Abstract
Investigation of immune cell differentiation and function is limited by shortcomings of suitable and scalable experimental systems. Here we show that an estrogen–regulated form of HOXB8 that is retrovirally delivered into mouse bone marrow cells can be used along with FLT3 ligand to conditionally immortalize early hematopoietic progenitor cells (Hoxb8–FL). Hoxb8–FL cells have lost self–renewal capacity and megakaryocyte/ erythroid lineage potential, but sustain myeloid and lymphoid potential. Hoxb8–FL cells differentiate in vitro and in vivo into different myeloid and lymphoid cell types, including macrophages, granulocytes, dendritic cells and B– and T–lymphocytes, which are phenotypically and functionally indistinguishable from their primary counterparts. Quantitative in vitro cell lineage potential assays implicate that myeloid and B–cell potential of Hoxb8–FL cells is comparable to primary lymphoid–primed multipotent progenitors, while T–cell potential is comparatively reduced. Given the simplicity and unlimited proliferative capacity of Hoxb8–FL cells, this system provides unique opportunities to investigate cell differentiation and immune cell functions.
INTRODUCTION
The evolutionary conserved, clustered family of Hox genes encodes 39 DNA–binding transcription factors in mammals which control many aspects of embryonic development and hematopoiesis1. In hematopoiesis, Hox genes are preferentially expressed in immature progenitor cells and hematopoietic stems cells (HSC), and are down–regulated during cell differentiation and maturation2. A substantial body of evidence suggests that one important Hox gene function is the regulation of cell differentiation, specifically an increase in cell self–renewal and an arrest of cell differentiation1. This property has been used experimentally to establish stably growing, homogenous hematopoietic progenitor cell lines through retrovirus–mediated expression of certain Hox genes, such as Hoxa9 and Hoxb83. Fusing the coding sequence of Hoxa9 or Hoxb8 to the hormone binding domain of the estrogen receptor (Erhbd), activation of these Hox genes by estrogen and the addition of the growth factors SCF or GM–CSF leads to conditional immortalization of committed myeloid progenitor cells of the granulocyte and the monocyte lineage, respectively3. While providing a valuable experimental tool for the investigation of specific cell lineages, the data revealed that the specific activity of the growth factor, i.e. SCF vs. GM–CSF, in context with Hox gene expression can be used to establish progenitor cell lines committed to different lineages.
Given the critical role of FLT3 in DC generation4,5, we evaluated the possibility that FLT3 ligand (FLT3L) in combination with activated HOXB8 could be used to immortalize DC progenitor cells. Starting from largely unfractionated BM cell preparations transduced with a retroviral ERHBD–HOXB8 expression vector, we were indeed able to generate a stably growing, homogenous cell population in the presence of estrogen (to activate HOXB8) and FLT3L, which upon estrogen withdrawal differentiated into DC. However, more detailed work in vitro and in vivo revealed that these Hoxb8–FL cells do not represent committed DC precursor cells, but retain myeloid and lymphoid differentiation potential. Here we describe the establishment of this cell culture system and the phenotype and functional characteristics of Hoxb8–FL cells, providing an intriguing tool for the investigation of diverse aspects of myeloid and lymphoid cell differentiation and immune cell function.
RESULTS
Generation of Hoxb8–FL cells
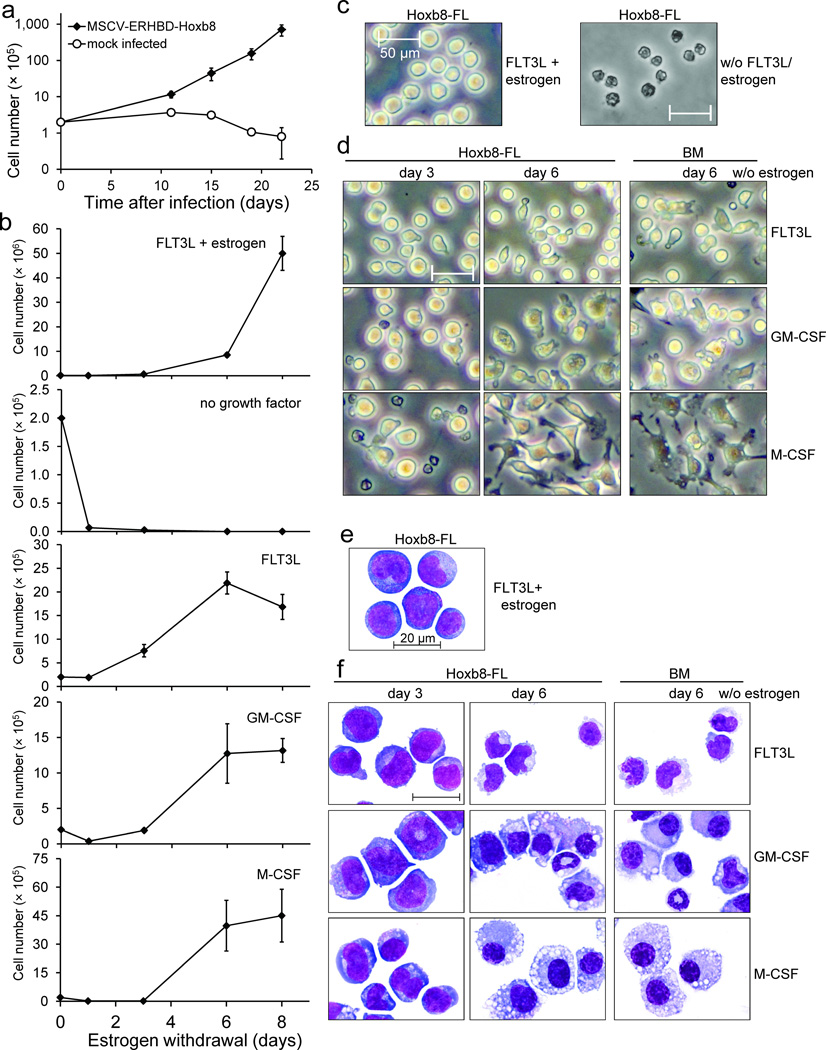
To test whether FLT3L could be used to conditionally immortalize a DC precursor, we infected BM cells, which were briefly expanded in vitro in medium containing IL–3, IL–6 and SCF, with a MSCV–based retrovirus expressing an estrogen–regulated ERHBD–HOXB8 construct (Supplementary Fig. 1), followed by cell culture in the presence of estrogen and FLT3L. In the absence of ERHBD–HOXB8 expressing virus, cells failed to expand and differentiated into typical FLT3L–driven DC as expected (Fig. 1a and see below)5. However, in the presence of activated HOXB8 and FLT3L, blast–like, stably growing cells expanded with exponential growth characteristics (Fig. 1). Growth and survival of these cells strictly depended on FLT3L (Fig. 1b, c). Hoxb8–FL cells could be grown for many weeks in culture without any apparent changes in growth characteristics and phenotype, and also could be subcloned (see below). FLT3L can thus be used to generate HOXB8–driven, growth factor dependent cell lines.
Figure 1. Growth and morphology of Hoxb8–FL cells.
(a) BM cells were infected with an MSCV–based, ERHBD–HOXB8 expressing retrovirus or mock infected and cultured in the presence of estrogen and FLT3L. Cell numbers were determined at time points indicated. Error bars represent standard deviation of cells grown in five individual wells. For viral construct and procedure see also Supplementary Figure 1.
(b) Medium of 2×105 exponentially growing Hoxb8–FL cells was exchanged by medium with indicated factors and cell numbers of live cells were determined at depicted time points. Mean cell numbers obtained after eight days of culture were: FLT3L, 1.6×106; GM–CSF, 1.3×106; M–CSF, 4.5×106; Error bars represent standard deviation of three Hoxb8–FL cell populations.
(c–f) β–estrogen– and FLT3L–containing medium of exponentially growing Hoxb8–FL cells was replaced by medium without growth factor (c), or with FLT3L, GM–CSF or M–CSF, as indicated, and cells were analyzed one day (c) and three and six days (d,f) later by phase contrast microscopy in cell culture (c, d) or after cytospin and May–Grünwald/ Giemsa staining by bright field microscopy (e, f). Unfractionated BM cells were cultured in parallel for six days under the same conditions as described for Hoxb8–FL cells and are shown for comparison. Scale bars: (c, d) = 50 µm, (e, f) = 20 µm.
Myeloid cell differentiation potential in vitro
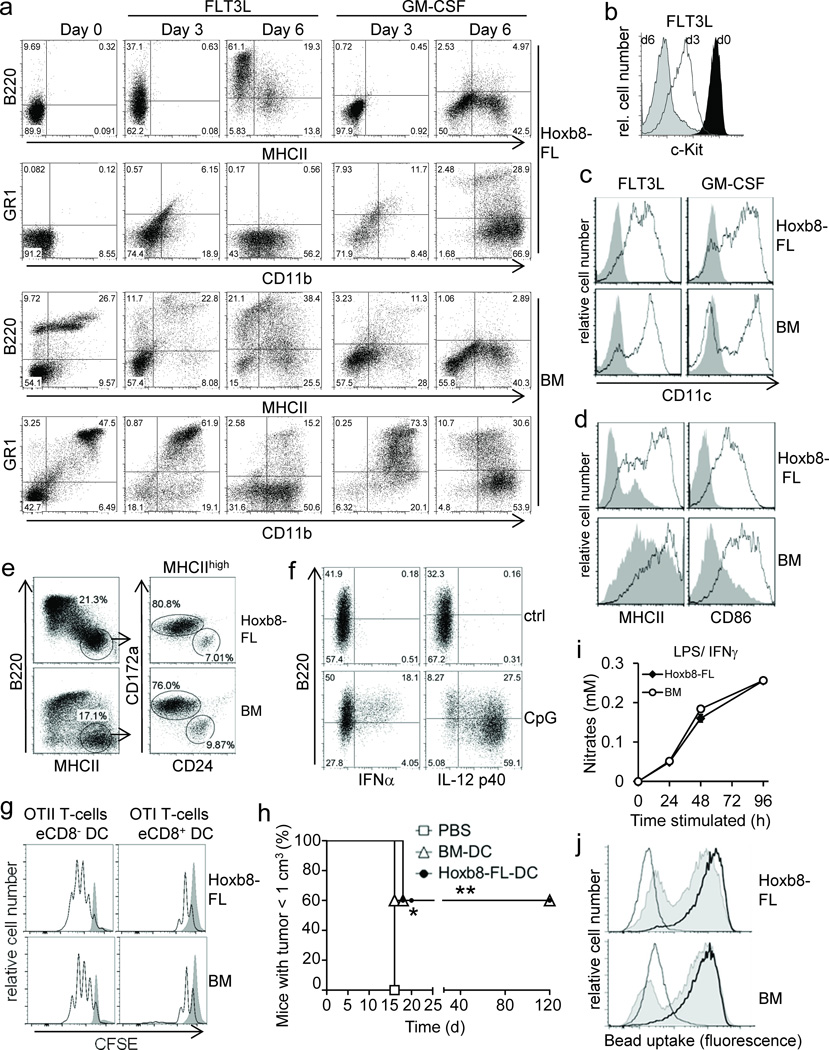
To characterize the cell fate of Hoxb8–FL cells during cell differentiation, we withdrew estrogen (to inactivate HOXB8) and analyzed growth and phenotype of cells obtained in the presence of FLT3L by microscopy and flow cytometry. Primary BM cells were used for comparison. Upon estrogen withdrawal, Hoxb8–FL cells continued to expand until around day six and stayed largely alive until day 8 (Fig. 1b). Phenotypic changes started to appear about three days after estrogen withdrawal with a decrease in the nucleus to cytoplasm ratio and a slightly increased expression of CD11b and B220 on some cells, and the down–regulation of c–Kit, which was found to be highly expressed on non–differentiated Hoxb8–FL cells (Fig. 1c–1f, 2a, 2b and see below). Six days after estrogen withdrawal, the cells displayed the typical phenotype of FLT3L–derived DC, i.e. a biphenotypic population of so–called conventional DC ((cDC) CD11b+, CD11c+, MHC class II (MHCII)+, B220− PDCA1−) and plasmacytoid DC ((pDC) CD11b− CD11c+, B220+, PDCA1+)(Fig. 2a, 2c and Supplementary Fig. 2).
Figure 2. Phenotype and function of Hoxb8–FL– and BM–derived myeloid cells.
(a–c) Hoxb8–FL cells or BM cells were cultured in the presence of FLT3L or GM–CSF and analyzed after three and six days (a,b) and six days (c) by flow cytometry. Day 0 corresponds to cells prior to differentiation. (d) Cells were differentiated with FLT3L for six days, followed by treatment with (open) or without (filled) CpG–DNA (1668) for 18 hours and flow cytometry analysis. (e) Cells were differentiated with FLT3L for ten days, followed by flow cytometry analysis. (f) Hoxb8–FL cells were differentiated with FLT3L for six days, treated with or without (ctrl) CpG–DNA (2216; CpG), followed by intracellular staining for IFNα and IL–12p40 and flow cytometry. (g) Cells were differentiated with FLT3L for nine days. Sorted eCD8+ DCs and eCD8– DCs were pulsed with OVA and co–incubated with CFSE–labeled naïve CD4 and CD8 T–cells from TCR–transgenic OTII and OTI mice. T–cell proliferation in the absence (filled) or presence of DC (open) was analyzed by flow cytometry after 72 hours. (h) Cells were differentiated with GM–CSF for nine days, pulsed with OVA for 4 hours and stimulated with CpG–DNA for 18 hours. Mice were immunized with cells or PBS subcutaneously and challenged one week later intra–dermally by OVA–expressing B16 cells. Mice were sacrificed when tumor size reached 1 cm3. n=5 mice per group. * = P < 0.05; ** = P < 0.003 (log–rank test). (d) Cells were differentiated with M–CSF, treated with LPS and IFNγ, and Nitrate levels in supernatants were determined. Error bars represent standard deviation of three biological replicates. (j) Cells were differentiated with M–CSF, incubated with FITC–labeled IgG–coated beads at 37°C for 2 h (solid) or 6 h (thick line), or at 4°C for 6 h (thin line), followed by flow cytometry analysis.
The Hoxb8–FL–derived cell population did not contain GR1high CD11c– granulocytes, which are contained in the input population of unfractionated BM and are only gradually lost during the in vitro cell culture (Fig. 2a). Treatment of Hoxb8–FL– and BM–derived DC with known maturation factors, such as the TLR9 agonist CpG–DNA, led to strong up–regulation of typical DC maturation markers, such as MHCII, CD86 (B7.2) and CD40 (Fig. 2d and data not shown) 6. Two major subtypes of splenic cDC have been characterized, i.e. CD8– and CD8+ DC, which correspond to in vitro–generated, FLT3L–driven CD11c+ CD172a+ CD24low cells (‘equivalent’ eCD8− DC) and CD11c+ CD172low CD24high cells (eCD8+ DC), respectively7. More detailed flow cytometry analysis demonstrated that Hoxb8–FL cells indeed generate these DC subtypes comparable to primary BM cells (Fig. 2e).
To see whether Hoxb8–FL cells might have myeloid potential beyond the DC lineage, we replaced FLT3L upon estrogen removal with GM–CSF or M–CSF, which support in vitro the generation of DC and granulocytes, and macrophages, respectively. After a short period of reduced cell growth and limited cell death upon estrogen withdrawal, both cytokines supported survival, expansion and eventually the generation of viable populations of differentiated cells (Fig. 1b,d,f). Cell differentiation became morphologically apparent after about three days (Fig. 1d,f). At day six, GM–CSF driven Hoxb8–FL cells exhibited the classic phenotype of GM–CSF–driven BM cells, characterized by a mixed population of DC (CD11b+, CD11c+, MHCII+, B220−) and granulocytes (GR1high CD11− MHCII−)(Fig. 2a,c). In contrast, M–CSF–cultured Hoxb8–FL cells exhibited the characteristic adherent morphology of macrophages with the typical surface expression of CD11b and lack of MHCII and GR1 (Fig. 1d,f and Supplementary Fig. 3). Similar to the FLT3L cultures, BM–derived cells still contained a small population of granulocytes (CD11b+, GR1+, MHCII−) (Supplementary Fig. 3). Taken together, Hoxb8–FL cells harbor differentiation potential for the major myeloid cell lineages.
Immune functions of Hoxb8–FL derived myeloid cells
To further establish the functionality of Hoxb8–FL–derived immune cells (Fig. 2d), we analyzed key immune functions of different myeloid cell types, including antigen–dependent T–cell activation and cytokine production by DC, and phagocytosis and nitric oxide (NO) production by M–CSF–driven macrophages. Upon CpG–DNA–mediated TLR9 activation, B220+ pDC produced robust levels of IFNα, while IL–12 p40 was preferentially expressed by cDC, both of which are key characteristics of respective cell types (Fig. 2f)8. Similarly, GM–CSF–driven, Hoxb8–FL–derived DC produced high levels of IL–6 and IL–12 upon CpG–DNA and LPS stimulation (Supplementary Fig. 4). To see if Hoxb8–FL–derived DC were able to process and present antigen, and promote T–cell activation, we exposed Hoxb8–FL– and BM–derived eCD8– and eCD8+ cDC to soluble Ovalbumin (OVA) as model antigen, followed by co–incubation with OVA–specific, MHCII– or MHCI–restricted T–cell receptor (TCR)–transgenic T–cells, i.e. OTII and OTI T–cells9,10, respectively. Hoxb8–FL–derived eCD8– cDC triggered proliferation of OTII T–cells, while eCD8+ cDC induced proliferation of OTI T–cells, comparable to BM–derived DC (Fig. 2g). To see if Hoxb8–FL–derived DC could also induce immunity in vivo, and also to test a possible application of Hoxb8–FL cells, we compared immunogenicity of Hoxb8-FL- and BM-derived DC in an antigen (OVA)–specific B16 melanoma model. Both, Hoxb8–FL– and BM–derived DC induced robust OTII– and OTI–T–cell activation in vitro, as expected (Supplementary Fig. 5). These cells were then used for vaccination of mice, followed by challenge with OVA–expressing B16 melanoma cells one week later. While all mock–treated control mice developed sizable tumors at around two weeks after challenge, mice vaccinated with DC derived from Hoxb8–FL cells or BM cells showed significant protection (Fig. 2h), demonstrating immunogenicity of respective DC.
NO release and phagocytosis were tested to evaluate macrophage functions. M–CSF–driven, Hoxb8–FL–derived macrophages produced NO levels upon treatment with LPS and IFNγ that were virtually indistinguishable from their primary counterparts (Fig. 2i). Likewise, phagocytosis of immunoglobulin G (IgG)–coated beads was very comparable between Hoxb8–FL– and BM–derived cells (Fig. 2j). Thus, Hoxb8–FL–derived DC and macrophages correspond phenotypically and functionally to BM–derived primary cells.
Myeloid and lymphoid differentiation potential in vivo
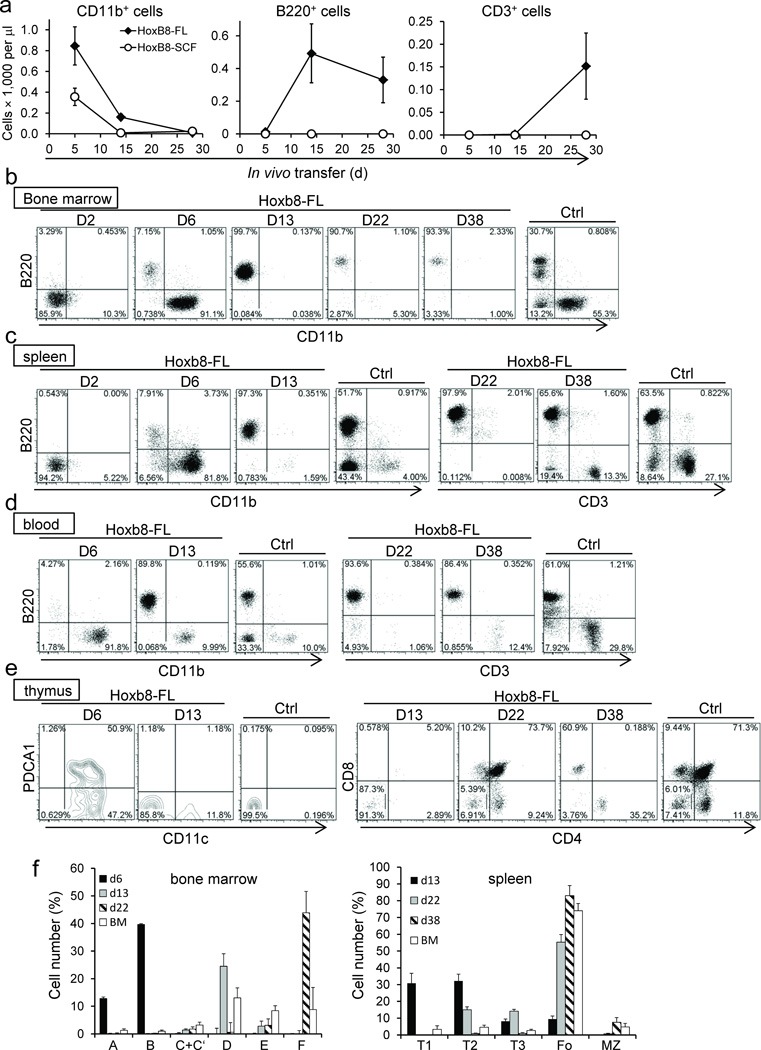
Given the known activity of FLT3L on very early hematopoietic progenitor cells, we further defined the lineage potential of Hoxb8–FL cells in vivo4,11. We transferred Hoxb8–FL cells established from CD45.1+ B6/SJL mice into lethally irradiated CD45.2+ C57Bl/6 mice along with unfractionated CD45.2+ BM cells and analyzed the appearance of mature cell types in the peripheral blood. In these experiments we included cell lines established based on Hoxb8 and SCF (Hoxb8–SCF), whose lineage potential has so far only been investigated in vitro3. Consistent with a largely granulocyte–restricted lineage potential in vitro, Hoxb8–SCF cells generated only CD11b+ myeloid cells, most of which were GR1high granulocytes (Fig. 3a and Supplementary Fig. 6a). One week later, Hoxb8–SCF–derived cells were largely lost due to the short half–life of granulocytes. In contrast, Hoxb8–FL cells generated a mixed population of CD11b+ cells, containing GR1high granulocytes and at least two additional populations with intermediate (GR1int) and GR1–negative (GR1−) phenotype (Supplementary Fig. 6a). The latter population was still detectable 14 days after transfer, but was largely absent at day 28 (Fig. 3a and Supplementary Fig. 6a,b). Intriguingly, Hoxb8–FL cells also generated B220+ CD11b– CD11c– B–lymphocytes and CD3+ T−lymphocytes, which were detectable 14 and 28 days after transfer, respectively (Fig. 3a and Supplementary Fig. 6b). To analyze the tissue–specific development of different immune cell types in more detail, we transferred Hoxb8–FL cells into lethally irradiated mice and analyzed their progeny in bone marrow, spleen, blood and thymus at different time points by flow cytometry. Two days after transfer, Hoxb8–FL–derived cells were detectable in high numbers in the BM and spleen, but not peripheral blood and thymus, and were largely negative for cell lineage markers (Supplementary Fig. 7, Fig. 3b – e and data not shown). At day six, CD11b+ myeloid cells dominated in BM, spleen and peripheral blood, and a smaller population of B220+ (CD11c−) B–cell progenitors appeared in the BM, which were absent in the spleen and peripheral blood (Fig. 3b – d and data not shown). These B–cell progenitors segregated almost exclusively into the Hardy Fractions A and B (Fr. A/B) of early Pre–Pro and Pro–B–cells (Fig. 3f, for B-cell gating strategy see Supplementary Fig. 8a,b)12. A small number of Hoxb8–FL–derived cells were also found in the thymus at this time point. These cells, however, did not appear to represent T–cell progenitors, but expressed CD11c and in part PDCA1, suggesting the immigration of Hoxb8–FL–derived DC to the thymus (Fig. 3e). At day 13, BM, spleen and peripheral blood were dominated by cells belonging to the B–cell lineage, while myeloid cells were merely detectable in the BM, and at reduced numbers in spleen, peripheral blood and thymus (Fig. 3b – e). B–cells in the BM segregated mostly into Hardy fraction D, while B–cells in the spleen displayed largely the phenotype of transitional T1 and T2 cells (Fig. 3f). The majority of Hoxb8–FL–derived cells at day 13 in the thymus displayed a lineage negative, CD4− CD8− CD25− CD44+ phenotype, characteristic of early thymus settling, double negative (DN) 1 (DN1) cells (Fig. 3e and Supplementary Fig. 9). At day 22, the BM contained only a small number of Hoxb8–FL–derived cells, which were identified almost exclusively as Fr. F (follicular) B–cells (Supplementary Fig. 7 and Fig. 3f). Likewise, the spleen contained primarily follicular B–cells, and some T2 and T3 transitional B–cells (Fig. 3c, f). No T–cells were found in the periphery at day 22, however, the thymus contained a composition of T–cells resembling the steady state of physiological T–cell development, i.e. CD4 CD8 double positive (DP) cells, mature CD4 or CD8 single positive (SP) cells, and a lower number of DN cells, which contained all DN stages (DN1–4)(Fig. 3e and Supplementary Fig. 9). At day 38, Hoxb8–FL cells had largely left the BM, but were still present in spleen, peripheral blood and thymus (Supplementary Fig. 7). Splenic B–cells had matured into follicular and marginal zone B–cells (Fig. 3f). Residual Hoxb8–FL–derived cells in the thymus displayed the mature phenotype of SP thymocytes, and mature T–cells were now also identified in spleen and peripheral blood (Fig. 3d,e). Of note, in contrast to myeloid cells and B–cells, Hoxb8–FL–derived T–cells reached only about 10–30% of physiological T–cell numbers found in untreated mice (Fig. 3a). This was not due to the irradiation protocol, which is known to influence T–cell maturation13, as transfer of Hoxb8–FL cells into non–irradiated IL–7R–deficient mice, which can support T–cell development without conditioning irradiation, showed comparably low T–cell reconstitution (Supplementary Figure 10a,b and see Discussion) 14. Taken together, Hoxb8–FL cells have myeloid and lymphoid lineage potential which they realize in vivo in a time–dependent manner. The loss of immature cells during time strongly suggests that Hoxb8–FL cells do not have self–renewal capacity.
Figure 3. In vivo differentiation of Hoxb8–FL cells.
(a) CD45.1+ Hoxb8–FL– and –Hoxb8–SCF cells were transferred into lethally irradiated CD45.2+ recipient mice and CD11b+, B220+, and CD3+ cells in the peripheral blood were quantified based on flow cytometry and complete blood cell counts. Respective cell populations are depicted as total cell numbers over time. Error bars represent standard deviation of three Hoxb8–FL populations. (b–f) CD45.1+ Hoxb8–FL cells were transferred into lethally irradiated CD45.2+ recipient mice and cells from BM (b), spleen (c), peripheral blood (d) and thymus (e) were analyzed by flow cytometry. Representative data obtained from six mice transferred with two independent Hoxb8–FL cell populations (three mice each per population) per time point are depicted. Tissue from untreated control mice (Ctrl) was used for comparison. Relative numbers of cells of the B–cell lineage in the BM and spleen were determined by flow cytometry as described in Supplementary Figure 8a,b (f). Error bars for Hoxb8–FL–derived cells represent standard deviation of six mice that were transferred with two populations of Hoxb8–FL cells (three mice each). Error bars of untreated mice represent standard deviation (n=6).
Characterization of Hoxb8–FL derived immune cells
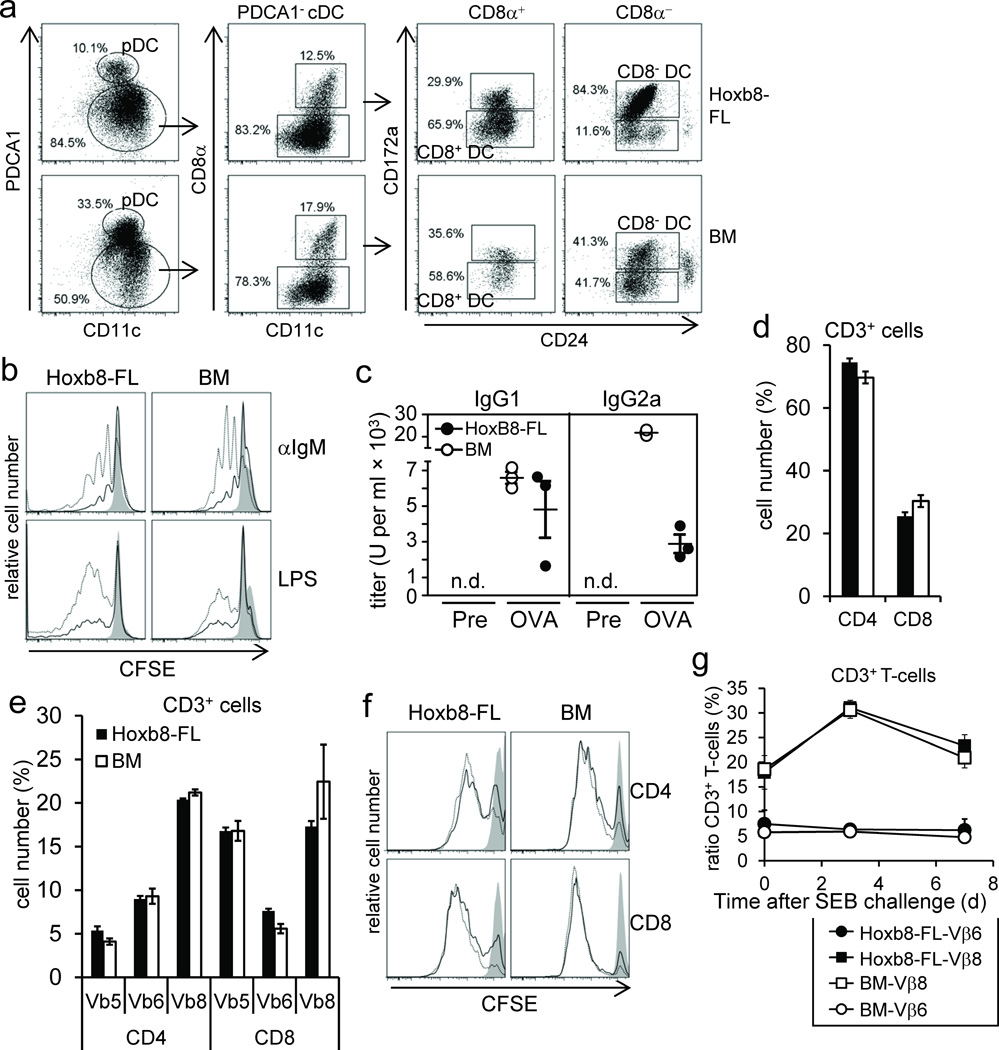
In vitro, Hoxb8–FL cells differentiated into different DC types (Fig. 2e). To see if Hoxb8–FL cells realize DC potential in vivo, we transferred Hoxb8–FL cells or unfractionated BM cells for comparison into lethally irradiated mice and purified CD11c+ and PDCA1+ cells from the spleen. Flow cytometry analysis showed that Hoxb8–FL cells indeed give rise to different defined DC types comparable to BM, i.e. pDC (CD11c+ PDCA1+), CD8+ cDC (CD11c+ PDCA1− CD8α+ CD172alow CD24high) and CD8− cDC (CD11c+ PDCA1− CD8α− CD172a+ CD24low) (Fig. 4a).
Figure 4. In vivo cell differentiation and function of Hoxb8–FL–derived lymphocytes.
(a) Hoxb8–FL cells or BM cells were transferred into lethally irradiated mice and splenic DC were analyzed by flow cytometry after seven days. (b) Hoxb8–FL cells and unfractionated BM cells were transferred into lethally irradiated mice. Seven weeks later, splenocytes were CFSE labeled and left untreated (filled) or stimulated for three days with anti–IgM antibodies (1 µg/ml (solid line), 10 µg/ml (dashed line)) or LPS (0.1 µg/ml (solid line), 0.5 µg/ml (dashed line), followed by flow cytometry analysis. (c) Hoxb8–FL cells or unfractionated BM cells were transferred into lethally irradiated µMT mice along with unfractionated BM helper cells from µMT mice, followed by intradermal injection of OVA plus CpG–DNA on day 21, 31 and 41 after transfer. Serum IgG1 and IgG2a antibody titers were determined by ELISA one day before immunization (Pre) and 10 days after the last immunization (OVA). n.d., not detectable. (d,e) Hoxb8–FL cells and BM cells were transferred into lethally irradiated mice. Seven weeks later, the percentage of CD3+CD4+ and CD3+CD8+ splenocytes (d), and the distribution of select TCR Vβ chains on CD3+CD4+ and CD3+CD8+ splenocytes (e) were analyzed by flow cytometry. Error bars represent standard deviation of three Hoxb8–FL populations. (f) Splenocytes described in (d,e) were CFSE labeled, stimulated for two days with plate–bound αCD3 antibodies (10 µg/ml: dashed line; 3 µ/ml: solid line; non–stim: filled) and αCD28 antibodies (10 µg/ml) and analyzed by flow cytometry. (g) CD45.1+ Hoxb8–FL cells and CD45.2+ BM cells were transferred into lethally irradiated mice. Five weeks later mice were challenged with SEB and the percentage of CD45.1+ CD3+T–cells and CD45.2+ CD3+ T–cells expressing TCR Vβ6– and TCR Vβ8–chains in the peripheral blood was determined by flow cytometry before and 3 and 7 days after SEB challenge. Mean and standard deviation of four mice are shown.
To characterize mature Hoxb8–FL–derived B– and T–cells, we performed in vivo and in vitro experiments. Comparable to BM–derived B–cells, antibody–mediated B–cell receptor crosslinking or TLR–stimulation induced cell proliferation (Fig. 4b). To analyze B–cell function in vivo, we transferred Hoxb8–FL cells, or unfractionated BM, into lethally irradiated B–cell–deficient (µMT) mice, which were immunized with OVA and CpG–DNA. As shown in Figure 4c, serum collected from Hoxb8–FL–reconstituted mice contained robust levels of OVA–specific IgG1 and IgG2a. While IgG1 levels were comparable to mice reconstituted with unfractionated BM, IgG2a levels were less strongly induced (Fig. 4c), possibly a consequence of less efficient T–cell reconstitution under these conditions.
Hoxb8–FL–derived T–cells were analyzed 5 weeks after transfer and showed an almost identical distribution of CD4– and CD8–single positive cells as BM–derived T–cells (Fig. 4d). Also, the repertoire of T–cells expressing certain T–cell receptor variableβ–chains (TCR Vβ) was very similar between Hoxb8–FL– and BM–derived T–cells, as was the proliferative response upon antibody–mediated T–cell receptor crosslinking and CD28 stimulation, at least at high antibody concentrations (Fig. 4e,f). To analyze T–cell activation in vivo, Hoxb8–FL cells and unfractionated BM cells were transferred into lethally irradiated mice. Five weeks after transfer, the mice were challenged with staphylococcal enterotoxin B (SEB), which activates selectively TCR Vβ8+ T–cells. Three days after SEB challenge, the pool of Hoxb8–FL– and BM–derived TCR Vβ8+ T–cells expanded about two–fold, and contracted close to baseline levels four days later (Fig. 4g). TCR Vβ6+–expressing T–cells were not affected by SEB injection, as expected. Thus, Hoxb8–FL–derived T–cells exhibit the typical biology of antigen–specific T–cell populations in vivo, i.e. expansion and contraction, reflecting initial cell proliferation and subsequent cell death.
Taken together, Hoxb8–FL cells differentiate in vivo into the major defined, splenic DC population and into lymphocytes that correspond phenotypically and functionally to primary BM–derived cells.
Lymphoid potential of Hoxb8–FL cells in vitro
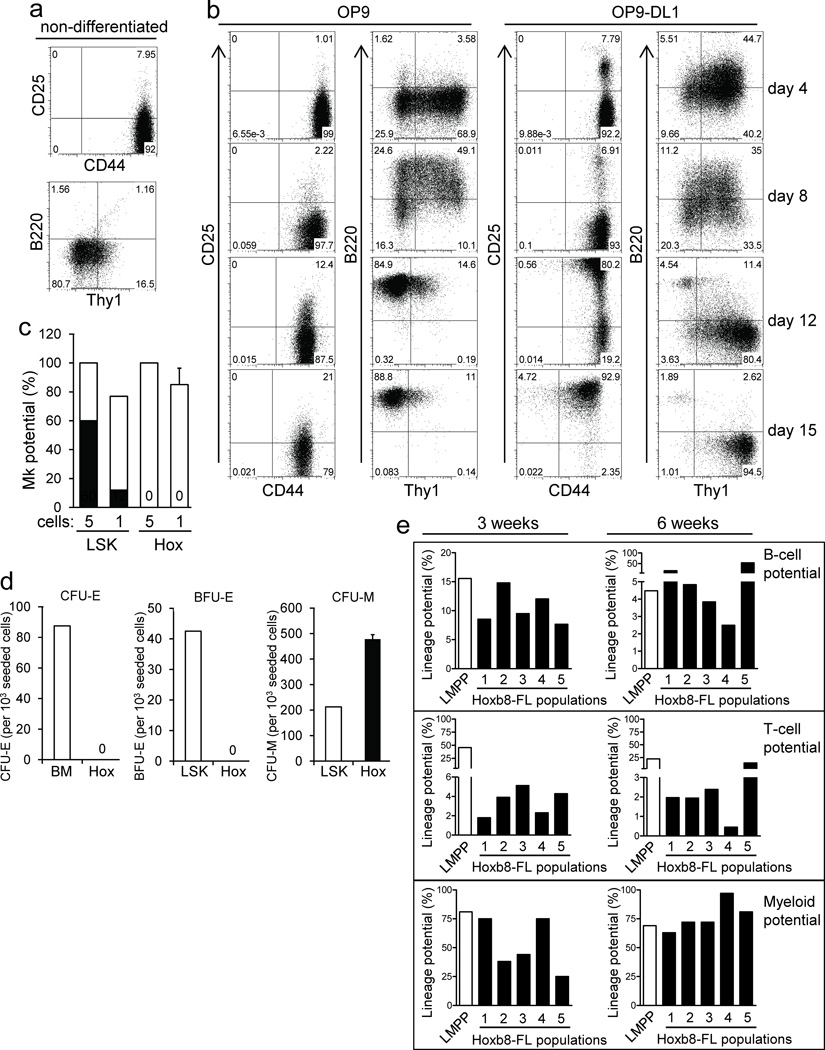
Tissue-specific lymphocyte development in vivo can be recapitulated in vitro using stromal cell lines, e.g. OP9 cells and the co–factors Interleukin (IL)–7 and FLT3L15. For T–cell development, additional Notch signaling is required, which can be provided by expression of the Notch ligand Delta–like 1 in OP9 cells (OP9–DL1)16. To see whether the lymphocyte potential of Hoxb8–FL cells could be recapitulated in vitro, we co–cultured Hoxb8–FL cells with OP9 and OP9–DL1 cells and analyzed their phenotype at various time points. Non–differentiated Hoxb8–FL cells expressed high levels of CD44, but no detectable or very low levels of CD25, B220 and Thy1 (Fig. 5a). Already four days upon co–culture of Hoxb8–FL cells on either OP9 or OP9–DL1 cells, Thy1 was up–regulated. Up–regulation was transient in case of OP9–cocultured cells and was followed by homogenous, high B220 expression, indicative of pre/pro–B–cell development and commitment to the B–cell lineage (Fig. 5b)12. In case of OP9–DL1 cocultured cells, Thy1 expression further increased and was sustained over time, accompanied by strong up–regulation of CD25, resulting in the typical phenotype of CD44+CD25+, CD4− CD8− DN2 T–cell progenitors (Fig. 5b and data not shown)17. B220+ Thy1− B–cell progenitors were almost completely absent under these conditions, reflecting the characterized function of Notch1 signaling in restricting B–cell development18, 19. Under the specific cell culture conditions used, DN2 T–cell progenitors maintained this phenotype and did not develop into more mature T–cells (Supplementary Fig. 11a and data not shown). However, when adoptively transferred into lethally irradiated mice, these cells further matured into CD3+ CD4+/ CD8+ single positive T–cells (Supplementary Fig. 11b). These cells also generated CD11b+ myeloid cells, but not B220+ B–cells, which is consistent with more recent reports demonstrating the sequential loss of B–cell– and myeloid lineage potential of DN2 and DN3 thymocytes, respectively (Supplementary Fig. 11b)20, 21,22. Together, Hoxb8–FL cells recapitulate early phases of B– and T–cell development in vitro.
Figure 5. Hoxb8–FL cells realize and sustain multi–lineage potential in vitro, but lack MkE potential.
(a) and (b) Hoxb8–FL cells were analyzed by flow cytometry, either in their non–differentiated state (a), or after estrogen withdrawal and co–culture on OP9 cells or OP9–DL1 cells for indicated periods of time (b). (c) Hoxb8–FL cells (Hox) and BM–derived LSK cells (LSK) were seeded as single cells or five cells per well into medium supporting Mk development (see Methods). Plating efficiency (open bars) and number of Mk colonies (filled bars) are shown. Error bars represent standard deviation of three Hoxb8–FL populations. Columns representing LSK cells express means of duplicate plates. (d) Hoxb8–FL cells (Hox), unfractionated BM (BM) or LSK cells (LSK) were seeded in methylcellulose containing medium supplemented with EPO (CFU–E) or EPO, SCF, IL–3 and IL–6 (BFU–E, CFU–M) and formation of CFU–E, BFU–E and CFU–M was determined after two (CFU–E) and 14 days (BFU–E, CFU–M). Error bars represent standard deviation of three Hoxb8–FL populations. Columns representing BM and LSK cells express means of duplicate wells. (e) Five independent Hoxb8–FL cell populations were generated and B–cell, T–cell and myeloid potential was determined after three and six weeks of continuous cell culture.
Quantitative analysis of Hoxb8–FL lineage potential
The above shown kinetics of appearance and decline of Hoxb8–FL–derived immune cells in vivo strongly suggest the lack of self–renewal capacity, indicating that Hoxb8–FL cells either represent a functional equivalent of multipotent progenitor cells (MPP) or lymphoid–primed MPP (LMPP), a cell type defined as committed progenitor with myeloid and lymphoid potential, but loss of megakaryocyte/ erythroid (MkE) potential 23, 24. To determine MkE lineage potential we used clonal and near–clonal assays, as well as colony forming unit (CFU)–assays supporting megakaryocyte/ myeloid cell differentiation and erythroid/ myeloid cell differentiation, respectively. LSK cells and unfractionated BM cells (for CFU assays only) were used as positive control. LSK cells formed megakaryocytes with the expected frequency, while Hoxb8–FL cells did not show any megakaryocyte potential (Fig. 5c). Moreover, under conditions permissive for erythroid development only, i.e. in the presence of erythropoietin (EPO), Hoxb8–FL cells died and did not form any CFU–erythroid (E) or burst forming units (BFU)–E, while LSK cells gave rise to the expected number of colonies (Fig. 5d). Under conditions providing additional factors that support myeloid development, i.e. EPO plus SCF, IL–3 and IL–6, Hoxb8–FL cells formed exclusively CFU–macrophage (M), while LSK cells formed CFU–M in addition to CFU–E and BFU–E, as expected (Fig. 5d). In functional terms, Hoxb8–FL cells thus resemble LMPP, which sustain lymphoid and myeloid potential, but lack MkE potential.
Stability and homogeneity of Hoxb8–FL cells
To test stability and homogeneity of Hoxb8–FL cells in more detail, we generated five independent Hoxb8–FL populations, which were cultured continuously in vitro for six weeks, and the lineage potential was determined after three and six weeks by clonal assays (for myeloid potential) and limiting dilution assays (for B– and T–cell potential). BM–derived LMPP served as control. Three week–old cultures showed B–cell– and myeloid lineage potential that was comparable to LMPP, and also comparable amongst the different populations (Fig. 5e). Both lineage potentials were preserved after six weeks of culture, although B–cell potential showed some variability amongst the Hoxb8–FL populations (Fig. 5e). Hoxb8–FL cells displayed homogenously T–cell potential, which was however about 10–fold lower than the one obtained from LMPP. Still, also T–cell potential was preserved after six weeks of continuous cell culture. To evaluate the homogeneity of the populations generated, we subcloned four populations and tested the lineage potential of four of these clones in quantitative assays after six weeks of culture. All clones displayed relatively homogenous B–cell– and myeloid lineage potential that was comparable to the original populations and close to the lineage potential of LMPP (Supplementary Fig. 12). With the exception of two clones, also T–cell potential was either preserved, or even increased (Supplementary Fig. 12). Taken together, Hoxb8–FL cells represent a homogenous population of cells that maintain multi–lineage potential stably over a period of at least six weeks.
Phenotype and gene expression analysis of Hoxb8–FL cells
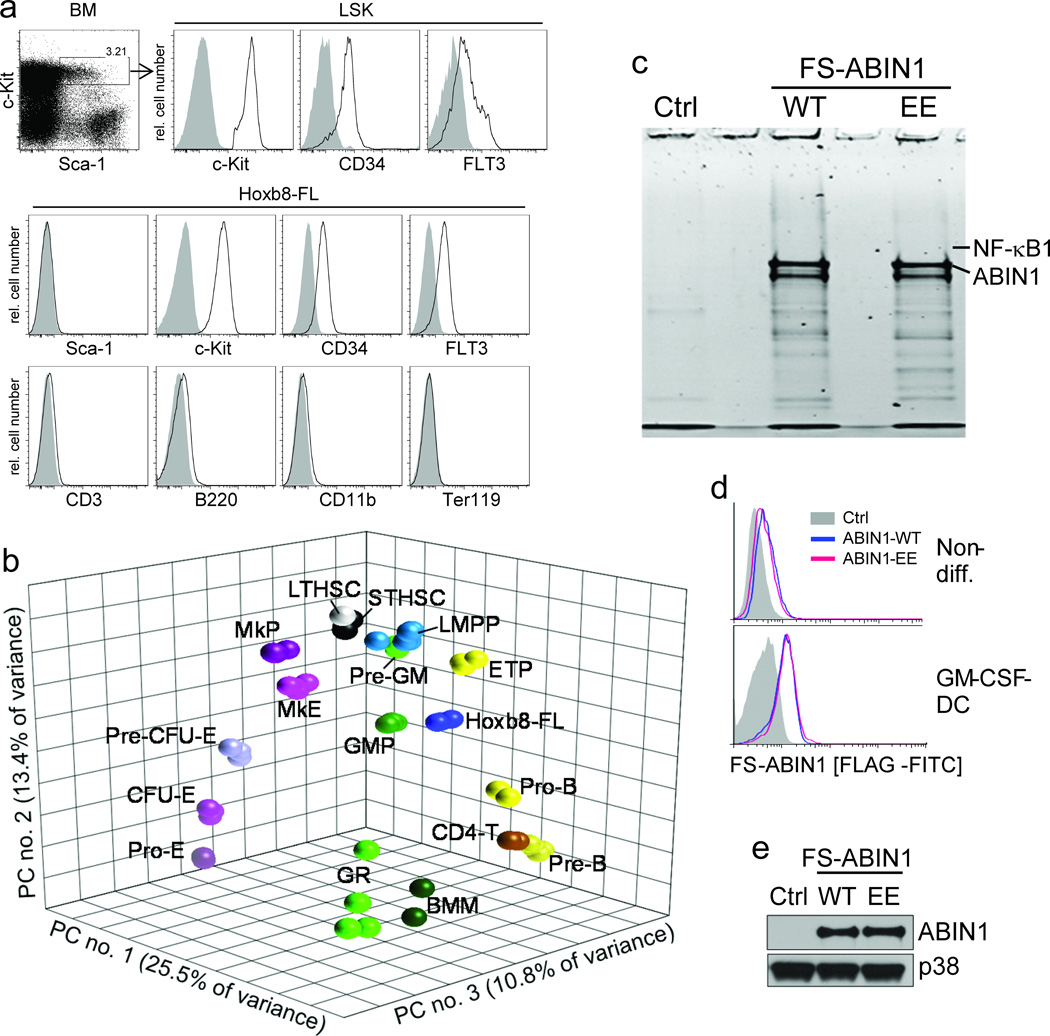
LMPP are phenotypically characterized as FLT3hi CD34+ LSK cells, and were opposed to FLT3–low LSK cells with MkE potential23. Comparable to LMPP, Hoxb8–FL cells were negative for hematopoietic lineage markers and expressed homogenously c–Kit, FLT3 and CD34; in contrast to LMPP cells, however, Hoxb8–FL cells did not express detectable levels of SCA–1 (Fig. 6a). To further refine the relation of Hoxb8–FL cells to other hematopoietic cells, we performed microarray–based genome–wide gene expression analyses, defined cell type–specific gene expression profiles by ANOVA and visualized those profiles by principal component analysis (PCA). The four independent Hoxb8–FL cell populations clustered closely together, reflecting their molecular similarity, but distant from the erythroid/ megakaryocytic lineage, and also distant from mature immune cells, i.e. granulocytes, macrophages and CD4+ T–cells, as expected (Fig. 6b). Interestingly, while Hoxb8–FL cells also cluster more distant to hematopoietic stem cells, and somewhat closer to LMPP (particularly in the first principal component), they cluster in between myeloid and lymphoid branches as extending from LMPP to GMP and myeloid cells, and from LMPP to ETP and lymphoid cells, respectively. This phenotype of Hoxb8-FL cells is consistent with their lack of erythroid/ megakaryocytic potential, their immature phenotype and their combined myeloid/ lymphoid lineage potential.
Figure 6. Phenotype, principal component analysis (PCA) and biochemical application of Hoxb8–FL cells.
(a) Lineage depleted BM cells and Hoxb8–FL cells were analyzed by flow cytometry. Open, specific antibodies; filled = isotype controls. Data are representative for at least five independent populations of Hoxb8–FL cells. (b) Three–dimensional PCA based on gene expression profiles of Hoxb8–FL cells (four independent populations), bone marrow–derived macrophages (BMM), granulocytes (GR) and purified hematopoietic progenitor cell populations. LTHSC, long–term hematopoietic stem cell; STHSC, short–term hematopoietic stem cell, LMPP, lymphoid–primed multipotent progenitor; ETP, early thymic progenitor; ProB, pro B–cell; PreB, pre B–cell; CD4T, CD4+ T–cell, Pre–GM, pre–granulocyte/ macrophage; GMP, granulocyte/macrophage progenitor; MkP, megakaryocyte progenitor; MkE, Megakaryocyte/ Erythrocyte progenitor; Pre CFU–E, pre–colony forming unit–erythrocytes; CFU–E, colony forming unit–erythrocytes; Pro–E, pro–Erythrocyte; GR, granulocyte. Each symbol represents an individual biological sample. PC, principal component. (c) Hoxb8–FL cells were generated from BM of ABIN1–deficient mice, reconstituted with epitope–tagged ABIN1 (FS–ABIN1), either wildtype (WT) or an ubiquitin–binding mutant (EE) or an empty control vector (Ctrl), differentiated into DC in the presence of GM–CSF for eight days, followed by tandem affinity purification of ABIN1. Purified proteins were separated by SDS/ PAGE, visualized by SYPRO Ruby (c) and identified by LC–MS (for peptide identifications see Supplementary Table 1). (d and e) ABIN1 expression was analyzed in non–differentiated cells (non–diff. (d)) and GM–CSF–driven DC (d, e) by intracellular (FLAG) staining and flow cytometry (d), and immuno–blotting with antibodies against ABIN1 (e).
An important aspect of Hoxb8–FL cells is their application in experimental settings that are not feasible or very difficult based on current technologies. One type of application is the proteomic analysis of rare cell types, e.g. DC, particularly from sparsely available mice, such as ABIN1–deficient mice, which die largely during embryonic development25,26. ABIN1 is an ubiquitin–binding protein with functions in different pathways, including the TLR–mediated C/EBPβ pathway and the TNFα–induced NF–κB and cell death pathways 26,27,25. Still, ABIN1’s molecular mechanism, particularly in immune cells, is largely unclear. To identify ABIN1–interacting proteins, we generated Hoxb8–FL cells from BM of ABIN1–deficient mice, and introduced epitope– (FS–) tagged forms of wildtype (WT) ABIN1 (FS–ABIN1) and an ubiquitin–binding mutant (FS–ABIN1–EE) by retroviral transduction. Stably growing, polyclonal populations and vector–transduced control cells were up–scaled and differentiated into 8×108 DC in the presence of GM–CSF, followed by tandem affinity purification (TAP) of ABIN1 and analysis of ABIN1–associated proteins by SDS/ PAGE and liquid–chromatography–coupled mass spectrometry (LC–MS) (Fig. 6c). ABIN1–expression was analyzed in parallel by intracellular staining and flow cytometry and immuno–blotting, demonstrating stable and homogenous expression of WT and mutant ABIN1 during differentiation (Fig. 6d,e). Several ABIN1–interacting proteins were identified, including NF–κB1 (p105), which was represented by a large number of unique peptides in both ABIN1–WT– and ABIN1–EE–purified samples, but not control samples (table 1). Overexpression of ABIN1 has been shown to counteract constitutive processing of NF–κB1, which is required to produce the transcriptionally active form of NF–κB1, i.e. p5028. As such, our data confirm interaction of ABIN1 and NF–κB1, and strongly suggest a physiological function of ABIN1 in the NF–κB pathway in DC.
DISCUSSION
A particularly remarkable aspect of the method described here is the simplicity of the procedure, which is based on largely unfractionated BM. Still, we generated similar cell lines starting with LSK cells which also showed myeloid and lymphoid lineage potential as investigated in a more limited set of experiments (Supplementary Fig. 13). We have established more than 90 Hoxb8–FL cell lines, varying systematically different aspects of the establishing procedure, including virus titer (multiplicity of infections 1 to 100) and viral vector (MSCV vs. lentivirus). Despite the different conditions used, the efficiency for generation of Hoxb8–FL cell lines was 100 % and all cell populations obtained showed uniformly a virtually identical phenotype of the Hoxb8–FL cells described in this paper.
With respect to phenotype and lineage potential, Hoxb8–FL cells resemble LMPP, but do not express SCA–1. Based on Pu.1–GFP reporter mice, Arinobu et al recently showed that LMPP express SCA–1 at the lower end of an expression– spectrum of LSK cells, indicating a gradual loss of SCA–1 expression along with the loss of multi–lineage potential and commitment to immune cell lineages29. Hoxb8–FL cells may therefore represent a cell type following strictly defined SCA–1+ LMPP during lineage commitment. This interpretation is also consistent with the PCA which clustered Hoxb8–FL cells distant of the megakaryocyte/ erythroid lineage and downstream of LMPP ‘in between’ myeloid and lymphoid branches generated by defined BM progenitor cells.
As noted, Hoxb8–FL cells generated B220+ B–cells in vivo at numbers comparable to endogenous B–cells, while T–cell reconstitution was more limited (Fig. 3a and Supplementary Fig. 10). Moreover, in–vitro assays assessing quantitatively lineage potential implicated reduced T–cell potential of Hoxb8–FL cells compared to LMPP, which exhibit the highest T–cell potential of currently defined progenitor cells23 (Fig. 5e). One possible explanation for these observations is therefore that Hoxb8–FL cells indeed have reduced T–cell potential, while maintaining myeloid and B–cell potential comparable to LMPP. This interpretation, however, is less compatible with observations related to thymic lineage commitment, where B–cell potential is reduced before restriction in myeloid lineage potential are apparent 20, 21,22. An alternative explanation is that the specific OP9–DL1–dependent cell culture conditions, which were optimized for LMPP, may be not ideal for Hoxb8–FL cells. Increasing the FLT3L concentration, or addition of other factors present in the BM environment (but not in cell culture), may raise the T–cell potential of Hoxb8–FL cells elicited in vitro. An alternative explanation for the relatively low number of T–cells generated in vivo is the known limitation of the thymus for access of thymus–settling progenitor cells from the blood, which has been estimated to be an extremely low number, possibly less than 10 cells per day30,31. This limitation, which is further subject to a periodic mode of regulation, referred to as ‘gating’32, 33, is likely to be particularly relevant for continuously differentiating Hoxb8–FL–derived cells, which are not replenished by self–renewing cells, thereby decreasing the temporal window of opportunity to enter the thymus. Interestingly, appearance of Hoxb8–FL cell–derived immature progenitor cells in the thymus was measurable at the earliest after day 6 of adoptive transfer, well after population of the BM, indicating that thymus settling occurs after, and potentially via, BM settling. This interpretation is consistent with data strongly suggesting that a cell type referred to as FLT3+ MPP, which phenotypically largely overlaps with LMPP, settle the thymus via transition of the BM34. If so, the BM environment, which is particularly conducive to B–cell development, may further reduce the number of potentially thymus–settling Hoxb8-FL-derived progenitors.
An important aspect of Hoxb8–FL cells are their wide range of possible applications, one of which is shown in the proteomic experiment based on ABIN1–deficient DC (Fig. 6c – d). Another important application of Hoxb8–FL cells is the investigation of factors implicated in cell differentiation, both in vitro and in vivo. Not least, Hoxb8–FL cells generated from normal or genetically modified mice, and possibly further modified, e.g. by retroviral transduction, can be differentiated in vitro into mature immune cells, e.g. DC, whose biology can be analyzed in vitro or in vivo, as illustrated in this paper by DC–mediated protection from B16 melanoma (Fig. 2h).
METHODS
Reagents and plasmids
Antibodies were used against B220 (RA3–6B2), CD4 (RM4–5), CD45.1 (A20), MHCII (M5/114.15.2), GR1 (Ly6G (RB6–8C5), CD11c (N418), IgM (eB121–15F9), CD25 (PC61.5), CD44 (IM7), Thy1.2 (53–2.1), CD34 (RAM34), FLT3 (A2F10), Sca–1 (D7), Ter119 (TER–119), CD19 (1D3), AA4.1 (AA4.1), CD21 (ebio8D9), CD24 (30–F1, for DC), CD24 (M1/69, for BM–B–cells), CD172a (P84), IgD (11–26)(all from eBiosciences); CD3 (145–2C11), CD8 (53–6.7), CD11b (M1/70), CD11c (HL3), c–Kit (2B8), Sca–1 (E13–161.7), IL–12p40 (C15.6), CD43 (S7), BP–1 (6C3), CD8α (53–6.7) (all from BD Biosciences); CD23 (30–F1, Genway), PDCA–1 (Miltenyi), IFNα (RMMA1, PBL), ABIN1 (1A11E3, Invitrogen). ELISA kits for IL–6 and IL–12 were from BD Biosciences. Griess assays for determination of nitric oxide concentration were done as described 35. CpG–DNA (1668) and CpG–DNA (2216) refer to the phosphothioate backbone containing oligonucleotides 1668 (1 µM, TCCATGACGTTCCTGATGCT) and 2216 (3µM, GGGGGACGATCGTCGGGGGG) (TIB Molbiol). Other agonists used were LPS (10ng/ml, E.coli 0127:B8 (Sigma–Aldrich)), IFNγ (10 ng/ml, Peprotech), β–estradiol (1 µM, Sigma–Aldrich). Recombinant mouse growth factors (SCF, FLT3L, TPO, IL–3, IL–6) were from Peprotech, human EPO was from Amgen. Mouse Hoxb8 cDNA was amplified by PCR using Wehi–3 cDNA as template, and was cloned into MSCVneo (BD Biosciences) downstream of a triple hemagglutinin (HA) epitope tag. The estrogen binding domain of the human estrogen receptor (ERHBD) was amplified by overlap extension PCR based on cDNA of the human estrogen receptor 1 (Open Biosystems, clone ID40128594), introducing the characterized G400V mutation, which renders the hormone binding domain insensitive to physiological concentrations of estrogen or phenol red contained in growth medium 36. The mutagenized ERHBD fragment was cloned in frame between triple HA–tag and Hoxb8, generating the final MSCV–ERHBD–HOXB8 vector. The correct sequence of the expression cassette was confirmed by DNA sequencing.
Virus production
The plasmids MSCV–ERHBD–HOXB8 and the ecotropic packaging vector pCL–Eco (Imgenex) were co–transfected into HEK293T cells using Lipofectamine 2000 (Invitrogen). 18 hours after transfection, the supernatant (SN) was replaced by fresh growth medium (DMEM (Invitrogen), supplemented with 10% (v/v) fetal calf serum (FCS, Hyclone), 50 mM 2–mercaptoethanol, antibiotics (penicillin G (100 IU/ml) and streptomycin sulfate (100 IU/ml) and Pyruvate (1mM)), followed by incubation for 24 hours and collection of virus–containing supernatant. Virus titers were determined on mouse embryonic fibroblast cells based on G418 resistance mediated by the retroviral vector.
Generation and cell culture of Hoxb8–FL and Hoxb8–SCF progenitor cell lines
BM cells were harvested by flushing femurs of 4–8 week old female C57BL/6J or B6.SJL mice with 10 ml RP–10 (RPMI 1640 (Invitrogen), supplemented with 10 % (v/v) fetal calf serum (FCS, Hyclone), 50 mM 2–mercaptoethanol and antibiotics (penicillin G (100 IU/ml) and streptomycin sulfate (100 IU/ml)), pelleted by centrifugation, resuspended in 4 ml RP–10, loaded on 3ml Ficoll–Paque (GE Healthcare) and separated by centrifugation at 450 g for 30 min. The entire supernatant was collected (discarding only 500 µl including the cell pellet), diluted with 45 ml PBS containing 1 % FCS (PBS/ FBS), pelleted at 800 g for 10 min, followed by resuspension in 10 ml RP–10, centrifugation at 450 g for 5min and resuspension at a concentration of 5 × 105 cells/ml in RP–10 containing recombinant mouse IL–3 (10 ng/ml), IL–6 (20 ng/ml) and 1% of cell culture supernatant from a SCF–producing B16 melanoma cell line (kindly provided by Mark Kamps (University of California, San Diego)), corresponding to the bioactivity of 250 ng/ml recombinant SCF (R&D Systems). After two days of cell culture, cells were collected, resuspended in progenitor outgrowth medium (POM), i.e. RP–10 supplemented with 1 µM β–estradiol and either cell culture supernatant from an FLT3L producing B16 melanoma cell line (generously provided by Ralph Steinman (Rockefeller University)) for generation of Hoxb8–FL cells (5 % final concentration) or the described SCF–producing cell line for generation of Hoxb8–SCF cells (1% final concentration). 2 × 105 cells were dispensed in 1 ml per well in a 12–well plate and infected with MSCV vectors (multiplicity of infection 5) by spin inoculation at 1500 g for 60 min in the presence of Lipofectamine (0.1 %, Invitrogen). After infection, cells were diluted by adding 1.5 ml POM for 24 hours, followed by removal and replacement of 2 ml of the cell culture medium. During the following cell culture period, cells were dispensed every 3–4 days in fresh medium and transferred into new wells. Once the cell populations were stably expanding, cells were kept at concentrations between 1 × 105 and 1.5 × 106 cells/ml medium. For consequent differentiation experiments, cells were washed twice with PBS/ FCS and resuspended in a concentration of 0.5–2 × 105 cells/ml in RP–10 containing specific growth factors as described for BM–derived DC and macrophages. For in vivo experiments, 5 × 106 cells were resuspended in 0.2 ml PBS/FBS (along with 2 × 105 unfractionated BM cells) and transferred into mice via tail vain injection.
Phagocytosis assay
Hoxb8–FL cells and BM cells were differentiated in the presence of M–CSF contained in L–cell conditioned medium for seven days, followed by incubation with FITC–labeled IgG–coated beads (1:100, Cayman Chemical, Phagocytosis Kit (IgG FITC)) either at 37° C or 4° C for 2 h and 6 h. Cells were washed with PBS and trypsin, detached with trypsin and analyzed by flow cytometry.
Mice and transfer of Hoxb8–FL cells and BM cells into mice
All mouse studies were carried out in accordance with protocols approved by the Institutional Animal Care and Use Committee at St. Jude Children’s Research Hospital. 5 × 106 CD45.1+ Hoxb8–FL cells (derived from B6/SJL mice), together with 2 × 105 CD45.2+ BM cells (derived from C57BL/6J mice), were collected by centrifugation, resuspended in 200µl PBS/ FCS and transferred via tail vain injection into C57BL/6J wildtype, Il7r−/−, or µMT mice (The Jackson Laboratory) that had been lethally irradiated at 950rad one day before cell transfer where indicated in legends. Hoxb8–FL– and BM–derived cells were differentiated based on CD45.1 and CD45.2 staining. ABIN1–deficient mice, which were generated using ES cells obtained from the German Gene Trap Consortium, have been described26. Peripheral blood was collected by retroorbital bleeding. Complete blood counts (CBC) were measured using the Forcyte Hematology System (Oxford Science).
Mouse immunization and analysis of serum IgG titers
5 × 106 Hoxb8–FL cells or unfractionated BM cells were transferred into lethally irradiated µMT mice along with unfractionated 2 × 105 BM helper cells from µMT mice, followed by intradermal injection of OVA (1µg) plus CpG–DNA (50µg) in 50 µl PBS at the base of the tail on day 21, 31 and 41 after transfer. Serum was collected one day before immunization and 10 days after the last immunization, and IgG1 and IgG2a antibody titers were determined by ELISA. Relative units were determined based on pooled, high–titer ‘standard’ serum obtained from wildtype mice that had been immunized with OVA plus Alum (IgG1) or OVA plus CpG–DNA (IgG2a).
Isolation of splenic DC
5 × 106 CD45.1+ Hoxb8–FL cells or unfractionated BM cells were transferred via tail vein injection into lethally irradiated CD45.2+ recipient mice along with 2 × 105 CD45.2+ BM cells. Seven days after transfer, spleens were removed, cut into small pieces using scissors to obtain a homogenous cell paste, resuspended in spleen dissociation medium (Stem Cell Technologies) and incubated for 30 min at room temperature, followed by incubation for another 5 min in the presence of EDTA (10 mM). Cells were passed through a 40µm mesh filter and purified by MACS using the Pan–DC isolation kit containing CD11c– and PDCA1–specific antibodies according to the manufacturer’s instructions (Miltenyi).
Evaluation of myeloid and lymphoid potential in bulk cultures in vitro
BM– and Hoxb8–FL–derived DC and macrophages were generated by cultivating unfractionated BM cells or Hoxb8–FL cells in Petri dishes for six days in standard growth medium (RPMI 1640 (Invitrogen, DC) or DMEM (Invitrogen, macrophages), supplemented with 10 % (v/v) FCS (Hyclone), 50 mM 2–mercaptoethanol and antibiotics (penicillin G (100 IU/ml) and streptomycin sulfate (100 IU/ml), Invitrogen) containing 5 % FLT3L or 2 % GM–CSF (DC), or 30 % L–cell–conditioned medium (macrophages) derived from growth factor producing cell lines. 5% FLT3L and 2% GM–CSF corresponds to the bioactivity of 70 ng/ml and 7 ng/ml of recombinant growth factors, respectively (FLT3L, R&D; GM–CSF, Becton Dickinson).
In FLT3L– and GM–CSF–driven cultures non–adherent cells were used for subsequent experiments. In cell cultures conditioned with L–cell supernatant, adherent cells were detached by trypsin treatment and used for subsequent experiments. For in vitro differentiation of lymphocyte progenitors, 2 × 105 Hoxb8–FL cells or LSK cells were cocultured on OP9 cells or OP9–DL1 cells that were maintained in alpha–minimum essential medium (Invitrogen) supplemented with 10 % fetal calf serum (Hyclone), penicillin/ streptomycin, and recombinant mouse IL–7 (Peprotech, 3ng/ml) and FLT3L (R&D Systems, 5 ng/ml) 16,37. Cells were enumerated, filtered through 100µm cell strainers (BD Falcon), and passaged onto fresh OP9 cells every 3–4 days. LSK cells were isolated from BM by sequential depletion of lineage positive cells by magnetic–activated cell sorting (MACS, Miltenyi, lineage cell depletion kit), followed by fluorescence–activated cell sorting based on antibodies against SCA–1 and c–Kit.
Evaluation of myeloid and lymphoid potential in clonal and limiting dilution assays in vitro
Myeloid potential of Hoxb8–FL cells and LMPP was assessed by clonal assays, seeding one cell per well in 96 flat bottom plates by FACS into 100 µl RP–10 containing FLT3L–SN (5 %) and L-cell–SN (30%). Formation of typical adherent, myeloid colonies was assessed by light microscopy on day 10 after seeding.
To determine B–cell and T–cell potential, irradiated (15 Gy) OP9 and OP9–DL1 cells were seeded in 50 µl Optimem–10 (Optimem (Invitrogen), supplemented as described for RP–10) into 96 well flat bottom plates (1.5 × 104 cells per well). After overnight culture, 50 µl Optimem–10 containing FLT3L, SCF and IL–7 was added to final concentrations of FLT3L–SN (1:50, equivalent to 30 ng/ml rec. FLT3L), SCF–SN (1:100, equivalent to 250 ng/ml rec. SCF) and IL–7 (20 ng/ml). 30, 5 or 1 cell per well of Hoxb8–FL cells or LMPP were seeded by FACS. 100 µl of fresh Optimem–10 containing FLT3L (1:50) and IL–7 (10 ng/ml) was added six days later, and also 13 days later (after removing 100 µl). Colony formation was evaluated by microscopy after 14–16 days (B–cells) and 17–20 days (T–cells) and Thy1– CD19+ CD11b– CD25– CD44+ and Thy1+ CD19– CD11b– CD25+ CD44+ cells were identified by flow cytometry as B– and T–cell progenitors, respectively. Lineage potential was calculated based on ELDA (Extreme Limiting Dilution Analysis) provided online (http://bioinf.wehi.edu.au/software/elda/)38.
Evaluation of in vitro megakaryocyte potential
Hoxb8–FL cells and LSK cells were seeded in minitray plates (Nunc) at a concentration of one or five cells per well (120 wells each) in 20 µl X–vivo 15 medium (Lonza) supplemented with FCS (10 % v/v, Hyclone), bovine serum albumin (0.5 %, Stemcell Technologies), SCF (50 ng/ml), FLT3L (50 ng/ml), TPO (50 ng/ml), EPO (5 U/ml) and IL–3 (20 ng/ml). Wells were scored for cell growth at different time points up to 8 days. Mk–containing colonies were identified by light microscopy and confirmed morphologically after transferring individual colonies to slides in a cytospin centrifuge and subsequent May–Grünwald/ Giemsa–staining.
Evaluation of in vitro erythroid/ macrophage potential
Colony forming cell assays were performed using methylcellulose containing IMDM media (Methocult, Stemcell Technologies) supplemented with 15 % FCS (v/v), 1 % bovine serum albumin, recombinant human (rh) insulin (10 ng/ml), human transferrin (iron saturated, 200 mg/ml) and different growth factors. To evaluate CFU–E formation, 2 × 102 Hoxb8–FL cells or 2 × 104 unfractionated BM cells were seeded into 35mm wells containing methylcellulose medium as described above, which was further supplemented with rh EPO (3 U/ml). Wells were analyzed microscopically for colony formation at different time points up to 10 days. BM–derived colonies were scored after two days. To evaluate BFU–E and CFU–M formation, 2 × 102 Hoxb8–FL cells or 2 × 102 LSK cells were seeded into 35mm wells containing methylcellulose medium as described above, which was further supplemented with rh EPO (3 U/ml), recombinant mouse (rm) IL–6 (10 ng/ml), rm IL–3 (10 ng/ml) and rm SCF (50 ng/ml). Wells were analyzed microscopically for colony formation at different time points and scored at day 14.
Fluorescence–activated cell sorting
Single cell suspensions of the spleen were prepared by straining tissues through a 100 µm cell strainer (Becton Dickinson). Peripheral blood was obtained by retro–orbital bleeding. To lyze red blood cells, 100 µl blood were treated with 1.2 ml Ammonium Chloride Solution (Stemcell Technologies) for 10 min on ice, followed by dilution with 10 volumes of PBS/ FBS and centrifugation for 5 min at 450g. Cells obtained from spleens or peripheral blood were blocked with antibodies against CD16/ CD32 (eBioscience), followed by staining for cell surface markers. For intracellular staining, DC that were first stained for B220 were fixed with 2% formaldehyde in PBS (20 min at 25°C), followed by incubation with FITC–labelled antibodies against IFNα (or isotype control) and PE–labelled IL–12p40 in PBS containing 0.5% saponin. Flow cytometry analysis was done using a FACSCalibur or FACSCanto II instrument (Becton Dickinson). Single cell sorting was done using a FACSAria instrument (Becton Dickinson).
DC–induced T–cell proliferation in vitro
DC were generated as described in Figure Legends and incubated with 400 µg/ml chicken Ovalbumin (OVA, Sigma–Aldrich) for 4 hours, washed twice and co–incubated in round–bottom 96 well plates at different concentrations with naïve (CD62Lhi CD44– CD25–) CD4– and CD8–positive T–cells isolated by FACS from spleens of OTI– and OTII–transgenic mice. In some cases, T–cells were labelled with CFSE according to the manufacturer’s instructions (Invitrogen). After co–incubation of DC and T–cells for three days, T–cell proliferation was either directly determined by flow cytometry (for CFSE–labelled cells), or after an eight–hour thymidine pulse in a scintillation beta–counter.
DC vaccination and melanoma (B16–OVA) mouse model
The cDNA encoding a cytoplasmic form of chicken Ovalbumin (OVA (AS 1–18/145–386)), kindly provided by W. Goebel and D. Schoeffler 39 was cloned into the MMLV–based retroviral expression vector pQCXIP (BD Clontech) and B16F1 melanoma cells were transduced with replication–deficient, amphotropic retrovirus virus obtained from transiently transfected HEK293T cells. Stably OVA–expressing B16F1 cells (B16–OVA) were selected using puromycin (10 µg/ml). Hoxb8–FL cells and BM cells were differentiated in the presence of GM–CSF for nine days, pulsed with OVA (400 µg/ml) for 4 hours, washed twice with PBS and stimulated with CpG–DNA for 18 hours. 6 week–old C57BL/6 mice were injected subcutaneously with 5 × 105 DC (as detailed in Figure legends) in 100 µl PBS in the right flank which had been treated by Nair crème to remove hair. One week later, 2×105 B16–OVA cells were injected intra–dermally (i.d.) in 50 µl PBS in the same area of the right flank, and tumor growth was monitored during time. Mice were sacrificed when the tumors size reached 1 cm3.
Microarray– and principal component–analysis (PCA) of hematopoietic cells
Total cellular RNA of four independent Hoxb8–FL cell lines and two BM–derived macrophage populations (BMM) was prepared using TRIzol (Invitrogen), purified using the RNeasy Mini kit (QIAGEN), processed and subjected to Affymetrix expression analysis based on MOE v430 chips according to the manufacturer’s instructions. Affymetrix MOE v430 CEL file data from Hoxb8-FL cells are available from NCBI GEO (GSE45759). Affymetrix MOE430v2 CEL file data were downloaded from three GEO sources (GSE14833 (progenitors), GSE10246 and GSE6506 (granulocytes)). RMA summarization was performed an all data using Partek Genomics Suite 6.6. To minimize confounding effects of cell culture and source, an ANOVA was applied to the probe sets of progenitor cells in order to extract genes which best defined cell types based on the matrix published by Tullio and colleagues40.8,690 probe sets that passed the Bonferroni correction at the 0.01 level were retained and used along with the same probe sets from the Hoxb8–FL cells, BMM and granulocytes for visualization by PCA (Partek Genomic Suite 6.6.). Gene arrays for hematopoietic progenitor cells were generated based on sorted populations using the following markers (generously provided by S. Zandi, Mikael Sigvardsson and David Bryder): LTHSC (Lin− Sca1+ Kit+ CD34− flt3−), STHSC (Lin− Sca1+ Kit+ CD34+ flt3−), LMPP (Lin– Sca1+ Kit+ CD34+ flt3+), ETP (Lin− Sca1+ Kit+ CD34+ flt3+ IL7−), proB (Lin− CD19− B220+ CD43+ IgM−), preB (Lin− CD19+ B220+ CD43− IgM−), CD4T (Ter119− Gr1− Mac1− Nk1.1− CD8− CD4+), PreGM (Lin− Sca1− Kit+ CD41−low CD16/32low CD150− CD34+ CD9low), GMP (Lin− Sca1– Kit+ CD41− CD16/32high CD150− CD34+), MkP (Lin− Sca1− Kit+ CD150+ CD41+ CD34−), MkE (Lin− Sca1− Kit+ CD150+ CD41− CD150+ Endoglin−), preCFUE (Lin− Sca1− Kit+ CD150+ CD41− CD150+ Endoglin+), CFUE (Lin− Sca1− Kit+ CD150+ CD41− CD150− Endoglin+ Ter119−), ProE (Lin− Sca1− Kit+ CD150+ CD41− CD150− Endoglin+ Ter119+).
Generation, reconstitution and differentiation of ABIN1–deficient Hoxb8–FL cells
Hoxb8–FL cells from BM of 4 week–old ABIN1–deficient mice were established as described above in detail. Three weeks after infection with the MSCV–ERHBD–Hoxb8 vector, cells were transduced with MSCV–Puro–based (BD Clontech) retroviral vectors expressing ABIN1-WT or an ubiquitin–binding deficient mutant (QQ477/478EE) of ABIN1 (ABIN1–EE), both containing triple FLAG– and One–STrEP–epitope tags (FS) in tandem orientation allowing tandem affinity purification. Two days after virus infection, transduced cells were selected using puromycin (10 µg/ml) and continuously maintained in culture for another 14 days. For subsequent differentiation, 3×107 cells were washed twice with PBS PBS/ FCS and resuspended at a concentration of 1 × 105 cells/ml in GM–CSF–conditioned RP–10 as described for generation of GM–CSF–derived DC. Cells were fed with GM–CSF–containing medium after three and five days. After eight days, 8×108 cells were harvested by centrifugation and snap freezing using dry ice/ methanol.
Tandem affinity purification
Frozen cell pellets were dissolved in lysis buffer ((LB) 20 mM Hepes/KOH (pH 7.5), 150 mM NaCl, 1.5 mM MgCl, 1 mM EDTA, 1 mM PMSF, 10% Glycerol, 1mM orthovanadate, 10 mM β–glycerophosphate, 5 mM 4–nitrophenyl–phosphate, 10 mM sodium fluoride, ‘complete protease inhibitors’ (Roche)) containing 0.5% NP40 for 20 min. Samples were cleared by centrifugation (10 min, 1500g, 4° C), and incubated with Strep–Tactin Superflow beads (IBA) for 90 min. Beads were collected by centrifugation, washed and proteins were eluted in 1 ml LB containing Desthiobiotin (5mM; Sigma–Aldrich) and 0.1% NP40. Proteins were re–captured on columns containing M2 FLAG beads (Sigma–Aldrich), washed and eluted at low pH (0.1 mM Glycine (pH 3.5) followed by TCA precipitation, separation on a 4–12 % Bis–Tris gel (BioRad) and staining with SYPRO–Ruby (Invitrogen).
Sample Preparation for MS Analysis
Individual gel lanes containing the immuno-purified samples were excised into 15 bands, each of which was cut into small plugs, washed with 50% (v/v) acetonitrile, and de-stained by repeated incubations in 100 mM ammonium bicarbonate (pH 8.0) containing 50% acetonitrile. Gel plugs were reduced (10 mM DTT, 1 h at 37 °C), alkylated (50 mM iodoacetamide, 45 min at room temperature in the dark), washed twice with 50% acetonitrile in 50 mM ammonium bicarbonate, dried in a SpeedVac (Savant), and rehydrated for 10 min in 10 µl of 0.2 µg/µl trypsin. Then 25 µl of ammonium bicarbonate (25 mM, pH 8.0) was added and incubated for ~12 h at 37 °C, followed by peptide extraction using 5–10 µl of 0.2 % formic acid. The peptide-containing solution was transferred to a sample vial for liquid chromatography tandem MS (LC-MS/MS) analysis.
Electrospray Ionization Orbitrap mass spectrometry (MS) Analysis
LC–MS/MS analysis was performed using a ThermoFisher LTQ Orbitrap XL mass spectrometer in line with a nanoAcquity ultra–performance LC system (Waters Corporation). Tryptic peptides were loaded onto a “precolumn” (Symmetry C18, 180–µm i.d. × 20 mm, 5–µm particle) (Waters Corporation) which was connected through a zero dead–volume union to the analytical column (BEH C18, 75–µm i.d. × 100 mm, 1.7–µm particle) (Waters Corporation). The peptides were eluted over a gradient (0−70% B in 60 min, 70−100% B in 10 min, where B = 70% (vol/vol) acetonitrile, 0.2% formic acid) at a flow rate of 250 nL/min and were introduced online into the Orbitrap mass spectrometer using electrospray ionization. Data–dependent scanning was incorporated to select the 10 most abundant ions [one microscan per spectrum; precursor isolation width 2.0 Da; 35% collision energy; 30–ms ion activation; 30 s exclusion duration; 15 s repeat duration; a repeat count of 2] from a full–scan mass spectrum for fragmentation by collision–activated dissociation.
Database Search
Product ions (b/y–type ions) were queried in an automated database search against a protein database [Swissprot 2012_10 (538259 sequences; 191113170 residues, Mus musculus subset)] by the Mascot search algorithm. The following residue modifications were allowed in the search: carbamidomethylation on cysteine (fixed modification) and oxidation on methionine (variable modification). Mascot was searched with a precursor ion tolerance of 100 ppm and a fragment ion tolerance of 0.7 Da and using the automatic decoy database–searching tool in Mascot. The identifications from the automated search were verified by manual inspection of the raw data.
Microscopy
Images were obtained using the microscopes Axio Scope.A1 and Axiovert 40 CFL (Zeiss) for May–Grünwald/ Giemsa-stained cells and live cells, respectively. Software used was Axiovision 4 (Zeiss).
Supplementary Material
ACKOWLEDGMENTS
We thank the staff of the Animal Resource Center, the Flow Cytometry and Cell Sorting Resource and the Proteomics & Mass Spectrometry laboratory at the St. Jude Children’s Research Hospital, and Liying Chi for their excellent technical assistance. We also thank H. Hochrein (Bavarian Nordic) for helpful discussion related to DC biology. This work was supported by NIH/NIAID grant AI083443 to H.H., the NIH/NCI grant P30 CA2175 and the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS V.R., R.W., V.C. and H.H. planned experiments, V.R., R.W., J.Z., V.C. and H.H. performed experiments, A.H., D.F., V.C., V.R. and H.H. analyzed data, V.R. and H.H. wrote manuscript, H.H. conceived project.
REFERENCES
- 1.Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007;26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 2.Pineault N, Helgason CD, Lawrence HJ, Humphries RK. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp Hematol. 2002;30:49–57. doi: 10.1016/s0301-472x(01)00757-3. [DOI] [PubMed] [Google Scholar]
- 3.Wang GG, et al. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat Methods. 2006;3:287–293. doi: 10.1038/nmeth865. [DOI] [PubMed] [Google Scholar]
- 4.McKenna HJ, et al. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95:3489–3497. [PubMed] [Google Scholar]
- 5.Gilliet M, et al. The development of murine plasmacytoid dendritic cell precursors is differentially regulated by FLT3-ligand and granulocyte/macrophage colony-stimulating factor. J Exp Med. 2002;195:953–958. doi: 10.1084/jem.20020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinman RM. Dendritic cells: understanding immunogenicity. Eur J Immunol. 2007;37(Suppl 1):s53–s60. doi: 10.1002/eji.200737400. [DOI] [PubMed] [Google Scholar]
- 7.Naik SH, et al. Cutting edge: generation of splenic CD8+ and CD8- dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures. J Immunol. 2005;174:6592–6597. doi: 10.4049/jimmunol.174.11.6592. [DOI] [PubMed] [Google Scholar]
- 8.Hemmi H, Kaisho T, Takeda K, Akira S. The roles of Toll-like receptor 9, MyD88, and DNA-dependent protein kinase catalytic subunit in the effects of two distinct CpG DNAs on dendritic cell subsets. J Immunol. 2003;170:3059–3064. doi: 10.4049/jimmunol.170.6.3059. [DOI] [PubMed] [Google Scholar]
- 9.Hogquist KA, et al. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 10.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 11.Sitnicka E, et al. Key role of flt3 ligand in regulation of the common lymphoid progenitor but not in maintenance of the hematopoietic stem cell pool. Immunity. 2002;17:463–472. doi: 10.1016/s1074-7613(02)00419-3. [DOI] [PubMed] [Google Scholar]
- 12.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zlotoff DA, et al. Delivery of progenitors to the thymus limits T-lineage reconstitution after bone marrow transplantation. Blood. 2011;118:1962–1970. doi: 10.1182/blood-2010-12-324954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prockop SE, Petrie HT. Regulation of thymus size by competition for stromal niches among early T cell progenitors. J Immunol. 2004;173:1604–1611. doi: 10.4049/jimmunol.173.3.1604. [DOI] [PubMed] [Google Scholar]
- 15.Carlyle JR, et al. Identification of a novel developmental stage marking lineage commitment of progenitor thymocytes. J Exp Med. 1997;186:173–182. doi: 10.1084/jem.186.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 17.Godfrey DI, Kennedy J, Suda T, Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol. 1993;150:4244–4252. [PubMed] [Google Scholar]
- 18.Pui JC, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 19.Sultana DA, Bell JJ, Zlotoff DA, De Obaldia ME, Bhandoola A. Eliciting the T cell fate with Notch. Semin Immunol. 2010;22:254–260. doi: 10.1016/j.smim.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wada H, et al. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 21.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 22.Luc S, et al. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat Immunol. 2012;13:412–419. doi: 10.1038/ni.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Yang L, et al. Identification of Lin(−)Sca1(+)kit(+)CD34(+)Flt3- short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. 2005;105:2717–2723. doi: 10.1182/blood-2004-06-2159. [DOI] [PubMed] [Google Scholar]
- 25.Oshima S, et al. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature. 2009;457:906–909. doi: 10.1038/nature07575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J, et al. A20-binding inhibitor of NF-kappaB (ABIN1) controls Toll-like receptor-mediated CCAAT/enhancer-binding protein beta activation and protects from inflammatory disease. Proc Natl Acad Sci U S A. 2011;108:E998–E1006. doi: 10.1073/pnas.1106232108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mauro C, et al. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J Biol Chem. 2006;281:18482–18488. doi: 10.1074/jbc.M601502200. [DOI] [PubMed] [Google Scholar]
- 28.Cohen S, Ciechanover A, Kravtsova-Ivantsiv Y, Lapid D, Lahav-Baratz S. ABIN-1 negatively regulates NF-kappaB by inhibiting processing of the p105 precursor. Biochem Biophys Res Commun. 2009;389:205–210. doi: 10.1016/j.bbrc.2009.08.074. [DOI] [PubMed] [Google Scholar]
- 29.Arinobu Y, et al. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Wallis VJ, Leuchars E, Chwalinski S, Davies AJ. On the sparse seeding of bone marrow and thymus in radiation chimaeras. Transplantation. 1975;19:2–11. doi: 10.1097/00007890-197501000-00002. [DOI] [PubMed] [Google Scholar]
- 31.Spangrude GJ, Scollay R. Differentiationof hematopoietic stem cells in irradiated mouse thymic lobes Kinetics and phenotype of progeny. J Immunol. 1990;145:3661–3668. [PubMed] [Google Scholar]
- 32.Zlotoff DA, et al. CCR7 and CCR9 together recruit hematopoietic progenitors to the adult thymus. Blood. 2010;115:1897–1905. doi: 10.1182/blood-2009-08-237784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foss DL, Donskoy E, Goldschneider I. The importation of hematogenous precursors by the thymus is a gated phenomenon in normal adult mice. J Exp Med. 2001;193:365–374. doi: 10.1084/jem.193.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serwold T, Ehrlich LI, Weissman IL. Reductive isolation from bone marrow and blood implicates common lymphoid progenitors as the major source of thymopoiesis. Blood. 2009;113:807–815. doi: 10.1182/blood-2008-08-173682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stuehr DJ, Nathan CF, Nitric oxide A. macrophage product responsible for cytostasis and respiratory inhibition in tumor target cells. J Exp Med. 1989;169:1543–1555. doi: 10.1084/jem.169.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tora L, et al. The cloned human oestrogen receptor contains a mutation which alters its hormone binding properties. EMBO J. 1989;8:1981–1986. doi: 10.1002/j.1460-2075.1989.tb03604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitt TM, Zuniga-Pflucker JC. T-cell development, doing it in a dish. Immunol Rev. 2006;209:95–102. doi: 10.1111/j.0105-2896.2006.00353.x. [DOI] [PubMed] [Google Scholar]
- 38.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Loeffler DI, Schoen CU, Goebel W, Pilgrim S. Comparison of different live vaccine strategies in vivo for delivery of protein antigen or antigen-encoding DNA and mRNA by virulence-attenuated Listeria monocytogenes. Infect Immun. 2006;74:3946–3957. doi: 10.1128/IAI.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Tullio A, et al. CCAAT/enhancer binding protein alpha (C/EBP(alpha))-induced transdifferentiation of pre-B cells into macrophages involves no overt retrodifferentiation. Proc Natl Acad Sci U S A. 2011;108:17016–17021. doi: 10.1073/pnas.1112169108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.