Abstract
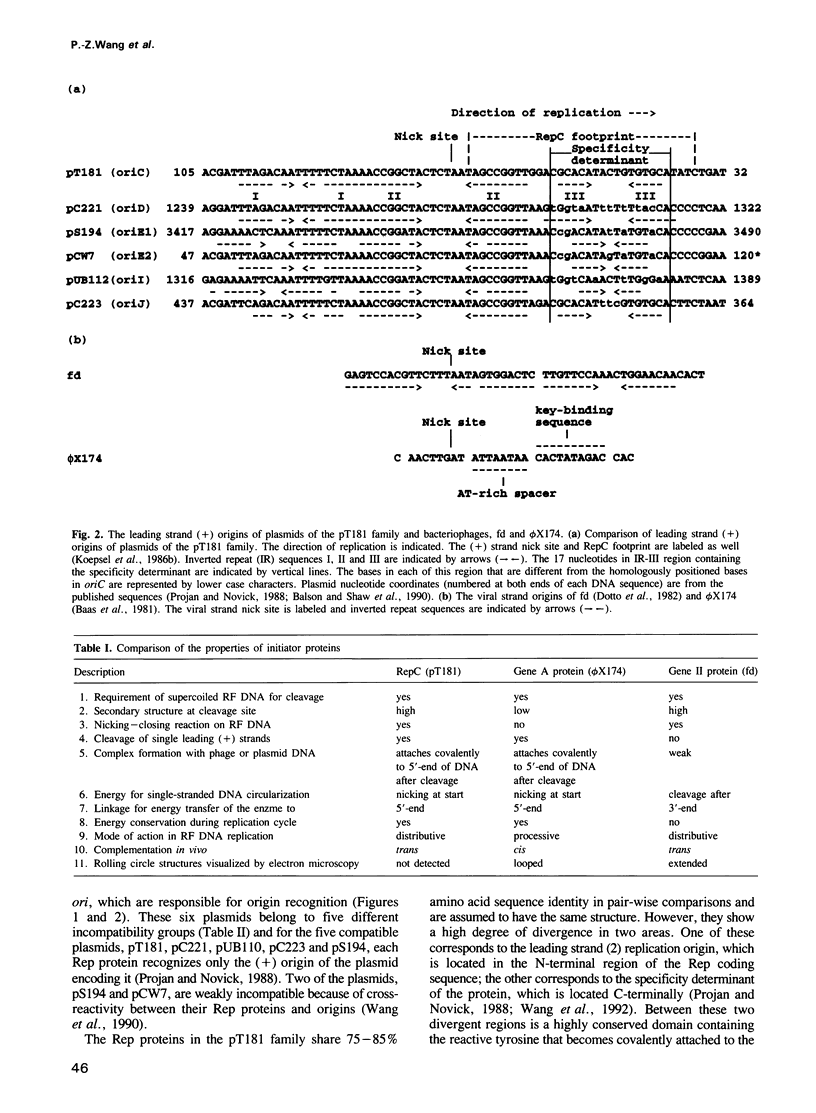
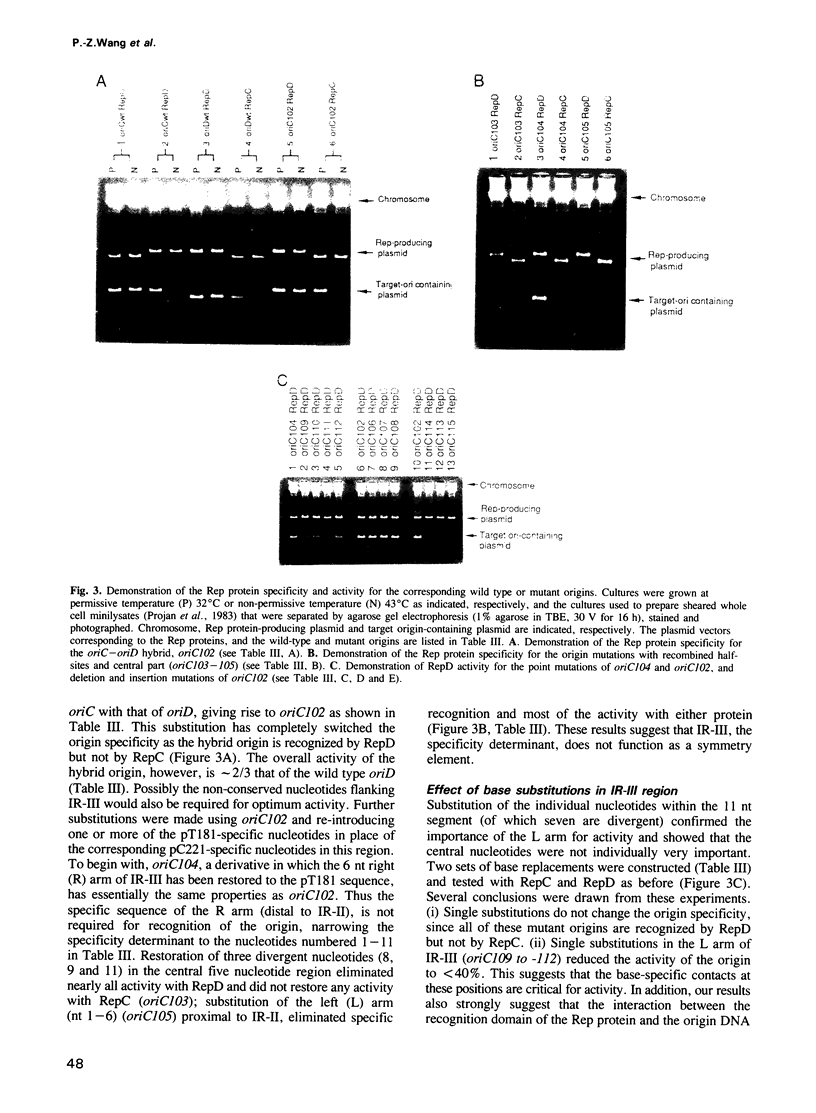
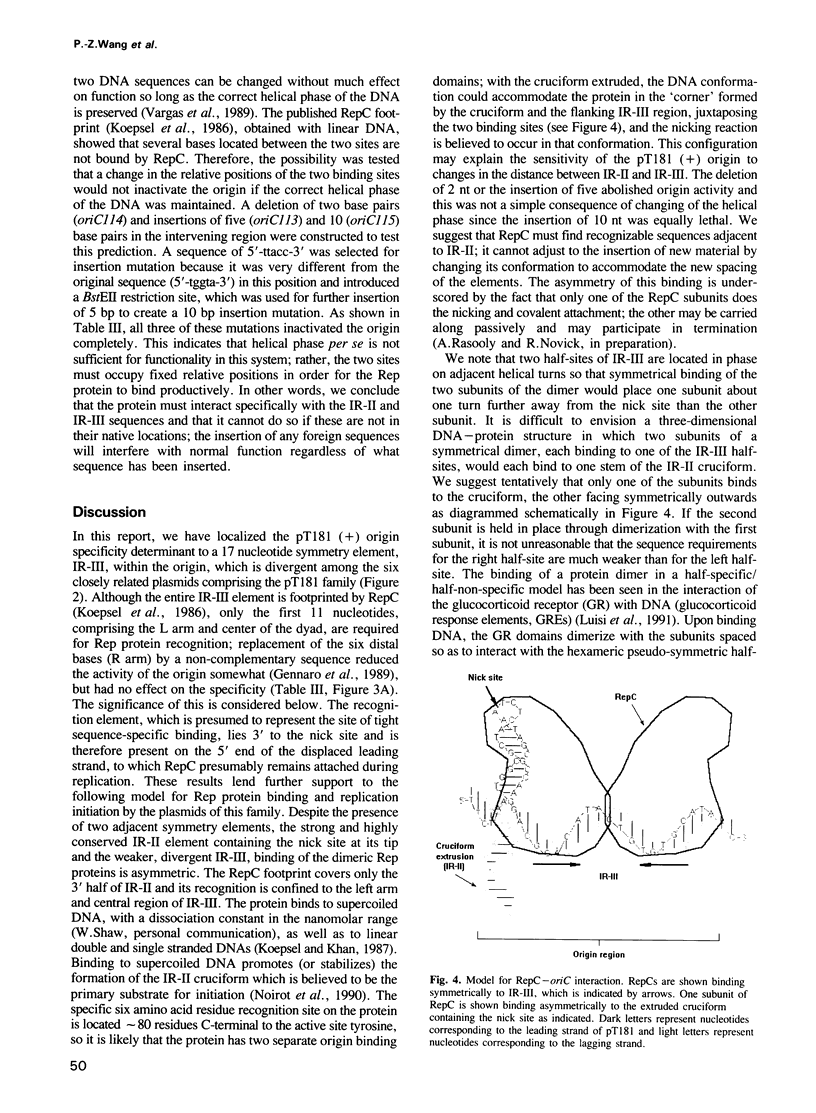
The leading strand replication origin of pT181 plasmids consists of two adjacent inverted repeat elements (IR-II and IR-III), which are involved in origin recognition by the initiator (Rep) protein. The conserved core element, IR-II, which contains the initiation nick site, is induced by Rep to form a cruciform structure, probably the primary substrate for the initiation of rolling circle replication. The divergent repeat, IR-III, constitutes the determinant of origin recognition specificity. We show here that the distal arm of IR-III is not required for sequence-specific recognition, whereas the proximal arm and central region are required. Since the initiator is dimeric, we presume that it binds symmetrically to IR-III. A unique type of DNA-protein interaction is proposed, in which the lack of sequence requirement for the distal arm is a consequence of binding to the adjacent IR-II, which thereby polarizes the stringency of binding to the two arms of IR-III. In addition, genetic evidence indicates that both the spacing and the phasing of IR-II to IR-III are crucial for function and that the central segment of IR-III may serve to position the two flanking half-sites for optimal interaction of Rep with IR-III.
Full text
PDF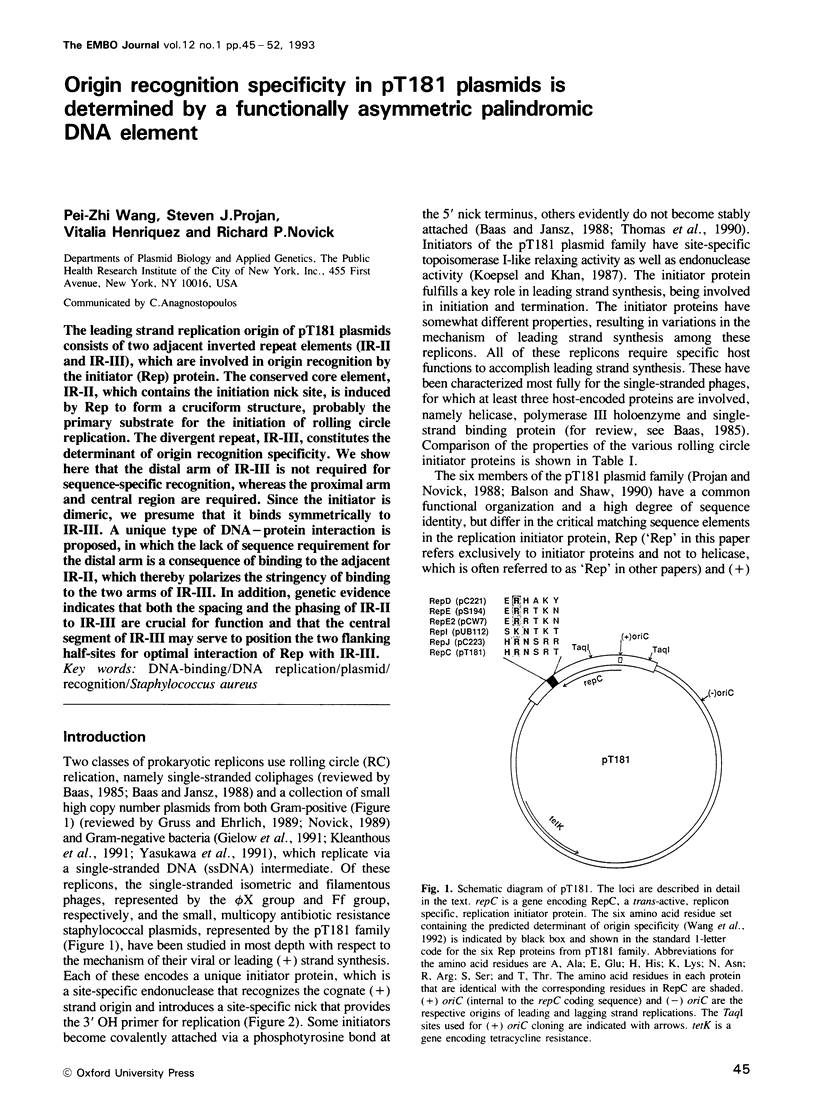




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baas P. D. DNA replication of single-stranded Escherichia coli DNA phages. Biochim Biophys Acta. 1985 Jun 24;825(2):111–139. doi: 10.1016/0167-4781(85)90096-x. [DOI] [PubMed] [Google Scholar]
- Baas P. D., Jansz H. S. Single-stranded DNA phage origins. Curr Top Microbiol Immunol. 1988;136:31–70. doi: 10.1007/978-3-642-73115-0_3. [DOI] [PubMed] [Google Scholar]
- Baas P. D., Teertstra W. R., van Mansfeld A. D., Jansz H. S., van der Marel G. A., Veeneman G. H., van Boom J. H. Construction of viable and lethal mutations in the origin of bacteriophage 'phi' X174 using synthetic oligodeoxyribonucleotides. J Mol Biol. 1981 Nov 15;152(4):615–639. doi: 10.1016/0022-2836(81)90120-0. [DOI] [PubMed] [Google Scholar]
- Balson D. F., Shaw W. V. Nucleotide sequence of the rep gene of staphylococcal plasmid pCW7. Plasmid. 1990 Jul;24(1):74–80. doi: 10.1016/0147-619x(90)90027-a. [DOI] [PubMed] [Google Scholar]
- Beato M. Gene regulation by steroid hormones. Cell. 1989 Feb 10;56(3):335–344. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988 Sep 23;54(7):915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- Carleton S., Projan S. J., Highlander S. K., Moghazeh S. M., Novick R. P. Control of pT181 replication II. Mutational analysis. EMBO J. 1984 Oct;3(10):2407–2414. doi: 10.1002/j.1460-2075.1984.tb02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Cohen S. N. High frequency transformation of Bacillus subtilis protoplasts by plasmid DNA. Mol Gen Genet. 1979 Jan 5;168(1):111–115. doi: 10.1007/BF00267940. [DOI] [PubMed] [Google Scholar]
- Dotto G. P., Horiuchi K., Zinder N. D. Initiation and termination of phage f1 plus-strand synthesis. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7122–7126. doi: 10.1073/pnas.79.23.7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennaro M. L., Iordanescu S., Novick R. P., Murray R. W., Steck T. R., Khan S. A. Functional organization of the plasmid pT181 replication origin. J Mol Biol. 1989 Jan 20;205(2):355–362. doi: 10.1016/0022-2836(89)90346-x. [DOI] [PubMed] [Google Scholar]
- Gielow A., Diederich L., Messer W. Characterization of a phage-plasmid hybrid (phasyl) with two independent origins of replication isolated from Escherichia coli. J Bacteriol. 1991 Jan;173(1):73–79. doi: 10.1128/jb.173.1.73-79.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989 Jun;53(2):231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanes S. D., Brent R. A genetic model for interaction of the homeodomain recognition helix with DNA. Science. 1991 Jan 25;251(4992):426–430. doi: 10.1126/science.1671176. [DOI] [PubMed] [Google Scholar]
- Iordanescu S. Specificity of the interactions between the Rep proteins and the origins of replication of Staphylococcus aureus plasmids pT181 and pC221. Mol Gen Genet. 1989 Jun;217(2-3):481–487. doi: 10.1007/BF02464921. [DOI] [PubMed] [Google Scholar]
- Iordănescu S. Temperature-sensitive mutant of a tetracycline resistance staphylococcal plasmid. Arch Roum Pathol Exp Microbiol. 1976 Jul;35(3):257–264. [PubMed] [Google Scholar]
- Iordănescu S. Three distinct plasmids originating in the same Staphylococcus aureus strain. Arch Roum Pathol Exp Microbiol. 1976 Jan-Jun;35(1-2):111–118. [PubMed] [Google Scholar]
- Khan S. A., Novick R. P. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid. 1983 Nov;10(3):251–259. doi: 10.1016/0147-619x(83)90039-2. [DOI] [PubMed] [Google Scholar]
- Kleanthous H., Clayton C. L., Tabaqchali S. Characterization of a plasmid from Helicobacter pylori encoding a replication protein common to plasmids in gram-positive bacteria. Mol Microbiol. 1991 Oct;5(10):2377–2389. doi: 10.1111/j.1365-2958.1991.tb02084.x. [DOI] [PubMed] [Google Scholar]
- Koepsel R. R., Khan S. A. Cleavage of single-stranded DNA by plasmid pT181-encoded RepC protein. Nucleic Acids Res. 1987 May 26;15(10):4085–4097. doi: 10.1093/nar/15.10.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepsel R. R., Khan S. A. Static and initiator protein-enhanced bending of DNA at a replication origin. Science. 1986 Sep 19;233(4770):1316–1318. doi: 10.1126/science.3749879. [DOI] [PubMed] [Google Scholar]
- Koepsel R. R., Murray R. W., Khan S. A. Sequence-specific interaction between the replication initiator protein of plasmid pT181 and its origin of replication. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5484–5488. doi: 10.1073/pnas.83.15.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudelka G. B., Carlson P. DNA twisting and the effects of non-contacted bases on affinity of 434 operator for 434 repressor. Nature. 1992 Jan 2;355(6355):89–91. doi: 10.1038/355089a0. [DOI] [PubMed] [Google Scholar]
- Kramer W., Drutsa V., Jansen H. W., Kramer B., Pflugfelder M., Fritz H. J. The gapped duplex DNA approach to oligonucleotide-directed mutation construction. Nucleic Acids Res. 1984 Dec 21;12(24):9441–9456. doi: 10.1093/nar/12.24.9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiswirth B. N., Löfdahl S., Betley M. J., O'Reilly M., Schlievert P. M., Bergdoll M. S., Novick R. P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983 Oct 20;305(5936):709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Luisi B. F., Xu W. X., Otwinowski Z., Freedman L. P., Yamamoto K. R., Sigler P. B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991 Aug 8;352(6335):497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- Moitoso de Vargas L., Kim S., Landy A. DNA looping generated by DNA bending protein IHF and the two domains of lambda integrase. Science. 1989 Jun 23;244(4911):1457–1461. doi: 10.1126/science.2544029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E. Inhibition of Tn554 transposition: deletion analysis. Plasmid. 1983 Nov;10(3):260–269. doi: 10.1016/0147-619x(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Noirot P., Bargonetti J., Novick R. P. Initiation of rolling-circle replication in pT181 plasmid: initiator protein enhances cruciform extrusion at the origin. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8560–8564. doi: 10.1073/pnas.87.21.8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Adler G. K., Projan S. J., Carleton S., Highlander S. K., Gruss A., Khan S. A., Iordanescu S. Control of pT181 replication I. The pT181 copy control function acts by inhibiting the synthesis of a replication protein. EMBO J. 1984 Oct;3(10):2399–2405. doi: 10.1002/j.1460-2075.1984.tb02146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Bouanchaud D. The problems of drug-resistant pathogenic bacteria. Extrachromosomal nature of drug resistance in Staphylococcus aureus. Ann N Y Acad Sci. 1971 Jun 11;182:279–294. doi: 10.1111/j.1749-6632.1971.tb30664.x. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Brodsky R. Studies on plasmid replication. I. Plasmid incompatibility and establishment in Staphylococcus aureus. J Mol Biol. 1972 Jul 21;68(2):285–302. doi: 10.1016/0022-2836(72)90214-8. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Staphylococcal plasmids and their replication. Annu Rev Microbiol. 1989;43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967 Sep;33(1):155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- Projan S. J., Carleton S., Novick R. P. Determination of plasmid copy number by fluorescence densitometry. Plasmid. 1983 Mar;9(2):182–190. doi: 10.1016/0147-619x(83)90019-7. [DOI] [PubMed] [Google Scholar]
- Projan S. J., Kornblum J., Moghazeh S. L., Edelman I., Gennaro M. L., Novick R. P. Comparative sequence and functional analysis of pT181 and pC221, cognate plasmid replicons from Staphylococcus aureus. Mol Gen Genet. 1985;199(3):452–464. doi: 10.1007/BF00330758. [DOI] [PubMed] [Google Scholar]
- Projan S. J., Novick R. Comparative analysis of five related Staphylococcal plasmids. Plasmid. 1988 May;19(3):203–221. doi: 10.1016/0147-619x(88)90039-x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. D., Balson D. F., Shaw W. V. In vitro studies of the initiation of staphylococcal plasmid replication. Specificity of RepD for its origin (oriD) and characterization of the Rep-ori tyrosyl ester intermediate. J Biol Chem. 1990 Apr 5;265(10):5519–5530. [PubMed] [Google Scholar]
- Wang P. Z., Projan S. J., Henriquez V., Novick R. P. Specificity of origin recognition by replication initiator protein in plasmids of the pT181 family is determined by a six amino acid residue element. J Mol Biol. 1992 Jan 5;223(1):145–158. doi: 10.1016/0022-2836(92)90722-v. [DOI] [PubMed] [Google Scholar]
- Weisblum B., Graham M. Y., Gryczan T., Dubnau D. Plasmid copy number control: isolation and characterization of high-copy-number mutants of plasmid pE194. J Bacteriol. 1979 Jan;137(1):635–643. doi: 10.1128/jb.137.1.635-643.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C. R., Baldwin N. J. Characterization and construction of molecular cloning vehicles within Staphylococcus aureus. J Bacteriol. 1978 Oct;136(1):402–413. doi: 10.1128/jb.136.1.402-413.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa H., Hase T., Sakai A., Masamune Y. Rolling-circle replication of the plasmid pKYM isolated from a gram-negative bacterium. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10282–10286. doi: 10.1073/pnas.88.22.10282. [DOI] [PMC free article] [PubMed] [Google Scholar]




