Abstract
The identification of mycobacteria is essential because tuberculosis (TB) and mycobacteriosis are clinically indistinguishable and require different therapeutic regimens. The traditional phenotypic method is time consuming and may last up to 60 days. Indeed, rapid, affordable, specific and easy-to-perform identification methods are needed. We have previously described a polymerase chain reaction-based method called a mycobacteria mobility shift assay (MMSA) that was designed for Mycobacterium tuberculosis complex (MTC) and nontuberculous mycobacteria (NTM) species identification. The aim of this study was to assess the MMSA for the identification of MTC and NTM clinical isolates and to compare its performance with that of the PRA-hsp65 method. A total of 204 clinical isolates (102 NTM and 102 MTC) were identified by the MMSA and PRA-hsp65. For isolates for which these methods gave discordant results, definitive species identification was obtained by sequencing fragments of the 16S rRNA and hsp65 genes. Both methods correctly identified all MTC isolates. Among the NTM isolates, the MMSA alone assigned 94 (92.2%) to a complex or species, whereas the PRA-hsp65 method assigned 100% to a species. A 91.5% agreement was observed for the 94 NTM isolates identified by both methods. The MMSA provided correct identification for 96.8% of the NTM isolates compared with 94.7% for PRA-hsp65. The MMSA is a suitable auxiliary method for routine use for the rapid identification of mycobacteria.
Keywords: nontuberculous mycobacteria, mycobacteria mobility shift assay, mycobacterial identification
The genus Mycobacterium comprises more than 150 species and subspecies (Euzéby 2012). The Mycobacterium tuberculosis complex (MTC) causes tuberculosis (TB), whereas nontuberculous mycobacteria (NTM) can cause mycobacteriosis. M. tuberculosis infection remains the leading cause of deaths related to infectious diseases worldwide, as this bacterium causes over eight million cases of TB and 1.4 million deaths annually (WHO 2012). NTM includes pathogenic, potentially pathogenic and saprophytic bacteria that are ubiquitous in the environment and have been associated with opportunistic diseases (Katoch 2004, Falkinham 2009). Indeed, in addition to multidrug-resistant strains of M. tuberculosis, NTM are becoming increasingly important as human pathogens. These mycobacteria are responsible for a variety of clinical manifestations, including pulmonary, lymphatic, soft tissues and disseminated infections, as well as nosocomial outbreaks related to the inadequate sterilisation of solutions and medical devices (Katoch 2004, Duarte et al. 2009). The infections caused by NTM are clinically indistinguishable from those caused by M. tuberculosis, although the therapeutic regimen differs between the MTC and NTM, even among different species of NTM. Thus, early identification followed by appropriate treatment is essential for the effective control of these diseases (Griffith et al. 2007).
The traditional identification of mycobacterial species is based on an analysis of phenotypic and biochemical characteristics using methods that are labour intensive, time consuming and poorly discriminative. These methods may take up 60 days and may result in ambiguous or erroneous identification. Molecular tests have been described as a rapid, highly discriminative and easy-to-perform alternative for mycobacterial identification. However, although easy to perform, commercially available tests based on DNA probes, such as the INNO-LiPA Mycobacteria Assay (Innogenetics, Belgium), AccuProbe (Gen-Probe, USA) and the GenoType Mycobacterium Assay (Hain Lifescience, Germany), are still too expensive, especially for developing countries. Direct gene sequencing is highly specific in discriminating mycobacterial species, but requires sophisticated equipment, hampering its use in routine laboratories (Leão et al. 2005, Neonakis et al. 2008).
Telenti et al. (1993) have described a molecular method based on restriction analysis of the hsp65 gene, named PRA-hsp65, which consists of the amplification of a 441 bp fragment of the hsp65 gene, followed by restriction analysis with the enzymes BstEII and HaeIII. The interpretation of the results is performed by following an algorithm available online (app.chuv.ch/prasite). The PRA-hsp65 method is rapid and simple and has the potential to be implemented in clinical laboratories (Telenti et al. 1993, Leão et al. 2005). In fact, this method has been employed for TB diagnosis in several reference clinical laboratories in Brazil (Chimara et al. 2008).
We have previously described a polymerase chain reaction (PCR)-based method called a mycobacteria mobility shift assay (MMSA) that was designed for MTC and NTM species identification (Waléria-Aleixo et al. 2000). Mycobacterial species identification by the MMSA is based on micro-heterogeneities in nucleotide sequences of the 16S rRNA gene. These micro-heterogeneities can be evaluated based on the mobility of heteroduplexes formed between 1,027 bp DNA fragments of the 16S rRNA mycobacterial gene from a test sample and from reference mycobacteria. Heteroduplexes are formed by mixing, denaturing and re-annealing PCR-amplified DNA from the test sample with amplified DNA from the reference mycobacteria and their motilities analysed on denaturing polyacrylamide gels. Here, we compare the MMSA with the PRA-hsp65 method.
SUBJECTS, MATERIALS AND METHODS
Clinical samples - A total of 204 (102 MTC and 102 NTM) mycobacterial isolates were included for identification by the MMSA and the PRA-hsp65 method. All isolates, which were grown in Löwenstein-Jensen or liquid medium (Mycobacteria Growth Indicator Tube - MGIT®, BD), were provided by the Central Public Health Laboratory of Santa Catarina State, Florianópolis, Brazil (TB reference laboratory in the state of Santa Catarina). The phenotypic distinction between the MTC and NTM was performed by macroscopic culture analysis (morphology and pigmentation), microscopic culture analysis (cord formation by isolates in liquid medium) and an inhibition test in para-nitrobenzoic acid medium (MS 2008).
This study was approved by the Human Research Ethical Committee of the Federal University of Santa Catarina under protocol 171/2001 on 17 December 2001. The agreement was forwarded to the Brazilian National Human Research Ethical Committee.
DNA extraction - DNA extraction from bacterial cultures was performed using 5 M guanidine (Lippke 1987). Briefly, a loopful of each culture was added to 1 mL of 5 M guanidine [5 M guanidine isothiocyanate (Sigma-Aldrich, USA); 0.1 M Tris (Promega, USA), pH 6.4; 0.5 M ethylenediamine tetraacetic acid (EDTA) (Bio-Rad, USA), pH 8.0 and 1.3% Triton X-100 (Amresco, USA)] and the mixture was shaken overnight at room temperature. In total, 50 μL of silica solution (Sigma, USA) was added and after homogenisation, the pellet was washed twice in washing solution (5 M guanidine isothiocyanate and 0.1 M Tris, pH 6.4), twice in 70% ethanol (Sigma-Aldrich) and once in acetone (Sigma-Aldrich). The pellet was dried at 56ºC and rehydrated in 35 µL of Tris-EDTA buffer (10 mM Tris and 1 mM EDTA, pH 7.4) for DNA elution. The eluted DNA was stored at -20ºC until use.
Standard bacterial strains and panel of reference plasmids for MMSA - To obtain a panel of reference plasmids for the MMSA, mycobacterial DNA extracted from standard cultures was used as a template for PCRs with primers F-285/Myc-264 to amplify a 1,027 bp fragment of the mycobacterial 16S rRNA gene. The mycobacterial standard cultures used were Mycobacterium avium (ATCC25291), Mycobacterium fortuitum (ATCC6841), Mycobacterium kansasii (ATCC12478), Mycobacterium gordonae, Mycobacterium abscessus, Mycobacterium terrae, M. tuberculosis H37Rv and Bacille Calmette-Guérin (BCG). The last two indistinctly represent the MTC and have identical 16S rRNA gene sequences (Böddinghaus et al. 1990). The M. gordonae, M. abscessus and M. terrae strains were kindly supplied by the Central Public Health Laboratory of Santa Catarina State. These strains were identified in the National Reference Laboratory for TB. Before use, our group sequenced the strains. DNA from Escherichia coli (ATCC25922) was used as a negative control. The amplicons were purified (QIAquick Gel Extraction Kit, Qiagen) and cloned into the pGEMT vector (Promega). All inserts were sequenced in both directions (MegaBACE 1000, GE Healthcare, Buckinghamshire) and the sequences were analysed using BLASTN to confirm the identity of the mycobacteria (Altschul et al. 1990).
MMSA - PCR amplification of a 1,027 bp highly conserved region of the 16S rRNA gene that is present in all mycobacteria (GenBank access X58890) was performed for all 204 isolates and standards using the primers F-285 (5’-AGAGTTTGATCCTGGCTCAG-3’) (Rogall et al. 1990, Waléria-Aleixo et al. 2000) and Myc-264 (3’-TGCACACAGGCCACAAGGGA-5’) (Böddinghaus et al. 1990, Waléria-Aleixo et al. 2000). PCR assays were performed in a final volume of 50 µL using 1.25 U of Taq DNA polymerase, 1.5 mM MgCl2, 0.2 mM of each deoxynucleotide triphosphates (all from Invitrogen), 10 pmol of each primer (Alpha DNA, Canada) and 1% dimethyl sulfoxide (Sigma-Aldrich). As a positive control, 0.3 ng of the pGEMT plasmid containing the 1,027 bp of the 16S rRNA gene from BCG was used. Two negative controls were included: one with no DNA template and one containing 0.5 ng of E. coli DNA. The PCR conditions were 94ºC for 1 min, 60ºC for 1 min and 72ºC for 1 min for 35 cycles and one final elongation cycle at 72ºC for 10 min. Aliquots of the PCR products were ana- lysed by 1% agarose gel electrophoresis and visualised by ethidium bromide staining under ultraviolet (UV) light.
The MMSA was performed as previously described (Waléria-Aleixo et al. 2000), with minor modifications, as follows. First, 6 µL of the PCR product of each test sample and 6 µL of the PCR product of a reference plasmid were added to a tube containing 1.2 µL of 10X annealing buffer [1 mM NaCl (Merck, Germany); 100 mM Tris-HCl, pH 7.8 (Sigma-Aldrich) and 20 mM EDTA (Sigma-Aldrich)]. The PCR products (unknown sample x reference plasmid) were mixed to the same concentration (maximum 450 ng and minimum 50 ng). The mixture was heated to 94ºC for 3 min, cooled in an ice bath and then electrophoresed in a denaturing 3% urea plus 5% polyacrylamide gel at 200 V (between 55-100 mA) in 1X tris/borate/EDTA buffer for 55 min. Gels were stained with ethidium bromide and photographed under UV light. Possible mixed infections can be observed by performing the MMSA on a pure sample, without mixing with plasmid.
Possible contamination of the PCR mixture with the undigested plasmids used as templates can be circumvented in the clinical laboratory by adopting good laboratory practices and by observing guidelines followed by researchers using PCR. These measures include dedicated rooms for the pre-PCR activities (sample preparation and PCR preparation) and the post-PCR activities (PCR execution and analysis). Each room must have its own separate set of equipment, including pipettors, reagents, pipettor tips and racks.
PRA-hsp65 - PCR amplification of a 441 bp fragment from the hsp65 gene was performed for all isolates using the primers TB11 (5’-ACCAACGATGGTGTGTCCAT-3’) and TB12 (5’-CTTGTCGAACCGCATACCCT-3’), as described (Telenti et al. 1993). The amplicons were digested separately with 10 U of BstEII (Promega) or HaeIII (Invitrogen, USA). The resulting restriction fragments were resolved by electrophoresis in a 3% agarose gel. Gels were stained with ethidium bromide and photographed under UV light. As references for DNA sizing, 50 bp and 25 bp ladders (Invitrogen) were used. The PRA-hsp65 patterns were analysed by visual comparison with the ladders and interpreted following an algorithm available at the PRASITE (app.chuv.ch/prasite).
DNA sequencing - All 16S rRNA and hsp65 PCR products of the isolates that presented discordant results between the two methods were sequenced using a DYEnamic® ET Dye Terminator Kit (GE Healthcare) for definite species identification. The cycle sequencing conditions were 95ºC for 25 sec and 35 cycles at 95ºC for 15 sec and 60ºC for 80 sec. The sequencing products were purified by 70% isopropanol (Sigma-Aldrich) precipitation and then analysed on MegaBACE 1000 DNA Analysis System (GE Healthcare). The trace files were initially submitted to the Phred/Phrap/Consed package (phrap.org) and the sequences were analysed using BLASTN to confirm the identity of the mycobacteria.
RESULTS
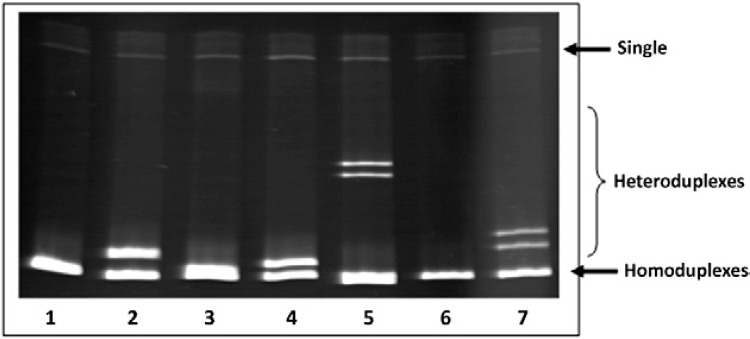
Fig. 1 shows an MMSA gel illustrating the homoduplexes formed when PCR products from the same species of mycobacterium are mixed, showing one band on the bottom of the gel (Lanes 1, 3, 6). In contrast, when there are distinct species in the mixture, heteroduplexes are formed, showing a shift in urea-PAGE that can be visualised as two or more bands, as observed in Lanes 2, 4, 5, 7.
Fig. 1. : visualisation of homoduplexes and heteroduplexes DNA bands in a mycobacteria mobility shift assay (MMSA) gel. The homoduplexes and heteroduplexes from MMSA performed with distinct mycobacterial species were resolved in a urea-polyacrylamide gel stained by ethidium bromide. Lane 1: Bacille Calmette-Guérin (BCG) only; 2: BCG + Mycobacterium avium; 3: M. avium only; 4: BCG + Mycobacterium kansasii; 5: BCG + Mycobacterium fortuitum; 6: M. fortuitum only; 7: BCG + Mycobacterium gordonae.
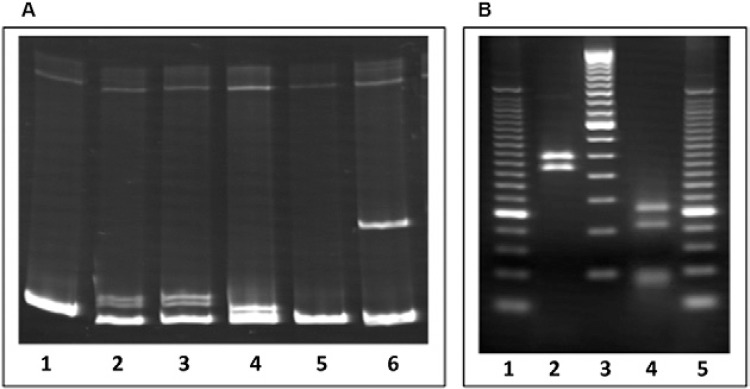
All 204 isolates were analysed by the MMSA method and by the PRA-hsp65 method. As an example, Fig. 2 shows the results for the identification of a mycobacterial culture (named ID59 here) as M. avium by both methods. In Fig. 2A, heteroduplex bands, which define non-identity between samples, are observed in Lanes 2-4, 6, in which ID59 was mixed with BCG, M. kansasii, M. fortuitum and M. gordonae, respectively. The mixture of ID59 plus M. avium formed a homoduplex band (Lane 5), indicative of a perfect match between these DNA sequences. Fig. 2B illustrates the restriction fragments of the hsp65 gene obtained using BstEII and HaeIII and analysis also identified this isolate as M. avium.
Fig. 2. : identification of a clinical sample (named ID59) as Mycobacterium avium by mycobacteria mobility shift assay (MMSA) (A) and PRA-hsp65 (B). A Lane 1: ID59 only; 2: ID59 + Bacille Calmette-Guérin; 3: ID59 + Mycobacterium kansasii; 4: ID59 + Mycobacterium gordonae; 5: ID59 + M. avium; 6: ID59 + Mycobacterium fortuitum; B Lane 1: 25 bp DNA ladder; 2: BstEII restriction fragments of hsp65 gene (235 bp, 210 bp, 0 bp); 3: 50 bp DNA ladder; 4: HaeIII restriction fragments of hsp65 gene (130 bp, 105 bp, 0 bp); 5: 25 bp DNA ladder.
Comparing the results obtained by the MMSA and PRA-hsp65 methods, we verified that both methods correctly identified all 102 MTC isolates (100% agreement).
Among the 102 NTM isolates, the MMSA method alone assigned 94 isolates (92.2%) to a complex, group or species, whereas the PRA-hsp65 method assigned 102 isolates (100%) to a species. Considering the 94 NTM isolates identified by the MMSA and PRA-hsp65, we observed 91.5% (86/94) agreement and these identifications were presumed to be correct (Table I).
TABLE I. Species identification of 86 isolates of nontuberculous mycobacteria with concordant results by both mycobacteria mobility shift assay (MMSA) and PRA-hsp65 methods.
| PRA-hsp65 | MMSA | Isolates n (%) |
|---|---|---|
| Mycobacterium avium | M. avium complex | 39 (45.3) |
| Mycobacterium intracellulare | M. avium complex | 6 (7) |
| Mycobacterium abscessus | M. chelonae-abscessus group | 13 (15.2) |
| Mycobacterium chelonae | M. chelonae-abscessus group | 3 (3.5) |
| Mycobacterium kansasii | M. kansasii | 6 (7) |
| Mycobacterium fortuitum | M. fortuitum group | 11 (12.8) |
| Mycobacterium porcinum/Mycobacterium peregrinum/Mycobacterium septicum | M. fortuitum group | 4 (4.6) |
| Mycobacterium gordonae | M. gordonae | 4 (4.6) |
Regarding the eight samples with inconclusive identification by the MMSA (Mycobacterium hiberniae, M. terrae, Mycobacterium szulgai, Mycobacterium kubicae and Mycobacterium simiae), it is fair to mention that none of these mycobacterial DNAs was included in our panel of reference plasmids for the MMSA.
Among the eight isolates with discordant identification, none of them presented a profile of mixed infection. DNA sequencing confirmed the MMSA results in five cases and the PRA-hsp65 results in three cases (Table II). Thus, the MMSA provided correct species identification of 96.8% of isolates identified by both methods (91/94) and PRA-hsp65 provided correct identification for 95% (97/102), with no significant statistical difference between these methods (p = 0.847, Fisher’s exact test).
TABLE II. Species identification by DNA sequencing of eight isolates with discordant results between mycobacteria mobility shift assay (MMSA) and PRA-hsp65 methods.
| Sample | PRA-hsp65 | MMSA | Sequencing | Correct method |
|---|---|---|---|---|
| ID07 | Mycobacterium bohemicum | Mycobacterium fortuitum complex | M. bohemicum | PRA-hsp65 |
| ID12 | Mycobacterium avium | Mycobacterium kansasii | M. kansasii | MMSA |
| ID24 | Mycobacterium szulgai | Mycobacterium terrae complex | M. szulgai | PRA-hsp65 |
| ID37 | M. bohemicum | M. fortuitum complex | Mycobacterium peregrinum | MMSA |
| ID45 | M. avium | M. kansasii | M. avium | PRA-hsp65 |
| ID54 | M. avium | M. fortuitum complex | M. fortuitum | MMSA |
| ID76 | M. bohemicum | M. fortuitum complex | M. peregrinum | MMSA |
| ID80 | M. avium | M. kansasii | M. kansasii | MMSA |
The NTM most commonly associated with clinically significant disease in Brazil (M. avium complex, M. fortuitum group, Mycobacterium chelonae-abscessus group and M. kansasii) represented 86.3% of the 102 NTM isolates in this study. The MMSA provided correct identification for 98.9% of these samples and PRA-hsp65 correctly identified 94.3% (p = 0.943, Fisher’s exact test). All of the inconclusive results provided by the MMSA corresponded to other species.
DISCUSSION
It is well known that the mycobacterial treatment protocol depends on the species isolated. Despite a considerable increase in the number of cases of and in knowledge about NTM infections, these infections still represent a diagnostic and therapeutic challenge (Griffith et al. 2007, Piersimoni & Scarparo 2009). Furthermore, although well recognised as agents of mycobacterial diseases in developed countries, the role of NTM in developing countries is not clear. Because TB is life threatening and usually endemic in these areas, patients are presumptively treated for TB. This strategy may be attributed to the limited resources and availability of laboratory facilities to culture and identify mycobacteria (Gopinath & Singh 2010, Satyanarayana et al. 2011, Maiga et al. 2012). Despite the advanced techniques available for mycobacterial identification, such as DNA sequencing and commercially available probe-based tests, these methods are still too expensive to be used routinely in Brazilian public laboratories. Thus, it is necessary to develop simple, rapid, low-cost and reliable methods that can be implemented in routine laboratories (Leão et al. 2005). Currently, reference laboratories in Brazil are trained to perform PRA-hsp65, but have the technical capacity to perform other methodologies in house.
The MMSA is a simple method to perform; does not require expensive and sophisticated equipment and, more importantly, enables a significant decrease in the turnaround time for the identification of mycobacterial species because identification can be achieved in only 2 h after PCR amplification. When compared with PRA-hsp65, the MMSA proved to be a rapid and reliable method to identify the MTC and NTM. This method provided correct species identification for 96.8% of isolates identified by both methods and PRA-hsp65 provided correct identification for 95%. All of the inconclusive results provided by the MMSA corresponded to species with low clinical significance that were not included as reference plasmids in the MMSA experiments, as previously mentioned.
The 16S rRNA gene sequences of the samples with incorrect identification by the MMSA were aligned with the reference sequences used in this method. When aligned with the M. fortuitum reference sequence used in the MMSA, ID07 showed 4.82% non-identity with this species. When aligned with the M. terrae reference sequence, ID24 (M. szulgai) showed 4% non-identity, whereas ID45 (M. avium) showed 1.8% non-identity with M. kansasii. Thus, the incorrect identifications provided by the MMSA were related to species that showed similarity greater than 95%.
Despite the large number of mycobacterial species described so far, their clinical significance still needs to be assessed (Esfahani et al. 2012, Shi et al. 2012). The main NTM species associated with clinically significant disease in Brazil are M. avium, Mycobacterium intracellulare, M. fortuitum, M. kansasii, M. abscessus and M. chelonae (Chimara et al. 2008, Pedro et al. 2008). When these species were evaluated in the present study, the MMSA provided correct identification for 98.9% of the samples. Another Brazilian study identified less typical NTM as a cause of lung disease, although these species were found at a minor frequency (de Lima et al. 2013).
Because different reports of mycobacterial species might be related to different geographical conditions and studied populations, the identification of other species could be important to specific areas. For example, Mycobacterium xenopi and Mycobacterium malmoense are commonly described in Europe (Grubek-Jaworska et al. 2009, Andréjak et al. 2010). Although the MMSA performed here was able to identify seven species and complexes (M. tuberculosis complex, M. avium complex, M. fortuitum group, M. kansasii, M. gordonae, M. terrae complex and M. abscessus group), this limitation can be overcome with the standardisation of other mycobacterial species as standards in the reaction. Indeed, the plasticity of the MMSA method for mycobacterial identification is remarkable.
PRA-hsp65 correctly identified 95% of the 102 NTM isolates. Similar results were reported by other studies that evaluated this method (Häfner et al. 2004, Chimara et al. 2008). The PRA-hsp65 method is rapid and simple to perform and the main drawbacks of its use are generally related to electrophoresis conditions and to the interpretation of band sizes when species present similar patterns (Leão et al. 2005). In the present study, all samples that were incorrectly identified by PRA-hsp65 were reviewed and the same patterns were observed. Moreover, it is possible to find undescribed hsp65 patterns. This method has been improved since its initial development and the revised algorithms allow the identification of more mycobacterial species (Telenti et al. 1993, da Silva-Rocha et al. 2002, Chimara et al. 2008). The MMSA also needs a degree of standardisation in electrophoresis conditions, but its simple band patterns are easy to interpret and no algorithm is required to differentiate between species. Furthermore, the use of restriction enzymes is not necessary.
The MMSA is not able to distinguish species of M. avium complex, M. abscessus group and M. fortuitum group. Mycobacterial species identification by the MMSA is based on micro-heterogeneities in 16S rRNA nucleotide sequences, but does not have sufficient sensitivity to detect differences between closely related species. The accurate identification of mycobacteria is becoming more complex, mainly because of the increase in the number of described species. This number is still increasing with the development of highly discriminatory techniques, such as DNA sequencing (Leão et al. 2005). Regarding clinical management, species belonging to the same complex require the same therapeutic regimen, except for M. abscessus and M. chelonae (Griffith et al. 2007). Therefore, despite its relatively low power to discriminate between closely related species, the MMSA could help clinicians in making therapeutic decisions about related diseases. The MMSA may be used as a screening assay and if necessary, NTM could further analysed by PRA-hsp65 or sequencing. Preliminary data showed that the MMSA also has the potential to identify mycobacteria directly from a clinical sample (data not shown). Indeed, this is an important feature of the MMSA because it is not necessary to isolate the bacteria to perform the test, which significantly reduces the turnaround time for mycobacterial identification.
Our results indicate that the MMSA is a reliable method for the identification of mycobacteria in culture isolates. Despite its relatively low discriminatory power, this disadvantage is compensated for by the clinical relevance of the MMSA’s results and the overall simplicity of the technique. We believe that this method has great potential to be implemented as a screening assay in diagnostic laboratories, mainly in TB-endemic areas, to assess the real role of NTM in these areas.
ACKNOWLEDGEMENTS
To the PDTIS-Fiocruz, for use of its facilities.
Footnotes
Financial support: CNPq
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andréjak C, Thomsen VO, Johansen S, Riis A, Benfiels TL, Duhaut P, Sorensen HT, Lescure F-X, Thomsen RW. Nontuberculous pulmonary mycobacteriosis in Denmark: incidence and prognostic factors. Am J Respir Crit Care Med. 2010;181:514–521. doi: 10.1164/rccm.200905-0778OC. [DOI] [PubMed] [Google Scholar]
- Böddinghaus B, Rogall T, Florhr T, Blocker H, Bottger EC. Detection and identification of mycobacteria by amplification of rRNA. J Clin Microbiol. 1990;28:1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimara E, Ferrazoli L, Ueky SY, Martins MC, Durham AM, Arbeite D, Leão SC. Reliable identification of mycobacterial species by PCR-restriction enzyme analysis (PRA)-hsp65 in reference laboratory and elaboration of a sequence-based extended algorithm of PRA-hsp65 patterns. BMC Microbiol. 2008;8:1–12. doi: 10.1186/1471-2180-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Rocha A, Werneck-Barreto AM, Dias-Campos CE, Silva M, Fonseca LV-B, Saad MH, Degrave WM, Suffys PN. Novel allelic variants of mycobacteria isolated in Brazil as determined by PCR-restriction enzyme analysis of hsp65. J Clin Microbiol. 2002;40:4191–4196. doi: 10.1128/JCM.40.11.4191-4196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima CAM, Gomes HM, Oelemann MAC, Ramos JP, Caldas PC, Campos CED, Pereira MAS, Montes FFO, Oliveira MSC, Suffys PN, Moura MMF. Nontuberculous mycobacteria in respiratory samples from patients with pulmonary tuberculosis in the state of Rondônia, Brazil. Mem Inst Oswaldo Cruz. 2013;108:457–462. doi: 10.1590/0074-0276108042013010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte RS, Lourenço MC, Fonseca LS, Leão SC, Amorim L, Rocha IL, Coelho FS, Viana-Niero C, Gomes KM, Silva MG, Lorena NS, Pitombo MB, Ferreira RM, Garcia MH, Oliveira GP, Lupi O, Vilaça BR, Serradas LR, Chebabo A, Marques EA, Teixeira LM, Dalcolmo M, Senna SG, Sampaio JL. Epidemic postsurgical infections caused by Mycobacterium massiliense. J Clin Microbiol. 2009;47:2149–2155. doi: 10.1128/JCM.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esfahani BN, Yazdi HR, Moghim S, Safaei HG, Esfahani HZ. Rapid and accurate identification of Mycobacterium tuberculosis complex and common non-tuberculous mycobacteria by multiplex real-time PCR targeting different housekeeping genes. Curr Microbiol. 2012;65:403–499. doi: 10.1007/s00284-012-0188-2. [DOI] [PubMed] [Google Scholar]
- Euzéby JP. List of prokaryotic names with standing in nomenclature. 2012 bacterio.cict.fr
- Falkinham JO. Surrounded by mycobacteria: nontuberculous mycobacteria in the human environment. J Appl Microbiol. 2009;107:356–367. doi: 10.1111/j.1365-2672.2009.04161.x. [DOI] [PubMed] [Google Scholar]
- Gopinath K, Singh S. Non-tuberculous mycobacteria in TB-endemic countries: are we neglecting the danger? e615PLoS Negl Trop Dis. 2010;4 doi: 10.1371/journal.pntd.0000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Hiott G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ, Winthrop K. An official ATS/IDSA statement: diagnosis, treatment and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- Grubek-Jaworska H, Walkiewicz R, Safianowska A, Nowacka-Mazurek M, Krenke R, Przybylowski T, Chazan R. Nontuberculous mycobacterial infections among patients suspected of pulmonary tuberculosis. Eur J Clin Microbiol Infect Dis. 2009;28:739–744. doi: 10.1007/s10096-008-0694-0. [DOI] [PubMed] [Google Scholar]
- Häfner B, Haag H, Geiss H-K, Nolte O. Different molecular methods for the identification of rarely isolated non-tuberculous mycobacteria and description of new hsp65 restriction fragment length polymorphism patterns 2004. Mol Cell Probes. 2004;18:59–65. doi: 10.1016/j.mcp.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Katoch VM. Infections due to non-tuberculous mycobacteria (NTM) Indian J Med Res. 2004;120:290–304. [PubMed] [Google Scholar]
- Leão SC, Bernardelli A, Cataldi A, Zumarraga M, Robledo J, Realpe T, Mejía GI, Telles MAS, Chimara E, Velazco M, Fernendez J, Rodrigues PA, Guerrero MI, León CI, Porras TB, Rastogi N, Goh KS, Suffys P, Rocha AS, Santos-Netto D, Ritacco V, López B, Barrera L, Palomino JC, Martin A, Portaels F. Multicenter evaluation of mycobacteria identification by PCR restriction enzyme analysis in laboratories from Latin America and the Caribbean. J Microbiol Methods. 2005;61:193–199. doi: 10.1016/j.mimet.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Lippke JA, Strzempko MN, Raia FF, Simon SL. Isolation of intact high-molecular-weight DNA by using guanidine isothiocyanate. Appl Environ Microbiol. 1987;53:2588–2589. doi: 10.1128/aem.53.10.2588-2589.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiga M, Siddiqui S, Diallo S, Diarra B, Traoré B, Shea YR, Zelazny AM, Dembele BPP, Goita D, Kassambara H, Hammond AS, Polis MA, Tounkara A. Failure to recognize nontuberculous mycobacteria leads to misdiagnosis of chronic pulmonary tuberculosis. e36902PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0036902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MS - Ministério da Saúde Brasil Manual nacional de vigilância laboratorial da tuberculose e outras micobactérias. 1. MS/Secretaria de Vigilância em Saúde/ Departamento de Vigilância Epidemiológica; Brasília: 2008. 436 [Google Scholar]
- Neonakis IK, Gitti Z, Kramboviti E, Spandidos DA. Molecular diagnostic tools in mycobacteriology. J Microbiol Methods. 2008;75:1–11. doi: 10.1016/j.mimet.2008.05.023. [DOI] [PubMed] [Google Scholar]
- Pedro HSP, Pereira MIF, Goloni MRA, Ueki SYM, Chimara E. Isolamento de micobactérias não-tuberculosas em São José do Rio Preto entre 1996 e 2005. J Bras Pneumol. 2008;34:950–955. doi: 10.1590/s1806-37132008001100010. [DOI] [PubMed] [Google Scholar]
- Piersimoni C, Scarparo C. Extrapulmonary infections associated with nontuberculous mycobacteria in immunocompetent persons. Emerg Infect Dis. 2009;15:1351–1358. doi: 10.3201/eid1509.081259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogall T, Wolters J, Flohr T, Bottger EC. Towards a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int J Syst Bacteriol. 1990;40:323–330. doi: 10.1099/00207713-40-4-323. [DOI] [PubMed] [Google Scholar]
- Satyanarayana G, Heysell SK, Scully KW, Houpt ER. Mycobacterial infections in a large Virginia hospital, 2001-2009. BMC Infect Dis. 2011;11:113–117. doi: 10.1186/1471-2334-11-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi XC, Liu XQ, Xie XL, Xu YC, Zhao ZX. Gene chip array for differentiation of mycobacterial species and detection of drug resistance. Chin Med J. 2012;125:3292–3297. [PubMed] [Google Scholar]
- Telenti A, Marchesi F, Balz M, Bally F, Böttger T. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waléria-Aleixo A, Kroon EG, Campos MAS, Margutti-Pinto ME, Bonjardim CA, Ferreira PCP. Heteroduplex mobility assay for rapid, sensitive and specific detection of mycobacteria. Diagn Microbiol Infect Dis. 2000;36:225–235. doi: 10.1016/s0732-8893(00)00112-7. [DOI] [PubMed] [Google Scholar]
- WHO - World Health Organization Global tuberculosis report 2012. WHO; Geneva: 2012. 98 [Google Scholar]


