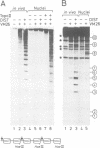
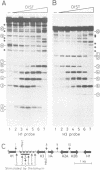
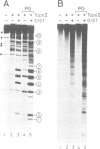
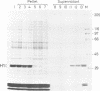
Abstract
Histone H1 preferentially and cooperatively binds scaffold-associated regions (SARs) in vitro via specific interactions with the numerous short A + T-rich tracts (A-tracts) contained in these sequences. Selective titration of A-tracts by the oligopeptide distamycin abolishes this interaction and results in a redistribution of H1. Similarly, treatment of intact cells and isolated nuclei with distamycin specifically enhances cleavage of internucleosomal linkers of SARs by topoisomerase II and restriction enzymes. The increased accessibility of these linkers is thought to result from the unfolding (or opening) of the chromatin fiber and to be due to a reduced occupancy by histone H1. Chromatin extraction and H1 assembly experiments support this view. We discuss a model whereby open, H1-depleted chromatin regions may be generated by titration of A-tracts by putative distamycin analogues; this local opening may spread to adjacent regions assuming highly cooperative H1-H1 interactions in chromatin.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi Y., Käs E., Laemmli U. K. Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J. 1989 Dec 20;8(13):3997–4006. doi: 10.1002/j.1460-2075.1989.tb08582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi Y., Luke M., Laemmli U. K. Chromosome assembly in vitro: topoisomerase II is required for condensation. Cell. 1991 Jan 11;64(1):137–148. doi: 10.1016/0092-8674(91)90215-k. [DOI] [PubMed] [Google Scholar]
- Bond B. J., Davidson N. The Drosophila melanogaster actin 5C gene uses two transcription initiation sites and three polyadenylation sites to express multiple mRNA species. Mol Cell Biol. 1986 Jun;6(6):2080–2088. doi: 10.1128/mcb.6.6.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutlag D. L. Molecular arrangement and evolution of heterochromatic DNA. Annu Rev Genet. 1980;14:121–144. doi: 10.1146/annurev.ge.14.120180.001005. [DOI] [PubMed] [Google Scholar]
- Clark D. J., Thomas J. O. Salt-dependent co-operative interaction of histone H1 with linear DNA. J Mol Biol. 1986 Feb 20;187(4):569–580. doi: 10.1016/0022-2836(86)90335-9. [DOI] [PubMed] [Google Scholar]
- Cockerill P. N., Garrard W. T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986 Jan 31;44(2):273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- De Bernardin W., Losa R., Koller T. Formation and characterization of soluble complexes of histone H1 with supercoiled DNA. J Mol Biol. 1986 Jun 5;189(3):503–517. doi: 10.1016/0022-2836(86)90320-7. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. The formation and function of DNase I hypersensitive sites in the process of gene activation. J Biol Chem. 1988 Dec 25;263(36):19259–19262. [PubMed] [Google Scholar]
- Ericsson C., Grossbach U., Björkroth B., Daneholt B. Presence of histone H1 on an active Balbiani ring gene. Cell. 1990 Jan 12;60(1):73–83. doi: 10.1016/0092-8674(90)90717-s. [DOI] [PubMed] [Google Scholar]
- Fox K. R., Howarth N. R. Investigations into the sequence-selective binding of mithramycin and related ligands to DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8695–8714. doi: 10.1093/nar/13.24.8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. R., Waring M. J. DNA structural variations produced by actinomycin and distamycin as revealed by DNAase I footprinting. Nucleic Acids Res. 1984 Dec 21;12(24):9271–9285. doi: 10.1093/nar/12.24.9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X. L., Patel D. J. Solution structure of the chromomycin-DNA complex. Biochemistry. 1989 Jan 24;28(2):751–762. doi: 10.1021/bi00428a051. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986 Aug 15;46(4):521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laemmli U. K. Improved methods for the isolation of individual and clustered mitotic chromosomes. Exp Cell Res. 1987 Nov;173(1):85–98. doi: 10.1016/0014-4827(87)90334-x. [DOI] [PubMed] [Google Scholar]
- Gasser S. M., Laroche T., Falquet J., Boy de la Tour E., Laemmli U. K. Metaphase chromosome structure. Involvement of topoisomerase II. J Mol Biol. 1986 Apr 20;188(4):613–629. doi: 10.1016/s0022-2836(86)80010-9. [DOI] [PubMed] [Google Scholar]
- Griffith J., Bleyman M., Rauch C. A., Kitchin P. A., Englund P. T. Visualization of the bent helix in kinetoplast DNA by electron microscopy. Cell. 1986 Aug 29;46(5):717–724. doi: 10.1016/0092-8674(86)90347-8. [DOI] [PubMed] [Google Scholar]
- Gross D. S., Garrard W. T. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Nucleosomes: regulators of transcription. Trends Genet. 1990 Dec;6(12):395–400. doi: 10.1016/0168-9525(90)90299-l. [DOI] [PubMed] [Google Scholar]
- Hsieh T., Brutlag D. Sequence and sequence variation within the 1.688 g/cm3 satellite DNA of Drosophila melanogaster. J Mol Biol. 1979 Dec 5;135(2):465–481. doi: 10.1016/0022-2836(79)90447-9. [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Käs E., Laemmli U. K. Highly preferential nucleation of histone H1 assembly on scaffold-associated regions. J Mol Biol. 1989 Dec 5;210(3):573–585. doi: 10.1016/0022-2836(89)90133-2. [DOI] [PubMed] [Google Scholar]
- Johnson K. R., Lehn D. A., Reeves R. Alternative processing of mRNAs encoding mammalian chromosomal high-mobility-group proteins HMG-I and HMG-Y. Mol Cell Biol. 1989 May;9(5):2114–2123. doi: 10.1128/mcb.9.5.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakaka R. T., Thomas J. O. Chromatin structure of transcriptionally competent and repressed genes. EMBO J. 1990 Dec;9(12):3997–4006. doi: 10.1002/j.1460-2075.1990.tb07621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Käs E., Izaurralde E., Laemmli U. K. Specific inhibition of DNA binding to nuclear scaffolds and histone H1 by distamycin. The role of oligo(dA).oligo(dT) tracts. J Mol Biol. 1989 Dec 5;210(3):587–599. doi: 10.1016/0022-2836(89)90134-4. [DOI] [PubMed] [Google Scholar]
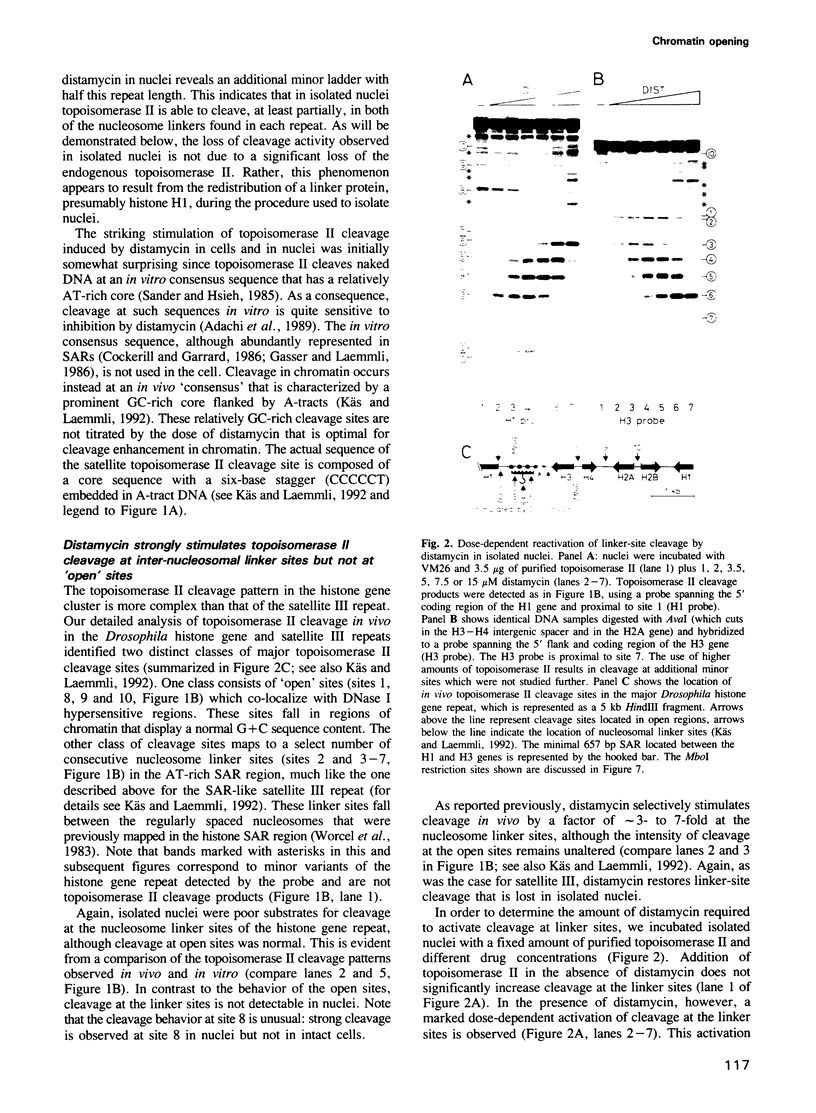
- Käs E., Laemmli U. K. In vivo topoisomerase II cleavage of the Drosophila histone and satellite III repeats: DNA sequence and structural characteristics. EMBO J. 1992 Feb;11(2):705–716. doi: 10.1002/j.1460-2075.1992.tb05103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künzler P., Stein A. Histone H5 can increase the internucleosome spacing in dinucleosomes to nativelike values. Biochemistry. 1983 Apr 12;22(8):1783–1789. doi: 10.1021/bi00277a007. [DOI] [PubMed] [Google Scholar]
- Lennard A. C., Thomas J. O. The arrangement of H5 molecules in extended and condensed chicken erythrocyte chromatin. EMBO J. 1985 Dec 16;4(13A):3455–3462. doi: 10.1002/j.1460-2075.1985.tb04104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinger L., Varshavsky A. Protein D1 preferentially binds A + T-rich DNA in vitro and is a component of Drosophila melanogaster nucleosomes containing A + T-rich satellite DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7152–7156. doi: 10.1073/pnas.79.23.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis C. D., Laemmli U. K. Higher order metaphase chromosome structure: evidence for metalloprotein interactions. Cell. 1982 May;29(1):171–181. doi: 10.1016/0092-8674(82)90101-5. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Gasser S. M., Laemmli U. K. Scaffold attachment of DNA loops in metaphase chromosomes. J Mol Biol. 1988 Mar 5;200(1):101–109. doi: 10.1016/0022-2836(88)90336-1. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M. E., Laemmli U. K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984 Nov;39(1):223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Radic M. Z., Lundgren K., Hamkalo B. A. Curvature of mouse satellite DNA and condensation of heterochromatin. Cell. 1987 Sep 25;50(7):1101–1108. doi: 10.1016/0092-8674(87)90176-0. [DOI] [PubMed] [Google Scholar]
- Reeves R., Nissen M. S. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem. 1990 May 25;265(15):8573–8582. [PubMed] [Google Scholar]
- Renz M. Preferential and cooperative binding of histone I to chromosomal mammalian DNA. Proc Natl Acad Sci U S A. 1975 Feb;72(2):733–736. doi: 10.1073/pnas.72.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M., Hsieh T. S. Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985 Feb 25;13(4):1057–1072. doi: 10.1093/nar/13.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
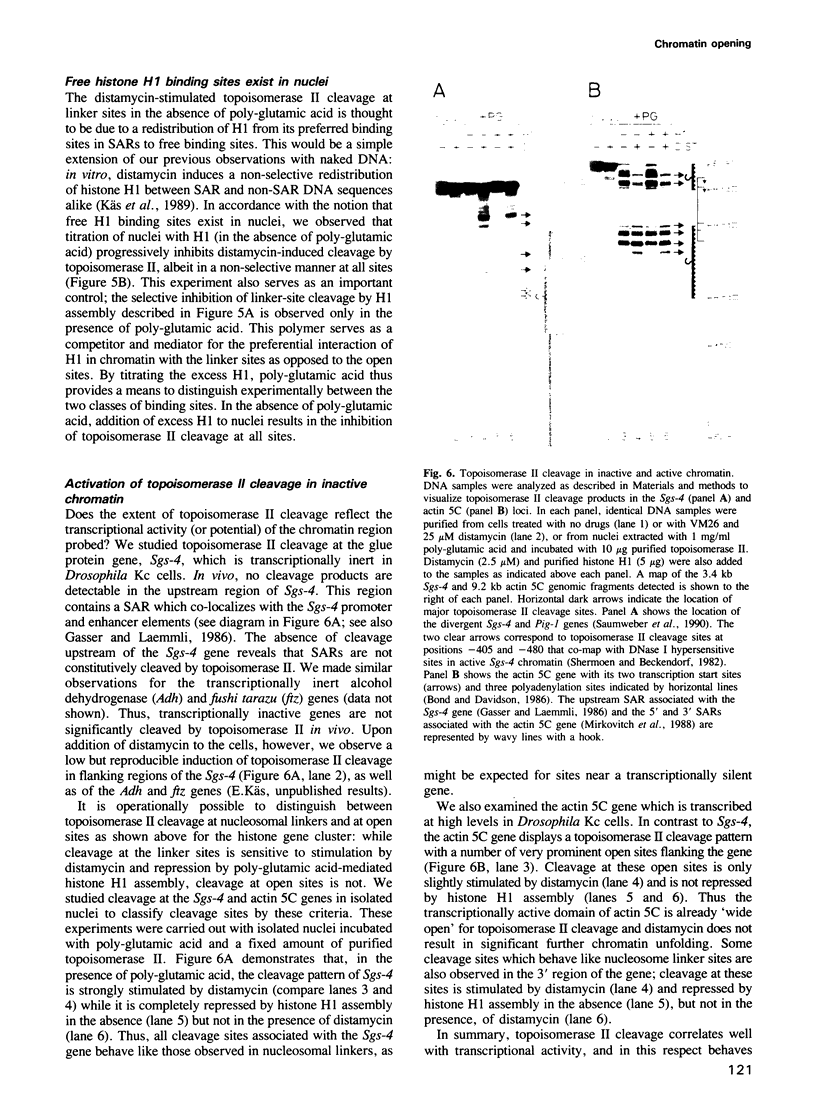
- Saumweber H., Frasch M., Korge G. Two puff-specific proteins bind within the 2.5 kb upstream region of the Drosophila melanogaster Sgs-4 gene. Chromosoma. 1990 Apr;99(1):52–60. doi: 10.1007/BF01737289. [DOI] [PubMed] [Google Scholar]
- Schlissel M. S., Brown D. D. The transcriptional regulation of Xenopus 5s RNA genes in chromatin: the roles of active stable transcription complexes and histone H1. Cell. 1984 Jul;37(3):903–913. doi: 10.1016/0092-8674(84)90425-2. [DOI] [PubMed] [Google Scholar]
- Sen D., Crothers D. M. Influence of DNA-binding drugs on chromatin condensation. Biochemistry. 1986 Apr 8;25(7):1503–1509. doi: 10.1021/bi00355a005. [DOI] [PubMed] [Google Scholar]
- Shermoen A. W., Beckendorf S. K. A complex of interacting DNAase I-hypersensitive sites near the Drosophila glue protein gene, Sgs4. Cell. 1982 Jun;29(2):601–607. doi: 10.1016/0092-8674(82)90176-3. [DOI] [PubMed] [Google Scholar]
- Shimamura A., Sapp M., Rodriguez-Campos A., Worcel A. Histone H1 represses transcription from minichromosomes assembled in vitro. Mol Cell Biol. 1989 Dec;9(12):5573–5584. doi: 10.1128/mcb.9.12.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A., Künzler P. Histone H5 can correctly align randomly arranged nucleosomes in a defined in vitro system. Nature. 1983 Apr 7;302(5908):548–550. doi: 10.1038/302548a0. [DOI] [PubMed] [Google Scholar]
- Stein A., Mitchell M. Generation of different nucleosome spacing periodicities in vitro. Possible origin of cell type specificity. J Mol Biol. 1988 Oct 20;203(4):1029–1043. doi: 10.1016/0022-2836(88)90127-1. [DOI] [PubMed] [Google Scholar]
- Strauss F., Varshavsky A. A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell. 1984 Jul;37(3):889–901. doi: 10.1016/0092-8674(84)90424-0. [DOI] [PubMed] [Google Scholar]
- Tazi J., Bird A. Alternative chromatin structure at CpG islands. Cell. 1990 Mar 23;60(6):909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Khabaza A. J. Cross-linking of histone H1 in chromatin. Eur J Biochem. 1980 Dec;112(3):501–511. doi: 10.1111/j.1432-1033.1980.tb06113.x. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Chromomycin, mithramycin, and olivomycin binding sites on heterogeneous deoxyribonucleic acid. Footprinting with (methidiumpropyl-EDTA)iron(II). Biochemistry. 1983 May 10;22(10):2373–2377. doi: 10.1021/bi00279a011. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. Assembly and propagation of repressed and depressed chromosomal states. Cell. 1985 Oct;42(3):705–711. doi: 10.1016/0092-8674(85)90267-3. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Winter E., Varshavsky A. A DNA binding protein that recognizes oligo(dA).oligo(dT) tracts. EMBO J. 1989 Jun;8(6):1867–1877. doi: 10.1002/j.1460-2075.1989.tb03583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe A. P. Dominant and specific repression of Xenopus oocyte 5S RNA genes and satellite I DNA by histone H1. EMBO J. 1989 Feb;8(2):527–537. doi: 10.1002/j.1460-2075.1989.tb03407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worcel A., Gargiulo G., Jessee B., Udvardy A., Louis C., Schedl P. Chromatin fine structure of the histone gene complex of Drosophila melanogaster. Nucleic Acids Res. 1983 Jan 25;11(2):421–439. doi: 10.1093/nar/11.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Zlatanova J. Histone H1 and the regulation of transcription of eukaryotic genes. Trends Biochem Sci. 1990 Jul;15(7):273–276. doi: 10.1016/0968-0004(90)90053-e. [DOI] [PubMed] [Google Scholar]