispecies biofilms in devices, such as flow cells and chemostats, on static support matrices, including enamel chips, hydroxyapaptite disks or glass slides [90]. Of the various culture systems available, flow-cell models in conjunction with confocal laser scanning microscopy (CLSM) and fluorescent in situ hybridization (FISH), or immunofluorescence staining have been widely used for to characterize individual species in in vitro grown biofilms of oral bacteria [91, 92]. Multiple staining of biofilms by FISH enable species differentiation [93], although, the methods of this technique could introduce structural artifacts [94, 95], present a challenge in differentiating closely related bacterial species with conserved 16S rRNA genes [96], and can create some difficulty in enabling the probes to penetrate the biofilms. Fluorescently labeled antibodies specific for individual bacterial species have also been used to identify the characteristics and distribution of individual species within complex biofilms [92, 97]. Various classic nucleic acid stains have been used to document the spatial arrangement of bacteria in biofilms [9, 98–101] that have been created through a co-aggregation model of bacterial accretion in biofilms [102, 103]. These types of studies have shown that P. gingivalis can assemble into a heterotypic microbial complex with F. nucleatum and S. gordonii [104]. Additional observations in this model indicated that this community provided necessary physiologic support for P. gingivalis growth, and that various proteins in P. gingivalis were up-regulated in response to the community dynamics, although other data indicated that an analysis of global gene expression showed adaptive responses of P. gingivalis to biofilm growth included a down regulation of genes involved in growth and metabolic activity [105, 106]. In studies of multispecies biofilms, A. naeslundii was identified in the inner portion of the multilayered biofilm, demonstrating its ability to attach directly to the acquired salivary pellicle and likely contributing important ecological effects on the developing biofilm [107]. A recent example of these types of interbacterial processes that appear to occur in pathogenic oral biofilms are based upon the capacity of P. gingivalis to undermines critical components of innate immunity, including altering functional levels of effector molecules, as well as receptors and subsequent intracellular signaling pathways required for effective host responses. These authors suggested that these “subversive activities” of the periodontal pathogen could promote the adaptive fitness of the pathogenic biofilm communities and enable the chronicity of the inflammatory response resulting in the tissue destruction of periodontitis [108].
Recent reports on oral biofilms have continued to emphasize the bacteria side of this equation. Palmer [55] described emerging concepts of the physiological basis for spatial distribution in natural oral biofilms that could contribute to modeling these phenomena with outcomes from other ecosystems [109]. Kolenbrander [110] reported on the findings of highly selective interspecies recognition between initial colonizers with early and middle colonizers to form multispecies communities that grow on saliva in helping to create the complexity and structure of oral biofilms. Xiao and colleagues [111] emphasize that all biofilms harbor a microbial-derived extracellular-matrix of exopolysaccharides (EPS) that are formed on host and bacterial surfaces and provide binding sites for microorganisms. This accumulated EPS enmeshes the microbial cells and affects the 3D architecture and the population shifts during evolution of the biofilms, as well as altering metabolic activities of the constituent microorganisms and influencing the microenvironment. These authors also reported that the EPS-matrix helped to create spatial heterogeneities by enabling the formation of microcolonies within the mesh network. Regarding the potential for complex biofilms to alter host cell responses compared to their individual component bacteria, recent results show the importance of pathogens in controlling gene expression of a healthy oral microbial ecology leading to a different portfolio of bacteria products that could interact with the host cells [85]. A recent review emphasized that important interactions among bacteria occur within the oral biofilm communities [71]. These interactions include planktonic bacterial cells directly attaching to surfaces of the oral cavity or indirectly bind to other bacterial cells that have already colonized. These co-aggregation properties of select bacterial species are likely crucial for retention of bacteria on oral surfaces, and may facilitate stable bacterial colonization. These interactions then create an environment for metabolic communication, genetic exchange, production of inhibitory factors (e.g., bacteriocins, hydrogen peroxide, etc.), and quorum-sensing molecules that are regulatory factors determining the eventual bacterial composition. Finally, while not directly biofilm related a recent study reported that under reduced oxygen tension (ie. 2% oxygen) selected oral bacteria, eg. Tannerella forsythia, P. gingivalis, and Prevotella intermedia elicited elevated levels cytokines/chemokines with elevated levels in a low oxygen environment [112]. This type of knowledge has also increased our understanding of unique features of biofilms that appear to increase the resistance of the bacteria to chemical anti-infectives/antimicrobials and bacteriostatic/bactericidal antibiotics [113–115]. Additionally, the literature supports that the bacteria in biofilms are also more resistant to the phagocytic activities of neutrophils and macrophages [38, 116–118]. However, there have been few publications that have attempted to examine the capacity of these 3-dimensional organized biofilm structures to stimulate host cells with regards to inflammation and innate immunity.
These model systems and in situ characterization of the structural organization of biofilms [119] have various methodological constraints and using them to examine the interactions of viable host cells with these biofilms has been minimal. A few studies have been reported that examine the capacity of oral bacteria in biofilms to alter host response profiles. Human beta-defensin-2 mRNA expression was significantly upregulated in gingival epithelial cells by challenge with Streptococcus mitis-biofilms compared to biofilms of S. mutans, while IL-8 and 5-lipoxygenase genes transcripts were significantly elevated with the S. mutans biofilms. RNAase-7, another antimicrobial peptides, mRNA expression was significantly higher in epithelial cells stimulated with both streptococcal biofilms [120]. An additional study suggested that the levels of these various gene transcripts are also related to to different stages of biofilm formation [121]. The results supported that biofilms can elicit unique profiles of responses in gingival epithelial cells. Recently, Guggenheim and colleagues used a hydroxyapatite disc model to prepare oral multispecies biofilms and used these to challenge epithelial cell cultures [122, 123]. While numerous species were used to create the biofilms, of the 9 species used, P. gingivalis and F. nucleatum made up a rather small proportion of the overall microbial composite at a level approximating 1% of the total, that is, these biofilms were vastly dominated by a very limited number of species. Thus, while the architecture of these very complex biofilms was described using confocal scanning laser microscopy, it remains undetermined the density of the bacteria that interacted with the individual cells, nor the species that may have been primary participants in this process. These reports did document a range of host responses that occurred under aerobic conditions and identified a range of mediators from the epithelial cells including IL-1β, IL-6, IL-8 [122] and RANKL/OPG [123]. The primary findings were an apparent increase in apoptosis and degradation of Il-1, IL-6 and IL-8 cytokines that were elicited from the epithelial cells likely related to proteases produced by P. gingivalis.
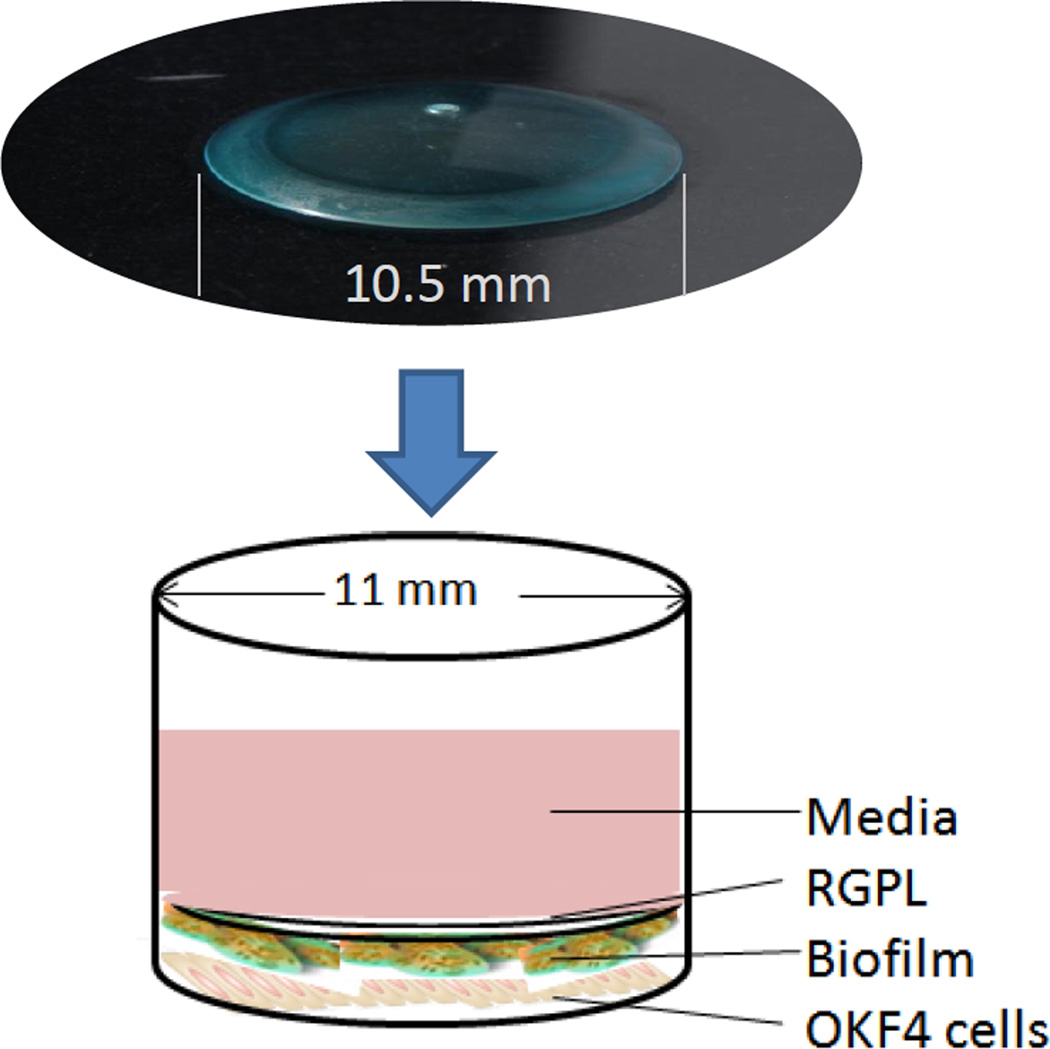
Thus, while these reports continue to lay the groundwork for more detailed understanding of the microbiological parameters that contribute to creation, evolution, and sustenance of oral biofilms there remain numerous questions regarding the stimulatory capacity of individual bacteria in biofilms, how they compare with planktonic stimuli, and how these responses would be altered in the presence of a complex multispecies biofilm. For this reason, our lab developed an in vitro model growing biofilms on gas permeable hard contact lenses (RGPLs), which allow substratum bound epithelial cells, or other host cell types to interact the biofilms (Fig. 1). We demonstrated the characteristics of single and multispecies biofilms created using this novel experimental model. Bacterial species belonging to early and late colonizing groups were successfully established as single- or three-species biofilms with each group comprising Streptococcus gordonii, S. oralis and S. sanguinis (healthy biofilm) or S. gordonii, Actinomyces naeslundii and Fusobacterium nucleatum (transition or gingivitis biofilm) or S. gordonii, F. nucleatum and Porphyromonas gingivalis (pathogenic biofilm). QPCR analysis revealed substantial differences in the magnitude of bacterial numbers in the single and multispecies biofilms, and using novel SYTO red, blue and green fluorochromes and CLSM to document distribution of the species in the multispecies biofilms. This novel model is also providing some seminal data on oral biofilm-host cell interactions with respect to characteristics of the cellular responses to various biofilms.
Figure 1.
Schematic of the RGPL model of biofilm stimulation of host cells. The RGPL are 10.5 mm in diameter with the culture plate wells at 11 mm. The biofilms are grown on the convex site of the RGPL and then inverted on top of the epithelial cells cultures with direct contact between the biofilms and host cells.
Extensive literature exists concerning responses from host cell types (eg. epithelial cells, fibroblasts, immune cells) that function in the oral cavity, when challenged in vitro with individual planktonic bacteria or components isolated from individual species [124, 125]. However, in vivo, it would be expected that numerous host cells, particularly epithelial cells, interact with the oral bacteria growing as complex biofilms. Also, these studies are generally performed under optimized and controlled in vitro conditions of nutrients, pH, and oxygen tension. However, it is clear that the apical aspect of the periodontitis subgingival sulcus is characterized by altered constituents resulting from the localize inflammatory response [126, 127], an elevated pH [128, 129], a hypoxic microenvironment [130, 131], and host antimicrobial factors, eg. antimicrobial peptides, antibodies, immune cells [132–135], that should substantially affect the interactions and responses of both the bacteria and the host cells (Fig. 2). Thus, we performed an initial study using the RGPL to create bacterial biofilms and determined the production of a biomarker of classic biomarker of host inflammatory responses, interleukin 8 (IL-8), by epithelial cells. This was chosen as a major constituent of the local inflammatory response that targets chemoattraction of neutrophils into the site [136, 137]. We demonstrated that with the exception of P. gingivalis, generally monospecies biofilms of each oral bacterial species elicited significantly greater IL-8 mRNA levels than the planktonic bacteria of the same species. Both A. naeslundii and F. nucleatum appeared most active for IL-8 induction, with the greatest levels observed with F. nucleatum biofilm challenge. Of the streptococcal species only S. gordonii, biofilm or planktonic cells, induced measurable levels of secreted IL-8, as has been reported previously [138, 139]. P. gingivalis, T. denticola, and T. forsythia have been reported to suppress the production of this chemokine [140], although, P. gingivalis strains that adhere to and invade epithelial cells did induce IL-8 mRNA [141]. Our studies found that neither P. gingivalis biofilms nor planktonic challenge resulted in detectable IL-8 in epithelial cell supernatants, likely reflecting gingipain degradation of the molecule [142]. Both A. naeslundii and F. nucleatum biofilms elicited significant elevations in IL-8 compared to challenge with comparable levels of planktonic bacteria. F. nucleatum has been routinely reported to up-regulate IL-8 mRNA and production of IL-8 by epithelial cells [139]. These findings provide a basis for utilization of this RGPL biofilm model system to explore multispecies biofilms [143] and their capacity to trigger a portfolio of host response molecules from epithelial cells. From this initial study it became clear that this model system can also be used to examine other host cell responses to mono- or multispecies biofilms targeting gene and product expression levels related to inflammation, innate immunity, and even adaptive immunity. We believe that this system would also be amenable to evaluating the response of the bacteria to these interactions, either affected by direct host cell contact or in response to a milieu of host derived factors that could alter the microbial physiology and/or cell communications, eg. quorum sensing, between the various bacterial species.
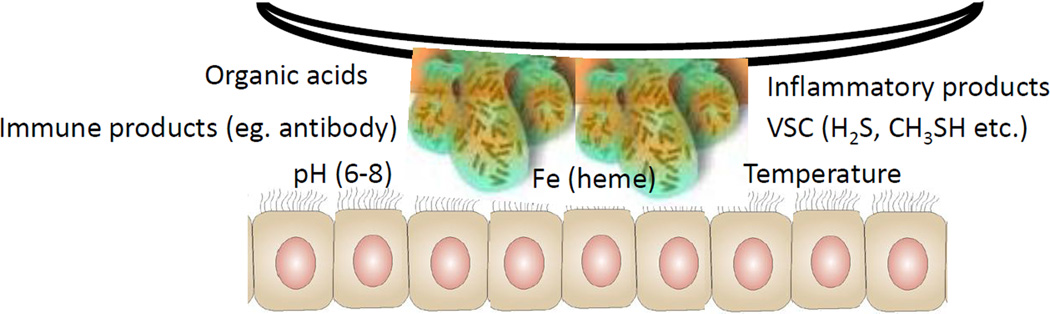
Figure 2.
Schematic of the biofilms interacting with epithelial cells monolayers. The model can be manipulated to alter a range of microenvironmental features that can help to reflect observations of the subgingival sulcus milieu in periodontitis related to host and microbial features affecting the environment.
We extended these finding using this novel model of bacterial biofilms to stimulate oral epithelial cells and profiled cytokines and chemokines that would contribute to the local inflammatory environment in the periodontium. Monospecies biofilms were developed with S. sanguinis, S. oralis, S. gordonii, A. naeslundii, F. nuclearum, and P. gingivalis on the RGPL. The human oral epithelial cells were challenged under anaerobic conditions and production of the cytokines, IL1, IL-6, and TGF, and chemokines Gro-1, IL-8, Fractalkine, MIP-1, and IP-10 was determined. P gingivalis biofilms significantly inhibited the production of all of these cytokines and chemokines, except MIP-1. Generally, the biofilms of all species inhibited Gro-1, TGF, and Fractalkine production, while F. nucleatum biofilms stimulated significant increases in IL-1, IL-6, IL-8, and IP-10. A naeslundii biofilms induced elevated levels of IL-6, IL-8 and IP-10. The oral streptococcal species in biofilms or planktonic forms were poor stimulants for any of these mediators from the epithelial cells. These results showed that oral bacterial biofilms elicit a substantially different profile of responses compared to planktonic bacteria of the same species. As importantly, some of these oral species were highly stimulatory when in biofilms. This supported the hypothesis that the biofilm bacteria interact with host cell receptors to trigger pathways of responses that appear quite divergent from individual planktonic bacteria.
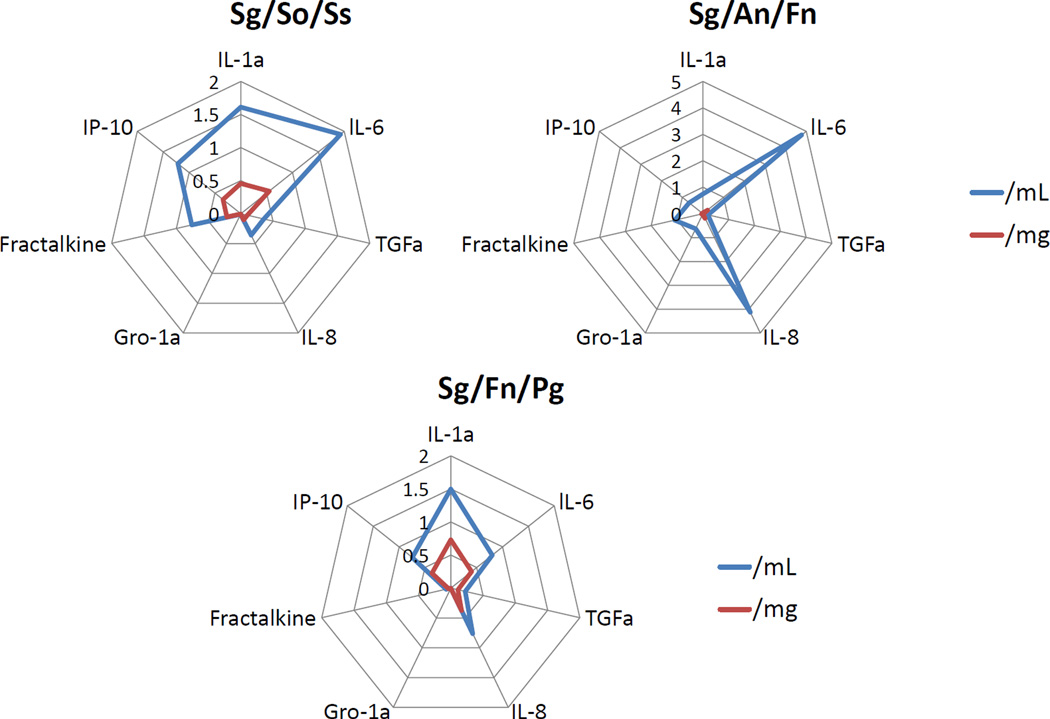
The next logical step in assessing the value of this model was based upon the recognition that the oral microbial ecology is comprised of hundreds of bacterial species that co-exist as multispecies biofilms in the oral cavity. We constructed, using the RGPL model, multispecies biofilms to stimulate cytokines/chemokines in the oral epithelial cells. We created three model biofilms. One composed of S. gordonii/ S. oralis/S. sanguinis representing the type of biofilm consistent with pioneer microorganisms and biofilms at healthy sites. A second model biofilm included S. gordonii/A. naeslundii/F. nucleatum to represent the type of microbial biofilm that appears to emerge during gingival inflammation, and contributes to the ability of more pathogenic species to emerge during progression of periodontitis. Finally, we created a multispecies biofilm with S. gordonii/F. nucleatum/P. gingivalis to represent the type of bacterial interactions that might occur in triggering host cells at diseased sites. The findings of these experiments could be summarized into 3 primary concepts: (i) select multispecies biofilms stimulated a somewhat distinctive profile of cytokines/chemokines from the oral epithelial cells. Some of these patterns were consistent with the presumed character of the different biofilms, that is commensal and pathogenic species, early coloinizers and biofilms triggering proinflammatory responses; (ii) examples of analyte levels were found in which the responses to the multispecies biofilms was significantly greater that a simple composite of the individual bacterial biofilms, supporting that the existence of individual species within the organized complex biofilms present a different profile of stimuli to the host cells; and (iii) analysis of the analyte responses adjusted for the actual number of bacteria of each species in the multispecies demonstrated what appear to be responses to the 3-dimensional structure of the biofilms was significantly greater than simply the number of the species inhabiting the environment. This was demonstrated with specific biofilms and specific cytokine/chemokine responses (Fig. 3).
Figure 3.
Spider web graphics displaying the relative fold changes in epithelial production of cytokines/chemokines when challenged with multispecies biofilms of S. gordonii/S. oralis/S. sanguis (representing features of pioneer colonizers), S. gordonii/A. naeslundii/F. nucleatum (representing transition and bridging colonizers), and S. gordonii/F. nucleatum/P. gingivalis (representing a more pathogenic biofilm). The data were evaluated by examining levels of the mediators as a concentration in the supernatants (/mL), as well as calculated estimates of the mediator levels based upon the wet weight of all of the bacteria in the biofilms (/mg).
Expanding literature continues to emphasize the importance of biofilms in medical and dental infections, and posits that an important feature of the biofilms is their complex structure and their enhanced resistance to therapeutics and host responses molecules and cells. Various reports have begun to delineate the molecular mechanisms that can occur which contribute to unique features of biofilms at the prokaryotic and eukaryotic cell levels [8, 39, 61, 71, 74, 110]. However, most of these have focused on very proscribed systems of monobacterial infections interacting with host cells. These molecular studies have attempted to better understand the altered transcriptome of the bacteria and focused on detection and intracellular signaling pathways in the mammalian cells with the bacteria. Little information has been provided examining the biology of multispecies biofilms, particularly in trying to elucidate how the host cells discriminate commensal from pathogenic microorganisms and how this recognition is translated intracellularly. Moreover, there are negligible data detailing these recognition and signaling pathways with a complex mixture of bacteria, with or without the overlying architectural features of biofilms. Nevertheless, it could be envisioned that an array of potential mechanisms could come into play in biofilms stimulating host cell responses. While clearly not an exhaustive list, the following could be considered:
The tertiary structure of the biofilms enables different biomolecules and/or bacterial components to interact with surface receptors on host cells (Fig. 4).
The contribution of quantity, as well as quality of the individual microbial constituents could influence the resulting response milieu. Thus, a large contingent of commensal bacteria could be expected to modulate an aggressive inflammatory response towards a small number of pathogenic species.
Certain bacteria within the biofilm microenvironment could up- or down-regulate host cell surface receptor expression, eg. TLRs, either enhancing or minimizing the capacity of the cells to recognize and respond to other bacteria.
Certain bacteria within the biofilm matrix, either through direct binding to host cells or release of products within the local microenvironment, could alter targeted cytokine production, which is an important autocrine signaling activity for host cell homeostasis.
Expression of a different portfolio of genes by the bacteria due to biofilm organization could trigger different responses by the host cells. As an example, recently, Frias-Lopez and Duran-Pinedo [85] reported using in vitro biofilms that the presence of oral pathogens altered patterns of gene expression of commensal bacteria including stimulation of transcription of protein-encoding genes and small noncoding RNAs.
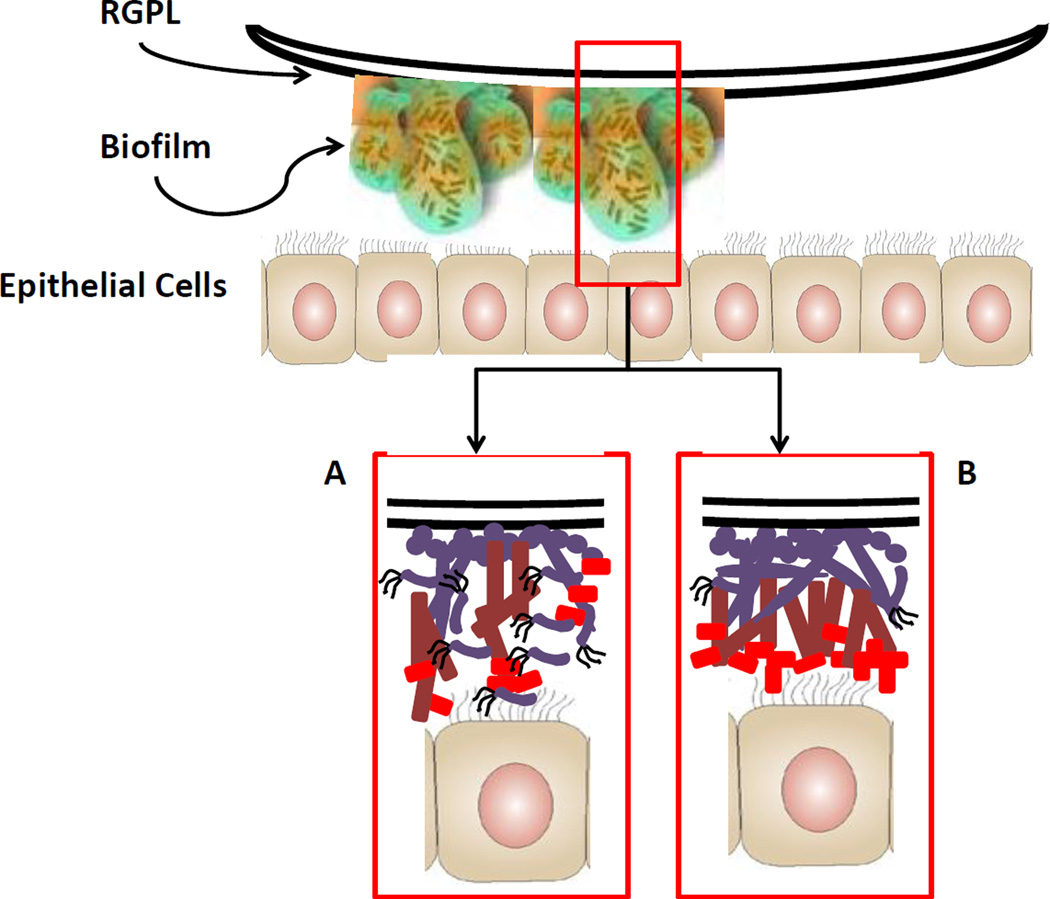
Figure 4.
Schematic of biofilm cells interacting with epithelial cells. It is clear that the multispecies biofilms create 3-dimensional towers of microcolonies; however the more detailed characteristics of the arrangement of the oral bacteria within these towers remains undefined. Slice A of the biofilm models the multispecies in which multiple members of the complex can actually directly interact with the epithelial cells, likely resulting in some composite of responses that reflect the unique features of the species. Coincident in this model would be the concept that certain species, while critical to the biofilm, are actually buried in the 3-D architecture and contribute minimally to host cell responses. Slice B of the biofilm presents a fundamentally different architecture in these towers with a more stratified arrangement of the multispecies, likely based upon co-aggregation capabilities and metabolic symbiosis. In this case, the primary response of the epithelial cells may actually reflect that species inhabiting the surface of the biofilms and not reflect as much a composite of the mixture of species that are present.
Thus, we have only scratched the surface in understanding at the molecular level the nuances of host-bacterial interactions that occur between biofilms and resident and inflammatory cells in the oral cavity.
We believe that this RGPL model system will enable us to explore variations in biofilm-host cells interactions under differing environmental conditions, eg. oxygen level, presence of volatile sulfur compounds and organic acids, and elevated pH that would be expected to occur in periodontal disease. While our studies have focused on a targeted set of mediators that could act as an important communication system connecting the external subgingival environment with the internal immune system components, the repertoire of mediators and intracellular signaling pathways that could/must be affected by the multispecies biofilm challenge needs to be better delineated. Moreover, monolayer epithelial cell cultures do not fully reflect the range of differentiation and maturation characteristics the epithelial cells express in vivo [144]. This novel biofilm model should also enable evaluation of variations in response profiles using organotypic models of epithelia, other host cell types, and host adaptive immune molecules that would interact with the biofilms within the subgingival milieu. All of these efforts have enhanced our knowledge of the complex host-bacterial interactions, but much remains to be elucidated before we understand how the host response system can differentiate the “trees from the forest”.
Host cell interactions with medically important pathogenic biofilms is reviewed
Models of multispecies oral biofilms are reviewed.
Innate immune responses of host cells to complex biofilms are reviewed.
Effect of interbacterial interactions within 3-D biofilms on host responses
Acknowledgements
This work was supported by U.S.P.H.S. grant DE 018177 from the National Institute of Dental and Craniofacial Research. We thank Dr. Karen Novak for support in developing some of the concepts of the in vitro modeling of cellular responses to the biofilms. We also acknowledge the technical contribution of Jason Stevens in using Luminex to detect the range of analytes. We also thank Jonathan Bott and Aaron Rose for their valuable help with the study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Beikler T, Flemmig TF. Oral biofilm-associated diseases: trends and implications for quality of life, systemic health and expenditures. Periodontol 2000. 2011;55:87–103. doi: 10.1111/j.1600-0757.2010.00360.x. [DOI] [PubMed] [Google Scholar]
- 2.Petersen PE, Bourgeois D, Ogawa H, Estupinan-Day S, Ndiaye C. The global burden of oral diseases and risks to oral health. Bull World Health Organ. 2005;83:661–669. [PMC free article] [PubMed] [Google Scholar]
- 3.Livorsi DJ, Stenehjem E, Stephens DS. Virulence factors of gram-negative bacteria in sepsis with a focus on Neisseria meningitidis. Contrib Microbiol. 2011;17:31–47. doi: 10.1159/000324008. [DOI] [PubMed] [Google Scholar]
- 4.Costerton JW, Montanaro L, Arciola CR. Biofilm in implant infections: its production and regulation. Int J Artif Organs. 2005;28:1062–1068. doi: 10.1177/039139880502801103. [DOI] [PubMed] [Google Scholar]
- 5.Fey PD, Olson ME. Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 2010;5:917–933. doi: 10.2217/fmb.10.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol. 2009;11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 7.Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- 8.Huang R, Li M, Gregory RL. Bacterial interactions in dental biofilm. Virulence. 2011;2:435–444. doi: 10.4161/viru.2.5.16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolenbrander PE, Palmer RJ, Jr, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 10.Zijnge V, Ammann T, Thurnheer T, Gmur R. Subgingival biofilm structure. Front Oral Biol. 2012;15:1–16. doi: 10.1159/000329667. [DOI] [PubMed] [Google Scholar]
- 11.Marsh PD, Devine DA. How is the development of dental biofilms influenced by the host? J Clin Periodontol. 2011;38(Suppl 11):28–35. doi: 10.1111/j.1600-051X.2010.01673.x. [DOI] [PubMed] [Google Scholar]
- 12.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 13.Rybtke MT, Jensen PO, Hoiby N, Givskov M, Tolker-Nielsen T, Bjarnsholt T. The implication of Pseudomonas aeruginosa biofilms in infections. Inflamm Allergy Drug Targets. 2011;10:141–157. doi: 10.2174/187152811794776222. [DOI] [PubMed] [Google Scholar]
- 14.Ford PJ, Raphael SL, Cullinan MP, Jenkins AJ, West MJ, Seymour GJ. Why should a doctor be interested in oral disease? Expert Rev Cardiovasc Ther. 2010;8:1483–1493. doi: 10.1586/erc.10.109. [DOI] [PubMed] [Google Scholar]
- 15.Lasa I, Del Pozo JL, Penades JR, Leiva J. Bacterial biofilms and infection. An Sist Sanit Navar. 2005;28:163–175. doi: 10.4321/s1137-66272005000300002. [DOI] [PubMed] [Google Scholar]
- 16.Cernohorska L, Votava M. Biofilms and their significance in medical microbiology. Epidemiol Mikrobiol Imunol. 2002;51:161–164. [PubMed] [Google Scholar]
- 17.Cos P, Tote K, Horemans T, Maes L. Biofilms: an extra hurdle for effective antimicrobial therapy. Curr Pharm Des. 2010;16:2279–2295. doi: 10.2174/138161210791792868. [DOI] [PubMed] [Google Scholar]
- 18.Keller D, Costerton JW. Oral biofilm: entry and immune system response. Compend Contin Educ Dent. 2009;30:24–32. quiz 4, 6. [PubMed] [Google Scholar]
- 19.Schaudinn C, Gorur A, Keller D, Sedghizadeh PP, Costerton JW. Periodontitis: an archetypical biofilm disease. J Am Dent Assoc. 2009;140:978–986. doi: 10.14219/jada.archive.2009.0307. [DOI] [PubMed] [Google Scholar]
- 20.Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence. 2011;2:445–459. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pang JM, Layre E, Sweet L, Sherrid A, Moody DB, Ojha A, et al. The polyketide Pks1 contributes to biofilm formation in Mycobacterium tuberculosis. J Bacteriol. 2012;194:715–721. doi: 10.1128/JB.06304-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulka K, Hatfull G, Ojha AK. Growth of Mycobacterium tuberculosis biofilms. J Vis Exp. 2012 doi: 10.3791/3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinnebusch BJ, Erickson DL. Yersinia pestis biofilm in the flea vector and its role in the transmission of plague. Curr Top Microbiol Immunol. 2008;322:229–248. doi: 10.1007/978-3-540-75418-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hostacka A, Ciznar I, Stefkovicova M. Temperature and pH affect the production of bacterial biofilm. Folia Microbiol (Praha) 2010;55:75–78. doi: 10.1007/s12223-010-0012-y. [DOI] [PubMed] [Google Scholar]
- 25.Harjai K, Khandwahaa RK, Mittal R, Yadav V, Gupta V, Sharma S. Effect of pH on production of virulence factors by biofilm cells of Pseudomonas aeruginosa. Folia Microbiol (Praha) 2005;50:99–102. doi: 10.1007/BF02931455. [DOI] [PubMed] [Google Scholar]
- 26.Moriarty TF, Elborn JS, Tunney MM. Effect of pH on the antimicrobial susceptibility of planktonic and biofilm-grown clinical Pseudomonas aeruginosa isolates. Br J Biomed Sci. 2007;64:101–104. doi: 10.1080/09674845.2007.11732766. [DOI] [PubMed] [Google Scholar]
- 27.He X, Ahn J. Differential gene expression in planktonic and biofilm cells of multiple antibiotic-resistant Salmonella Typhimurium and Staphylococcus aureus. FEMS Microbiol Lett. 2011;325:180–188. doi: 10.1111/j.1574-6968.2011.02429.x. [DOI] [PubMed] [Google Scholar]
- 28.Murray TS, Egan M, Kazmierczak BI. Pseudomonas aeruginosa chronic colonization in cystic fibrosis patients. Curr Opin Pediatr. 2007;19:83–88. doi: 10.1097/MOP.0b013e3280123a5d. [DOI] [PubMed] [Google Scholar]
- 29.Hill D, Rose B, Pajkos A, Robinson M, Bye P, Bell S, et al. Antibiotic susceptabilities of Pseudomonas aeruginosa isolates derived from patients with cystic fibrosis under aerobic, anaerobic, and biofilm conditions. J Clin Microbiol. 2005;43:5085–5090. doi: 10.1128/JCM.43.10.5085-5090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Secor PR, James GA, Fleckman P, Olerud JE, McInnerney K, Stewart PS. Staphylococcus aureus Biofilm and Planktonic cultures differentially impact gene expression, mapk phosphorylation, and cytokine production in human keratinocytes. BMC Microbiol. 2011;11:143. doi: 10.1186/1471-2180-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirker KR, Secor PR, James GA, Fleckman P, Olerud JE, Stewart PS. Loss of viability and induction of apoptosis in human keratinocytes exposed to Staphylococcus aureus biofilms in vitro. Wound Repair Regen. 2009;17:690–699. doi: 10.1111/j.1524-475X.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guenther F, Stroh P, Wagner C, Obst U, Hansch GM. Phagocytosis of staphylococci biofilms by polymorphonuclear neutrophils: S. aureus and S. epidermidis differ with regard to their susceptibility towards the host defense. Int J Artif Organs. 2009;32:565–573. doi: 10.1177/039139880903200905. [DOI] [PubMed] [Google Scholar]
- 33.Stroh P, Gunther F, Meyle E, Prior B, Wagner C, Hansch GM. Host defence against Staphylococcus aureus biofilms by polymorphonuclear neutrophils: oxygen radical production but not phagocytosis depends on opsonisation with immunoglobulin G. Immunobiology. 2011;216:351–357. doi: 10.1016/j.imbio.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Gunther F, Wabnitz GH, Stroh P, Prior B, Obst U, Samstag Y, et al. Host defence against Staphylococcus aureus biofilms infection: phagocytosis of biofilms by polymorphonuclear neutrophils (PMN) Mol Immunol. 2009;46:1805–1813. doi: 10.1016/j.molimm.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 35.Mittal R, Sharma S, Chhibber S, Harjai K. Evaluation of tumour necrosis factor-alpha and interleukin-1beta in an experimental pyelonephritis model induced with planktonic and biofilms cells of Pseudomonas aeruginosa. Can J Infect Dis Med Microbiol. 2009;20:e35–e42. doi: 10.1155/2009/810791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mittal R, Sharma S, Chhibber S, Harjai K. Effect of macrophage secretory products on elaboration of virulence factors by planktonic and biofilm cells of Pseudomonas aeruginosa. Comp Immunol Microbiol Infect Dis. 2006;29:12–26. doi: 10.1016/j.cimid.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Aujla SJ, Dubin PJ, Kolls JK. Th17 cells and mucosal host defense. Semin Immunol. 2007;19:377–382. doi: 10.1016/j.smim.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jesaitis AJ, Franklin MJ, Berglund D, Sasaki M, Lord CI, Bleazard JB, et al. Compromised host defense on Pseudomonas aeruginosa biofilms: characterization of neutrophil and biofilm interactions. J Immunol. 2003;171:4329–4339. doi: 10.4049/jimmunol.171.8.4329. [DOI] [PubMed] [Google Scholar]
- 39.Diaz PI. Microbial diversity and interactions in subgingival biofilm communities. Front Oral Biol. 2012;15:17–40. doi: 10.1159/000329669. [DOI] [PubMed] [Google Scholar]
- 40.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teles FR, Teles RP, Uzel NG, Song XQ, Torresyap G, Socransky SS, et al. Early microbial succession in redeveloping dental biofilms in periodontal health and disease. J Periodontal Res. 2012;47:95–104. doi: 10.1111/j.1600-0765.2011.01409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Curtis MA, Zenobia C, Darveau RP. The relationship of the oral microbiotia to periodontal health and disease. Cell Host Microbe. 2011;10:302–306. doi: 10.1016/j.chom.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kebschull M, Papapanou PN. Periodontal microbial complexes associated with specific cell and tissue responses. J Clin Periodontol. 2011;38(Suppl 11):17–27. doi: 10.1111/j.1600-051X.2010.01668.x. [DOI] [PubMed] [Google Scholar]
- 44.Huang C, Altimova Y, Strange S, Ebersole J. Polybacterial challenge effects on cytokine/chemokine production by macrophages and dendritic cells. Inflammation Research. 2011;60:119–125. doi: 10.1007/s00011-010-0242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Johansson A. Aggregatibacter actinomycetemcomitans Leukotoxin: A Powerful Tool with Capacity to Cause Imbalance in the Host Inflammatory Response. Toxins (Basel) 2011;3:242–259. doi: 10.3390/toxins3030242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kadowaki T, Takii R, Yamatake K, Kawakubo T, Tsukuba T, Yamamoto K. A role for gingipains in cellular responses and bacterial survival in Porphyromonas gingivalis-infected cells. Front Biosci. 2007;12:4800–4809. doi: 10.2741/2428. [DOI] [PubMed] [Google Scholar]
- 47.Belibasakis GN, Bostanci N, Reddi D. Regulation of protease-activated receptor-2 expression in gingival fibroblasts and Jurkat T cells by Porphyromonas gingivalis. Cell Biol Int. 2010;34:287–292. doi: 10.1042/CBI20090290. [DOI] [PubMed] [Google Scholar]
- 48.Madianos PN, Bobetsis YA, Kinane DF. Generation of inflammatory stimuli: how bacteria set up inflammatory responses in the gingiva. J Clin Periodontol. 2005;32(Suppl 6):57–71. doi: 10.1111/j.1600-051X.2005.00821.x. [DOI] [PubMed] [Google Scholar]
- 49.Kajiya M, Komatsuzawa H, Papantonakis A, Seki M, Makihira S, Ouhara K, et al. Aggregatibacter actinomycetemcomitans Omp29 is associated with bacterial entry to gingival epithelial cells by F-actin rearrangement. PLoS One. 2011;6:e18287. doi: 10.1371/journal.pone.0018287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakakibara J, Nagano K, Murakami Y, Higuchi N, Nakamura H, Shimozato K, et al. Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiology. 2007;153:866–876. doi: 10.1099/mic.0.29275-0. [DOI] [PubMed] [Google Scholar]
- 51.Ding PH, Wang CY, Darveau RP, Jin L. Porphyromonas gingivalis LPS stimulates the expression of LPS-binding protein in human oral keratinocytes in vitro. Innate Immun. 2012 doi: 10.1177/1753425912450348. [DOI] [PubMed] [Google Scholar]
- 52.Herath TD, Wang Y, Seneviratne CJ, Lu Q, Darveau RP, Wang CY, et al. Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity differentially modulates the expression of IL-6 and IL-8 in human gingival fibroblasts. J Clin Periodontol. 2011;38:694–701. doi: 10.1111/j.1600-051X.2011.01741.x. [DOI] [PubMed] [Google Scholar]
- 53.Jeon DI, Park SR, Ahn MY, Ahn SG, Park JH, Yoon JH. NOD1 and NOD2 stimulation triggers innate immune responses of human periodontal ligament cells. Int J Mol Med. 2012;29:699–703. doi: 10.3892/ijmm.2012.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang L, Zhou XD, Wang Q, Zhang L, Wang Y, Li XY, et al. Expression of TRAF6 and pro-inflammatory cytokines through activation of TLR2, TLR4, NOD1, and NOD2 in human periodontal ligament fibroblasts. Arch Oral Biol. 2011;56:1064–1072. doi: 10.1016/j.archoralbio.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 55.Palmer RJ., Jr Supragingival and subgingival plaque: paradigm of biofilms. Compend Contin Educ Dent. 2010;31:104–106. 8, 10 passim; quiz 24, 38. [PubMed] [Google Scholar]
- 56.Takeuchi H, Yamanaka Y, Yamamoto K. Morphological analysis of subgingival biofilm formation on synthetic carbonate apatite inserted into human periodontal pockets. Aust Dent J. 2004;49:72–77. doi: 10.1111/j.1834-7819.2004.tb00053.x. [DOI] [PubMed] [Google Scholar]
- 57.Faveri M, Figueiredo LC, Duarte PM, Mestnik MJ, Mayer MP, Feres M. Microbiological profile of untreated subjects with localized aggressive periodontitis. J Clin Periodontol. 2009;36:739–749. doi: 10.1111/j.1600-051X.2009.01449.x. [DOI] [PubMed] [Google Scholar]
- 58.Heller D, Varela VM, Silva-Senem MX, Torres MC, Feres-Filho EJ, Colombo AP. Impact of systemic antimicrobials combined with anti-infective mechanical debridement on the microbiota of generalized aggressive periodontitis: a 6-month RCT. J Clin Periodontol. 2011;38:355–364. doi: 10.1111/j.1600-051X.2011.01707.x. [DOI] [PubMed] [Google Scholar]
- 59.Colombo AP, Bennet S, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. Impact of Periodontal Therapy on the Subgingival Microbiota of Severe Periodontitis: Comparison between Good Responders and "Refractory" Subjects by the Human Oral Microbe Identification Microarray (HOMIM) J Periodontol. 2012 doi: 10.1902/jop.2012.110566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jakubovics NS, Kolenbrander PE. The road to ruin: the formation of disease-associated oral biofilms. Oral Dis. 2010;16:729–739. doi: 10.1111/j.1601-0825.2010.01701.x. [DOI] [PubMed] [Google Scholar]
- 61.Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8:471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 62.Ding AM, Palmer RJ, Jr, Cisar JO, Kolenbrander PE. Shear-enhanced oral microbial adhesion. Appl Environ Microbiol. 2010;76:1294–1297. doi: 10.1128/AEM.02083-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brooks W, Demuth DR, Gil S, Lamont RJ. Identification of a Streptococcus gordonii SspB domain that mediates adhesion to Porphyromonas gingivalis. Infect Immun. 1997;65:3753–3758. doi: 10.1128/iai.65.9.3753-3758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daep CA, Novak EA, Lamont RJ, Demuth DR. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect Immun. 2011;79:67–74. doi: 10.1128/IAI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nagata H, Iwasaki M, Maeda K, Kuboniwa M, Hashino E, Toe M, et al. Identification of the binding domain of Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase for Porphyromonas gingivalis major fimbriae. Infect Immun. 2009;77:5130–5138. doi: 10.1128/IAI.00439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Forsgren N, Lamont RJ, Persson K. Crystal structure of the variable domain of the Streptococcus gordonii surface protein SspB. Protein Sci. 2009;18:1896–1905. doi: 10.1002/pro.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hamada T, Kawashima M, Watanabe H, Tagami J, Senpuku H. Molecular interactions of surface protein peptides of Streptococcus gordonii with human salivary components. Infect Immun. 2004;72:4819–4826. doi: 10.1128/IAI.72.8.4819-4826.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mishra A, Wu C, Yang J, Cisar JO, Das A, Ton-That H. The Actinomyces oris type-2 fimbrial shaft FimA mediates co-aggregation with oral streptococci, adherence to red blood cells and biofilm development. Mol Microbiol. 2010 doi: 10.1111/j.1365-2958.2010.07252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li T, Khah MK, Slavnic S, Johansson I, Stromberg N. Different type 1 fimbrial genes and tropisms of commensal and potentially pathogenic Actinomyces spp. with different salivary acidic proline-rich protein and statherin ligand specificities. Infect Immun. 2001;69:7224–7233. doi: 10.1128/IAI.69.12.7224-7233.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takahashi N, Washio J, Mayanagi G. Metabolomics of supragingival plaque and oral bacteria. J Dent Res. 2010;89:1383–1388. doi: 10.1177/0022034510377792. [DOI] [PubMed] [Google Scholar]
- 71.Hojo K, Nagaoka S, Ohshima T, Maeda N. Bacterial interactions in dental biofilm development. J Dent Res. 2009;88:982–990. doi: 10.1177/0022034509346811. [DOI] [PubMed] [Google Scholar]
- 72.Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Sommer DD, et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One. 2012;7:e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schaudinn C, Gorur A, Keller D, Sedghizadeh PP, Costerton JW. Periodontitis: an archetypical biofilm disease. J Am Dent Assoc. 2009;140:978–986. doi: 10.14219/jada.archive.2009.0307. [DOI] [PubMed] [Google Scholar]
- 74.Davey ME, Costerton JW. Molecular genetics analyses of biofilm formation in oral isolates. Periodontol 2000. 2006;42:13–26. doi: 10.1111/j.1600-0757.2006.00052.x. [DOI] [PubMed] [Google Scholar]
- 75.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 76.Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 77.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Listgarten MA. A rationale for monitoring the periodontal microbiota after periodontal treatment. J Periodontol. 1988;59:439–444. doi: 10.1902/jop.1988.59.7.439. [DOI] [PubMed] [Google Scholar]
- 79.Listgarten MA. Direct microscopy of periodontal pathogens. Oral Microbiol Immunol. 1986;1:31–38. doi: 10.1111/j.1399-302x.1986.tb00314.x. [DOI] [PubMed] [Google Scholar]
- 80.Listgarten MA. The structure of dental plaque. Periodontol 2000. 1994;5:52–65. doi: 10.1111/j.1600-0757.1994.tb00018.x. [DOI] [PubMed] [Google Scholar]
- 81.Beighton D. Can the ecology of the dental biofilm be beneficially altered? Adv Dent Res. 2009;21:69–73. doi: 10.1177/0895937409335641. [DOI] [PubMed] [Google Scholar]
- 82.Paddick JS, Brailsford SR, Rao S, Soares RF, Kidd EA, Beighton D, et al. Effect of biofilm growth on expression of surface proteins of Actinomyces naeslundii genospecies 2. Appl Environ Microbiol. 2006;72:3774–3779. doi: 10.1128/AEM.72.5.3774-3779.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Periasamy S, Kolenbrander PE. Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of enamel. J Bacteriol. 2010;192:2965–2972. doi: 10.1128/JB.01631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gursoy UK, Pollanen M, Kononen E, Uitto VJ. Biofilm formation enhances the oxygen tolerance and invasiveness of Fusobacterium nucleatum in an oral mucosa culture model. J Periodontol. 2010;81:1084–1091. doi: 10.1902/jop.2010.090664. [DOI] [PubMed] [Google Scholar]
- 85.Frias-Lopez J, Duran-Pinedo A. Effect of periodontal pathogens on the metatranscriptome of a healthy multispecies biofilm model. J Bacteriol. 2012;194:2082–2095. doi: 10.1128/JB.06328-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Redanz S, Standar K, Podbielski A, Kreikemeyer B. A five-species transcriptome array for oral mixed-biofilm studies. PLoS One. 2011;6:e27827. doi: 10.1371/journal.pone.0027827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu J, Wu C, Huang IH, Merritt J, Qi F. Differential response of Streptococcus mutans towards friend and foe in mixed-species cultures. Microbiology. 2011;157:2433–2444. doi: 10.1099/mic.0.048314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Klein MI, Xiao J, Heydorn A, Koo H. An analytical tool-box for comprehensive biochemical, structural and transcriptome evaluation of oral biofilms mediated by mutans streptococci. J Vis Exp. 2011 doi: 10.3791/2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Klein MI, DeBaz L, Agidi S, Lee H, Xie G, Lin AH, et al. Dynamics of Streptococcus mutans transcriptome in response to starch and sucrose during biofilm development. PLoS One. 2010;5:e13478. doi: 10.1371/journal.pone.0013478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sissons CH. Artificial dental plaque biofilm model systems. Adv Dent Res. 1997;11:110–126. doi: 10.1177/08959374970110010201. [DOI] [PubMed] [Google Scholar]
- 91.Al-Ahmad A, Follo M, Selzer A-C, Hellwig E, Hannig M, Hannig C. Bacterial colonisation of enamel in situ investigated with fluorescence in situ hybridization (FISH) J Med Microbiol. 2009;58:1359–1366. doi: 10.1099/jmm.0.011213-0. [DOI] [PubMed] [Google Scholar]
- 92.Chalmers NI, Palmer RJ, Jr, Cisar JO, Kolenbrander PE. Characterization of a Streptococcus sp.-Veillonella sp. Community Micromanipulated from Dental Plaque. J Bacteriol. 2008;190:8145–8154. doi: 10.1128/JB.00983-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thurnheer T, Gmür R, Guggenheim B. Multiplex FISH analysis of a six-species bacterial biofilm. Journal of Microbiological Methods. 2004;56:37–47. doi: 10.1016/j.mimet.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 94.Fischer AH, Jacobson KA, Rose J, Zeller R. Fixation and permeabilization of cells and tissues. CSH Protoc. 2008;2008 doi: 10.1101/pdb.top36. pdb top36. [DOI] [PubMed] [Google Scholar]
- 95.Loqman MY, Bush PG, Farquharson C, Hall AC. A cell shrinkage artefact in growth plate chondrocytes with common fixative solutions: importance of fixative osmolarity for maintaining morphology. Eur Cell Mater. 2010;19:214–227. doi: 10.22203/ecm.v019a21. [DOI] [PubMed] [Google Scholar]
- 96.Dige I, Nilsson H, Kilian M, Nyvad B. In situ identification of streptococci and other bacteria in initial dental biofilm by confocal laser scanning microscopy and fluorescence in situ hybridization. Eur J Oral Sci. 2007;115:459–467. doi: 10.1111/j.1600-0722.2007.00494.x. [DOI] [PubMed] [Google Scholar]
- 97.Periasamy S, Kolenbrander PE. Aggregatibacter actinomycetemcomitans Builds Mutualistic Biofilm Communities with Fusobacterium nucleatum and Veillonella Species in Saliva. Infect Immun. 2009a;77:3542–3551. doi: 10.1128/IAI.00345-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Capestany CA, Kuboniwa M, Jung IY, Park Y, Tribble GD, Lamont RJ. Role of the Porphyromonas gingivalis InlJ protein in homotypic and heterotypic biofilm development. Infect Immun. 2006;74:3002–3005. doi: 10.1128/IAI.74.5.3002-3005.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shen Y, Stojicic S, Haapasalo M. Bacterial viability in starved and revitalized biofilms: comparison of viability staining and direct culture. J Endod. 2010;36:1820–1823. doi: 10.1016/j.joen.2010.08.029. [DOI] [PubMed] [Google Scholar]
- 100.Kolenbrander PE, Egland PG, Diaz PI, Palmer RJ., Jr Genome-genome interactions: bacterial communities in initial dental plaque. Trends Microbiol. 2005;13:11–15. doi: 10.1016/j.tim.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 101.Jun HK, Kang YM, Lee HR, Lee SH, Choi BK. Highly conserved surface proteins of oral spirochetes as adhesins and potent inducers of proinflammatory and osteoclastogenic factors. Infect Immun. 2008;76:2428–2438. doi: 10.1128/IAI.01128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kuboniwa M, Hendrickson E, Xia Q, Wang T, Xie H, Hackett M, et al. Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiology. 2009;9:98. doi: 10.1186/1471-2180-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Daep CA, Lamont RJ, Demuth DR. Interaction of Porphyromonas gingivalis with Oral Streptococci Requires a Motif That Resembles the Eukaryotic Nuclear Receptor Box Protein-Protein Interaction Domain. Infect Immun. 2008;76:3273–3280. doi: 10.1128/IAI.00366-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Periasamy S, Kolenbrander PE. Mutualistic biofilm communities develop with Porphyromonas gingivalis and initial, early, and late colonizers of enamel. J Bacteriol. 2009;191:6804–6811. doi: 10.1128/JB.01006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kuboniwa M, Hendrickson EL, Xia Q, Wang T, Xie H, Hackett M, et al. Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiol. 2009;9:98. doi: 10.1186/1471-2180-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lo AW, Seers CA, Boyce JD, Dashper SG, Slakeski N, Lissel JP, et al. Comparative transcriptomic analysis of Porphyromonas gingivalis biofilm and planktonic cells. BMC Microbiol. 2009;9:18. doi: 10.1186/1471-2180-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dige I, Raarup MK, Nyengaard JR, Kilian M, Nyvad B. Actinomyces naeslundii in initial dental biofilm formation. Microbiology. 2009;155:2116–2126. doi: 10.1099/mic.0.027706-0. [DOI] [PubMed] [Google Scholar]
- 108.Hajishengallis G, Krauss JL, Liang S, McIntosh ML, Lambris JD. Pathogenic microbes and community service through manipulation of innate immunity. Adv Exp Med Biol. 2012;946:69–85. doi: 10.1007/978-1-4614-0106-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, et al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 110.Kolenbrander PE. Multispecies communities: interspecies interactions influence growth on saliva as sole nutritional source. Int J Oral Sci. 2011;3:49–54. doi: 10.4248/IJOS11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xiao J, Klein MI, Falsetta ML, Lu B, Delahunty CM, Yates JR, 3rd, et al. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 2012;8:e1002623. doi: 10.1371/journal.ppat.1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Grant MM, Kolamunne RT, Lock FE, Matthews JB, Chapple ILC, Griffiths HR. Oxygen tension modulates the cytokine response of oral epithelium to periodontal bacteria. Journal of Clinical Periodontology. 2010;37:1039–1048. doi: 10.1111/j.1600-051X.2010.01622.x. [DOI] [PubMed] [Google Scholar]
- 113.Simoes M. Antimicrobial strategies effective against infectious bacterial biofilms. Curr Med Chem. 2011;18:2129–2145. doi: 10.2174/092986711795656216. [DOI] [PubMed] [Google Scholar]
- 114.Lazar V, Chifiriuc MC. Medical significance and new therapeutical strategies for biofilm associated infections. Roum Arch Microbiol Immunol. 2010;69:125–138. [PubMed] [Google Scholar]
- 115.Roberts AP, Mullany P. Oral biofilms: a reservoir of transferable, bacterial, antimicrobial resistance. Expert Rev Anti Infect Ther. 2010;8:1441–1450. doi: 10.1586/eri.10.106. [DOI] [PubMed] [Google Scholar]
- 116.Kristian SA, Birkenstock TA, Sauder U, Mack D, Gotz F, Landmann R. Biofilm formation induces C3a release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. J Infect Dis. 2008;197:1028–1035. doi: 10.1086/528992. [DOI] [PubMed] [Google Scholar]
- 117.Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, et al. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol. 2011;186:6585–6596. doi: 10.4049/jimmunol.1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Leid JG, Willson CJ, Shirtliff ME, Hassett DJ, Parsek MR, Jeffers AK. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J Immunol. 2005;175:7512–7518. doi: 10.4049/jimmunol.175.11.7512. [DOI] [PubMed] [Google Scholar]
- 119.Zijnge V, van Leeuwen MB, Degener JE, Abbas F, Thurnheer T, Gmur R, et al. Oral biofilm architecture on natural teeth. PLoS One. 2010;5:e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Eberhard J, Menzel N, Dommisch H, Winter J, Jepsen S, Mutters R. The stage of native biofilm formation determines the gene expression of human beta-defensin-2, psoriasin, ribonuclease 7 and inflammatory mediators: a novel approach for stimulation of keratinocytes with in situ formed biofilms. Oral Microbiol Immunol. 2008;23:21–28. doi: 10.1111/j.1399-302X.2007.00385.x. [DOI] [PubMed] [Google Scholar]
- 121.Eberhard J, Pietschmann R, Falk W, Jepsen S, Dommisch H. The immune response of oral epithelial cells induced by single-species and complex naturally formed biofilms. Oral Microbiol Immunol. 2009;24:325–330. doi: 10.1111/j.1399-302X.2009.00518.x. [DOI] [PubMed] [Google Scholar]
- 122.Guggenheim B, Gmur R, Galicia J, Stathopoulou P, Benakanakere M, Meier A, et al. In vitro modeling of host-parasite interactions: the 'subgingival' biofilm challenge of primary human epithelial cells. BMC Microbiology. 2009;9:280. doi: 10.1186/1471-2180-9-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Belibasakis GN, Meier A, Guggenheim B, Bostanci N. Oral biofilm challenge regulates the RANKL-OPG system in periodontal ligament and dental pulp cells. Microbial Pathogenesis. 2011;50:6–11. doi: 10.1016/j.micpath.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 124.Kebschull M, Papapanou PN. Periodontal microbial complexes associated with specific cell and tissue responses. Journal of Clinical Periodontology. 2011;38:17–27. doi: 10.1111/j.1600-051X.2010.01668.x. [DOI] [PubMed] [Google Scholar]
- 125.Teng YT. Protective and destructive immunity in the periodontium: Part 1--innate and humoral immunity and the periodontium. J Dent Res. 2006;85:198–208. doi: 10.1177/154405910608500301. [DOI] [PubMed] [Google Scholar]
- 126.Teles RP, Gursky LC, Faveri M, Rosa EA, Teles FR, Feres M, et al. Relationships between subgingival microbiota and GCF biomarkers in generalized aggressive periodontitis. J Clin Periodontol. 2010;37:313–323. doi: 10.1111/j.1600-051X.2010.01534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Teles R, Sakellari D, Teles F, Konstantinidis A, Kent R, Socransky S, et al. Relationships among gingival crevicular fluid biomarkers, clinical parameters of periodontal disease, and the subgingival microbiota. J Periodontol. 2010;81:89–98. doi: 10.1902/jop.2009.090397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Polland KE, Higgins F, Orchardson R. Salivary flow rate and pH during prolonged gum chewing in humans. J Oral Rehabil. 2003;30:861–865. doi: 10.1046/j.1365-2842.2003.01177.x. [DOI] [PubMed] [Google Scholar]
- 129.Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Noguchi K, Miwa Y, Sunohara M, Sato I. Analysis of vascular distribution and growth factors in human gingival tissue associated with periodontal probing depth. Okajimas Folia Anat Jpn. 2011;88:75–83. doi: 10.2535/ofaj.88.75. [DOI] [PubMed] [Google Scholar]
- 131.Xiao X, Li Y, Zhang G, Gao Y, Kong Y, Liu M, et al. Detection of bacterial diversity in rat's periodontitis model under imitational altitude hypoxia environment. Arch Oral Biol. 2012;57:23–29. doi: 10.1016/j.archoralbio.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 132.Ebersole JL, Cappelli D. Gingival crevicular fluid antibody to Actinobacillus actinomycetemcomitans in periodontal disease. Oral Microbiol Immunol. 1994;9:335–344. doi: 10.1111/j.1399-302x.1994.tb00283.x. [DOI] [PubMed] [Google Scholar]
- 133.Ebersole JL, Cappelli D, Steffen MJ, Willmann DE, O'Dell DS. Host response assessment in recurring periodontitis. J Clin Periodontol. 1996;23:258–262. doi: 10.1111/j.1600-051x.1996.tb02085.x. [DOI] [PubMed] [Google Scholar]
- 134.Benakanakere M, Kinane DF. Innate cellular responses to the periodontal biofilm. Front Oral Biol. 2012;15:41–55. doi: 10.1159/000329670. [DOI] [PubMed] [Google Scholar]
- 135.Lamster IB, Novak MJ. Host mediators in gingival crevicular fluid: implications for the pathogenesis of periodontal disease. Crit Rev Oral Biol Med. 1992;3:31–60. doi: 10.1177/10454411920030010501. [DOI] [PubMed] [Google Scholar]
- 136.Scott DA, Krauss J. Neutrophils in periodontal inflammation. Front Oral Biol. 2012;15:56–83. doi: 10.1159/000329672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jiemtaweeboon S, Shirasuna K, Nitta A, Kobayashi A, Schuberth HJ, Shimizu T, et al. Evidence that polymorphonuclear neutrophils infiltrate into the developing corpus luteum and promote angiogenesis with interleukin-8 in the cow. Reprod Biol Endocrinol. 2011;9:79. doi: 10.1186/1477-7827-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vernier A, Diab M, Soell M, Haan-Archipoff G, Beretz A, Wachsmann D, et al. Cytokine production by human epithelial and endothelial cells following exposure to oral viridans streptococci involves lectin interactions between bacteria and cell surface receptors. Infect Immun. 1996;64:3016–3022. doi: 10.1128/iai.64.8.3016-3022.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hasegawa Y, Mans JJ, Mao S, Lopez MC, Baker HV, Handfield M, et al. Gingival epithelial cell transcriptional responses to commensal and opportunistic oral microbial species. Infect Immun. 2007;75:2540–2547. doi: 10.1128/IAI.01957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bodet C, Chandad F, Grenier D. Inflammatory responses of a macrophage/epithelial cell co-culture model to mono and mixed infections with Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia. Microbes Infect. 2006;8:27–35. doi: 10.1016/j.micinf.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 141.Sandros J, Karlsson C, Lappin DF, Madianos PN, Kinane DF, Papapanou PN. Cytokine responses of oral epithelial cells to Porphyromonas gingivalis infection. J Dent Res. 2000;79:1808–1814. doi: 10.1177/00220345000790101301. [DOI] [PubMed] [Google Scholar]
- 142.Peyyala R, Kirakodu S, Novak KF, Ebersole JL. Epithelial interleukin-8 responses to oral bacterial biofilms. Clinical and vaccine immunology: CVI. 2011;18:1770–1772. doi: 10.1128/CVI.05162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Peyyala R, Kirakodu SS, Ebersole JL, Novak KF. Characterization of a Novel Model for Multispecies Biofilms Using Rigid Gas Permeable Lenses. Appl Environ Microbiol. 2011;77:3413–3421. doi: 10.1128/AEM.00039-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Seifi A, Weaver EM, Whipple ME, Ikoma M, Farrenberg J, Huang M-L, et al. The lytic activation of KSHV during keratinocyte differentiation is dependent upon a suprabasal position, the loss of integrin engagement, and calcium, but not the interaction of cadherins. Virology. 2011;410:17–29. doi: 10.1016/j.virol.2010.10.023. [DOI] [PubMed] [Google Scholar]