Abstract
Association mapping can quickly and efficiently dissect complex agronomic traits. Rapeseed is one of the most economically important polyploid oil crops, although its genome sequence is not yet published. In this study, a recently developed 60K Brassica Infinium® SNP array was used to analyse an association panel with 472 accessions. The single-nucleotide polymorphisms (SNPs) of the array were in silico mapped using ‘pseudomolecules’ representative of the genome of rapeseed to establish their hypothetical order and to perform association mapping of seed weight and seed quality. As a result, two significant associations on A8 and C3 of Brassica napus were detected for erucic acid content, and the peak SNPs were found to be only 233 and 128 kb away from the key genes BnaA.FAE1 and BnaC.FAE1. BnaA.FAE1 was also identified to be significantly associated with the oil content. Orthologues of Arabidopsis thaliana HAG1 were identified close to four clusters of SNPs associated with glucosinolate content on A9, C2, C7 and C9. For seed weight, we detected two association signals on A7 and A9, which were consistent with previous studies of quantitative trait loci mapping. The results indicate that our association mapping approach is suitable for fine mapping of the complex traits in rapeseed.
Keywords: Brassica napus, association mapping, SNP, seed quality, seed weight
1. Introduction
Rapeseed (Brassica napus L.; AACC, 2n = 38) is the one of the most important oil crops worldwide. Brassica napus is a recent allopolyploid species derived from natural interspecies hybridization between two phylogenetically close species, B. rapa (AA, 2n = 20) and B. oleracea (CC, 2n = 18) <10 000 years ago.1,2 Rapeseed has a short domestication history of only ∼400–500 years.3–5 During the breeding history of rapeseed, the most outstanding event is the introduction of two traits, zero seed erucic acid and low seed glucosinolate content (so called double-low, canola quality 00). However, many traits such as oil content, seed yield, disease resistance, etc. urgently need to be further improved in the modern cultivars. Molecular design breeding is currently one of the most available breeding methods. Screening-specific materials with some desired traits in a comparatively genetically diverse source of germplasm including old genotypes with high levels of erucic acid and seed glucosinolates and discovering the advantageous allelic variants by genetic analysis may advance molecular design breeding.
Genetic mapping of quantitative trait loci (QTL) in rapeseed is well established and has been employed to localize QTLs for quantitative traits, such as oil content,6 glucosinolate content,7 fatty acid composition of the seed oil,8 flowering time,9 yield components,10–13 etc. In all these studies, although QTL mapping is very successful in detecting QTL, the genetic variation in the mapping population is restricted to only the two parents, and the markers for detected QTL are not necessarily transferable to other materials. Furthermore, the ability of fine mapping a QTL in these studies is limited by the frequency of polymorphic loci between the two parents and requires a population consisting of several thousands of individuals, making it laborious and time-consuming to perform. There is, so far, no report on map-based cloning of a causal gene in a QTL of rapeseed.
Association mapping, also called linkage disequilibrium (LD) mapping, which directly studies statistical associations between genetic markers and phenotypes in natural populations, is an alternative to QTL mapping.14 An association mapping study utilizes the higher number of historical recombination events that have occurred throughout the entire evolutionary history of the mapping population, allowing fine-scale QTL mapping.14,15 With the rapid developments in genomics and dramatically decreasing cost of genotyping technology, association mapping has rapidly became a promising approach for the genetic dissection of complex traits. For a species with available genome sequences such as Arabidopsis thaliana, rice, maize, etc., genome-wide association studies (GWASs) have contributed to revealing rich genetic architectures of complex traits.16–21
Since rapeseed contains two homologous but divergent subgenomes A and C, which were shaped by whole-genome triplication followed by extensive diploidization,22,23 its genome structure is complex, which has hindered genomic research and high throughput discovery of high-quality molecular markers. The population structure, LD and association mapping in rapeseed were studied using amplified fragment length polymorphism and simple sequence repeat markers.24–29 However, due to the limited number of DNA markers and low-efficient genotyping technologies, a small number of lines or markers were used in these studies. Recent advances in sequencing and computational technology have enabled the discovery and efficient assay of large numbers of single-nucleotide polymorphism (SNP) markers in rapeseed.30,31 Delourme et al.32 analysed the genetic diversity and LD of a rapeseed collection of 313 inbred lines from different geographical origins using >4300 SNPs that were localized on an integrated map of rapeseed. The genomic research of Brassica genera has developed rapidly in recent years. The Brassica A genome sequence from B. rapa has been published,33 and Brassica C genome sequencing from B. oleracea has also been completed and will be published soon (http://www.ocri-genomics.org/bolbase/). Owing to the complicated genome of B. napus, genome sequencing has not yet been completed. Based on a high-density (∼21 K) SNP linkage map of rapeseed,31 Harper et al.34 refined the order and orientation of the A and C genome sequence scaffolds, constructed pseudomolecules representative of the 19 chromosomes of B. napus and successfully carried out associative transcriptomics of glucosinolate and erucic acid content in a population of 53 B. napus lines.
In this study, we genotyped a panel of 472 rapeseed accessions from all over the world using a 60K Brassica Infinium® SNP array recently developed by the international Brassica Illumina SNP consortium. By in silico mapping of the SNPs of the array to ‘pseudomolecules’ representative of the B. napus genome to obtain their hypothetical position, we performed a GWAS of seed weight and seed quality in B. napus. The SNPs significantly associated with traits were identified, and the candidate genes were confirmed or predicted.
2. Materials and methods
2.1. Plant materials
A set of 472 rapeseed inbred lines collected from the National Mid-term Gene Bank for Oil Crops of China were used for association analysis. According to the information from the gene bank and our own observations, the accessions were assigned to five different germplasm types, i.e. winter oilseed rape (OSR) (160), semi-winter OSR (200), spring OSR (110), spring fodder (1) and winter fodder (1). Based on their origins, 266 accessions originated from Asia, 128 from Western Europe, 20 from Oceania, 26 from North America and 32 from Eastern Europe (Supplementary Table S1). For SNP array quality control, 12 doubled haploid (DH) lines (Supplementary Table S2), a resynthesized B. napus SBn8, a cultivar Zhongshuang9 and their F1 hybrid named F1ZSSB were employed.
2.2. Experiment design and traits measurement
One representative plant of each accession of the association population was self-pollinated in the 2010/2011 winter–spring growing season, and the leaves of each plant were sampled to extract genomic DNA. The self-pollinated seeds of each accession were grown in the experimental farm at Wuhan (114.31°E, 30.52°N), China, in the 2011/2012 growing season. Each accession was grown in a plot with three rows and 10–12 plants in each row, with a distance of 0.2 m between plants within each row and 0.3 m between rows. Before flowering time, each plot was separated by nylon net with a 0.35-mm square hole to propagate seeds. Fatty acid analysis was performed by using gas liquid chromatography (GC) with a Model 6890N GC analyzer (Agilent Technologies, Inc., Wilmington, DE, USA), following the protocol as described.35 The erucic acid content was expressed as its percentage of total fatty acids in mature seeds.
The 472 accessions of the association population were grown using a randomized complete block design with three replications in the experimental farm at Wuhan, and Nanchang (116.27°E, 28.37°N), China, in the 2012/2013 growing season. Each accession was grown in a plot with five rows with the same plant density abovementioned. The weight of randomly selected 1000 well-filled, open-pollinated seeds of each plot was measured to represent seed weight for one replicate of each accession. Seed oil content and total glucosinolate content were estimated by near-infrared reflectance spectroscopy (NIRS). Approximately 3 g seeds per accession were measured using a Foss NIRS Systems 5000 instrument on a reflectance scanning mode.
As the traits of the association panel were investigated in multi-environments with three replications, an R script (www.eXtension.org/pages/61006) based on a linear model described by Merk et al.36 was used to obtain the best linear unbiased prediction (BLUP) of each trait of each line. The resulting values were used as phenotypes for the association analysis.
2.3. SNP genotyping and filtering
SNP genotyping was performed using the Brassica 60K Illumina® Infinium SNP array by Emei Tongde Co. (Beijing) according to the manufacture's protocol (http://www.illumina.com/technology/infinium_hd_assay.ilmn). The SNP data were clustered and called automatically using the Illumina BeadStudio genotyping software. Those SNPs with AA or BB frequency equal to zero, call frequency <0.8 or minor frequency <0.05 were excluded. The remaining SNPs were scrutinized visually and those SNPs that did not show three clearly defined clusters representing the three possible genotypes (AA, AB and BB) were also excluded.
2.4. In silico mapping of SNPs
The source sequences for designing SNP probes of the SNP array were used to perform a BLAST37 search against version 4 of the ‘pseudomolecules’ representative of the genome of B. napus (Supplementary Table S3). Only the top BLAST hits against the pseudomolecules were considered. BLAST matches to multiple loci, with the same top identity, were not considered to be mapped.
2.5. Genetic diversity, population structure and linkage disequilibrium analysis
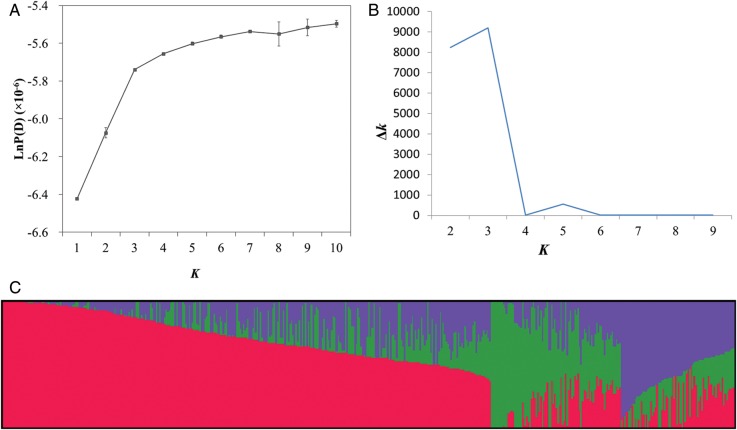
Polymorphism information content (PIC) of the SNPs were estimated using the PowerMarker version 3.51.38 The differences of PIC between linkage groups were assessed using one-way analysis of variance implemented in SPSS 9.0. All the SNPs were used to estimate the genetic relatedness between individuals by principal component analysis (PCA) using the GCTA tool.39 A total of 3900 SNPs [minor allele frequency (MAF) ≥ 0.2] evenly distributed across the whole genome were selected to perform the following population structure and relative kinship analysis. The software package STRUCTURE v2.3.440 was used to infer population structure. Five independent runs were performed with a K-value (the putative number of genetic groups) varying from 1 to 10, with the length of burnin period and the number of MCMC (Markov Chain Monte Carlo) replications after burnin both to 100 000 under the ‘admixture model’. The most likely k-value was determined by the log probability of data [LnP(D)] and an ad hoc statistic Δk based on the rate of change of LnP(D) between successive k as described by Evanno et al.41 The cluster membership coefficient matrices of replicate runs from STRUCTURE were integrated to get a Q matrix by the CLUMPP software42 and graphically displayed using the DISTRUCT software package.43 Accessions with the probability of membership >0.7 were assigned to corresponding clusters, and those <0.7 were assigned to a mixed group. Nei's genetic distance44 was estimated and used for constructing an unrooted neighbour-joining tree by the PowerMarker software, and the tree was visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). The relative kinship matrix comparing all pairs of the 472 accessions was calculated using the software package SPAGeDi.45 Negative values between two individuals were set to 0, as described by Yu et al.46 LD was estimated as the squared allele frequency correlations (r2) between all pairs of the SNPs, using the software TASSEL3.0.47
2.6. Genome-wide association analysis
Trait–SNP association analysis was performed using six models to evaluate the effects of population structure (Q, PC) and kinship (K): (i) naive—without controlling for Q and K, (ii) Q model, controlling for Q, (iii) PCA model, controlling for PC, the top 10 principal components were used as fixed effects, (iv) K model, controlling for K, (v) Q + K model, controlling for both Q and K and (vi) P + K model, controlling for both PC and K. The naive, Q and PCA models were performed using a general linear model; the K, Q + K and P + K models were performed using a mixed linear model with optimum compression and population parameters previously determined (P3D) variance component estimation in TASSEL 3.0.46,48 The distribution of observed −log10(p) for each SNP from marker–trait associations was compared with the expected distribution in a quantile–quantile plot. Significance of associations between SNPs and traits was based on threshold p < 2 × 10−6 (i.e. −log10(p) = 5.7), a stringent Bonferroni correction calculated by dividing 0.05 by the total number of SNPs in the analysis, 24 256. The significant association of more than one SNP was regarded as a true association. Furthermore, false discovery rates (FDRs) were calculated as [(m× P)/n] × 100%,49,50 where m is the total number of SNPs (i.e. 24 256), P is the p-value threshold for detecting significant association (i.e. 2 × 10−6) and n is the total number of significant associations per trait.
3. Results
3.1. Phenotypic variations of measured quantitative traits
Glucosinolate content, oil content and seed weight of the B. napus association panel comprising 472 accessions were measured for three replications in two locations. As the key genes responsible for the natural variation of the erucic acid content are known, this trait was investigated for only one replication in one location and used for evaluating the ability and accuracy of the GWAS of this panel. Extensive phenotypic variations were observed as shown in the descriptive statistics in Table 1. The erucic acid content, which varied from 0 to 53.7 with an average of 22.7, had the maximum coefficient of variation of 73.4%, whereas oil content in Nanchan, which varied from 33.4 to 52.6 with an average of 43.6, had the lowest coefficient of variation of 5.4%. Seed weight and glucosinolate content were quite consistent across different geographic location replicates with the correlation coefficient of 0.876 and 0.914, respectively, and oil content has the correlation coefficient of ∼0.775 (Supplementary Fig. S1).
Table 1.
Phenotypic variations for seed weight and seed quality in this B. napus panel
| Traits | Min ± SDa | Max ± SD | Mean ± SD | CV (%)b |
|---|---|---|---|---|
| Glucosinolate content (µmol/g)—Wuhan 2013 | 28.5 ± 2.0 | 142.8 ± 0.7 | 91.5 ± 0.3 | 29.4 |
| Glucosinolate content (µmol/g)—Nanchan 2013 | 30.1 ± 0.9 | 138.0 ± 3.6 | 90.1 ± 0.9 | 27.4 |
| Oil content (%)—Wuhan 2013 | 34.2 ± 0.8 | 51.4 ± 0.4 | 42.6 ± 0.3 | 5.9 |
| Oil content (%)—Nanchan 2013 | 33.4 ± 1.9 | 52.6 ± 1.5 | 43.6 ± 0.7 | 5.4 |
| Erucic acid content (%)—Wuhan 2012 | 0 | 53.7 | 22.7 | 73.4 |
| Seed weight (g)—Wuhan 2013 | 2.3 ± 0.1 | 5.9 ± 0.2 | 3.5 ± 0.0 | 14.2 |
| Seed weight(g)—Nanchan 2013 | 2.3 ± 0.1 | 5.2 ± 0.2 | 3.6 ± 0.1 | 13.8 |
aSD is an abbreviation of standard deviation, which was calculated based on the measured values of seeds from three replicated experimental blocks.
bCV is an abbreviation of coefficient of variation, which was estimated as the ratio of the standard deviation to the mean of all accessions.
3.2. SNP performance and quality
Calling SNP genotype data using the BeadStudio genotyping software generally produced three clear clusters, i.e. the AA homozygote, BB homozygote and AB heterozygote. Of the 52 157 SNPs in the array, 10 389 which had zero call frequency of AA or BB were excluded. Then, with a cut-off of missing data >0.2 and MAF <0.05, 1866 and 1428 SNPs were filtered, respectively, reducing the number of SNPs to 38 474. Those that remained were scrutinized visually and 29 027 SNPs with three clearly defined clusters were selected. Theoretically, the DH lines are homozygous throughout their genome, while the 12 DH lines derived from geographically different inbred lines of B. napus showed a heterozygosity rate of 1.8∼2.8%, which might be caused by miss-calling or homologous sequences and should be excluded. As a result, 2186 SNPs with the heterozygous genotype in any DH lines were filtered and 26 841 SNPs were finally selected.
To further control SNP quality, a parent/F1 triplet and four DNA duplicates were used to analyse the pedigree consistency and technical reproducibility, respectively. As a result, the allele calls were highly reproducible (Supplementary Table S4) with no or negligible inconsistencies between technical replicates. Furthermore, the parent/F1 triplet showed a pedigree inconsistency of <1%, confirming the high quality of the genotype calling of the selected SNPs in the array.
3.3. SNP in silico mapping and diversity
By BLAST analysis of the resource sequences of the SNPs in the array against ‘pseudomolecules’ representative of the genome of B. napus, 47 805 of 52 157 SNPs were in silico mapped (Supplementary Table S3). Among the selected 26 841 SNPs with fine performance, 24 256 were mapped (Supplementary Table S5 and Fig. S2). Linkage group A2 had the least markers of 506 with a marker density of one per 52.9 kb, and C4 had the most markers of 2104 with a marker density of one per 25.0 kb. The SNP diversity in the whole collection was expressed by a PIC value, and the results were shown in Supplementary Table S5 and summarized in Table 2. Except for linkage groups C5, C7 and C8, 75% of SNPs in each linkage group had PIC values over 0.25. The levels of polymorphism of linkage groups in the A subgenome (with the exception of A2, A4 and A5) were generally higher than those in the C subgenome. The mean PIC values of A8 and A9 were highest, nearly 0.32, whereas that of C7 was lowest, only 0.254.
Table 2.
Summary of the PIC values in different linkage groups of the B. napus
| Linkgae group | Number of SNPs | PICa value |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 0.05–0.1 | 0.1–0.15 | 0.15–0.2 | 0.2–0.25 | 0.25–0.3 | 0.3–0.35 | 0.35–0.4 | Average PICb | ||
| A1 | 1072 | 9 (0.8%) | 34 (3.2%) | 83 (7.7%) | 117 (10.9%) | 102 (9.5%) | 248 (23.1%) | 479 (44.7%) | 0.31 g |
| A2 | 506 | 2 (0.4%) | 34 (6.7%) | 25 (4.9%) | 46 (9.1%) | 115 (22.7%) | 130 (25.7%) | 154 (30.4%) | 0.294 cd |
| A3 | 1469 | 6 (0.4%) | 45 (3.1%) | 93 (6.3%) | 130 (8.8%) | 210 (14.3%) | 383 (26.1%) | 602 (41%) | 0.311 g |
| A4 | 1035 | 6 (0.6%) | 61 (5.9%) | 56 (5.4%) | 131 (12.7%) | 209 (20.2%) | 272 (26.3%) | 300 (29%) | 0.293 bcd |
| A5 | 1123 | 1 (0.1%) | 35 (3.1%) | 69 (6.1%) | 115 (10.2%) | 179 (15.9%) | 376 (33.5%) | 348 (31%) | 0.307 fg |
| A6 | 1099 | 3 (0.3%) | 36 (3.3%) | 58 (5.3%) | 91 (8.3%) | 168 (15.3%) | 317 (28.8%) | 426 (38.8%) | 0.312 g |
| A7 | 1427 | 11 (0.8%) | 50 (3.5%) | 70 (4.9%) | 126 (8.8%) | 217 (15.2%) | 382 (26.8%) | 571 (40%) | 0.31 g |
| A8 | 691 | 7 (1%) | 35 (5.1%) | 38 (5.5%) | 40 (5.8%) | 53 (7.7%) | 143 (20.7%) | 375 (54.3%) | 0.319 h |
| A9 | 1225 | 9 (0.7%) | 45 (3.7%) | 39 (3.2%) | 68 (5.6%) | 190 (15.5%) | 261 (21.3%) | 613 (50%) | 0.32 h |
| A10 | 805 | 8 (1%) | 33 (4.1%) | 38 (4.7%) | 72 (8.9%) | 104 (12.9%) | 193 (24%) | 357 (44.3%) | 0.312 g |
| C1 | 2012 | 1 (0%) | 73 (3.6%) | 124 (6.2%) | 238 (11.8%) | 298 (14.8%) | 1074 (53.4%) | 204 (10.1%) | 0.298 cde |
| C2 | 1292 | 2 (0.2%) | 23 (1.8%) | 54 (4.2%) | 116 (9%) | 397 (30.7%) | 321 (24.8%) | 379 (29.3%) | 0.3 de |
| C3 | 2201 | 29 (1.3%) | 77 (3.5%) | 159 (7.2%) | 307 (13.9%) | 352 (16%) | 516 (23.4%) | 761 (34.6%) | 0.298 cde |
| C4 | 2104 | 32 (1.5%) | 94 (4.5%) | 163 (7.7%) | 181 (8.6%) | 351 (16.7%) | 560 (26.6%) | 723 (34.4%) | 0.301 ef |
| C5 | 719 | 2 (0.3%) | 32 (4.5%) | 59 (8.2%) | 105 (14.6%) | 110 (15.3%) | 121 (16.8%) | 290 (40.3%) | 0.298 cde |
| C6 | 1539 | 28 (1.8%) | 52 (3.4%) | 101 (6.6%) | 136 (8.8%) | 403 (26.2%) | 417 (27.1%) | 402 (26.1%) | 0.296 cde |
| C7 | 2006 | 44 (2.2%) | 591 (29.5%) | 52 (2.6%) | 185 (9.2%) | 156 (7.8%) | 587 (29.3%) | 391 (19.5%) | 0.254 a |
| C8 | 1096 | 5 (0.5%) | 23 (2.1%) | 40 (3.6%) | 362 (33%) | 90 (8.2%) | 229 (20.9%) | 347 (31.7%) | 0.292 bc |
| C9 | 835 | 14 (1.7%) | 18 (2.2%) | 78 (9.3%) | 97 (11.6%) | 227 (27.2%) | 192 (23%) | 209 (25%) | 0.287 b |
aPIC is an abbreviation of polymorphism information content.
bValues followed by different letters in this column are significantly different at the level of 0.05.
3.4. Population structure, relative kinship and linkage disequilibrium
The population structure of the association panel was calculated using 3900 SNPs by STRUCTURE, and clustering inference performed with possible clusters (k) from 1 to 10 showed that the most significant change of likelihood occurred when K increased from 3 to 4, and the highest Δk value was observed at k = 3 (Fig. 1). Both parameters suggested that the 472 genotypes could be assigned into three groups. Using a probability of membership threshold of 70%, 164, 29 and 17 lines were assigned into the three groups, respectively. The remaining 262 lines were classified into a mixed group (Supplementary Table S1). Most of the lines in Group 1 were from Asia and belong to semi-winter OSR (Supplementary Table S6). European lines were the main component of the Groups 2 and 3, in which the larger part were winter OSR and spring OSR, respectively. The NJ phylogenetic tree based on Nei's genetic distances displayed three clear clades (Supplementary Fig. S3), corresponding to the three groups estimated by STRUCTURE. The lines belong to mixed groups distributed across the whole tree. The PCA based on the 24 256 genome-wide SNPs showed that the first two principal components explained 11.7 and 5.8% of the genetic variance, respectively (Supplementary Fig. S4). The three major groups identified using STRUCTURE (i.e. except for the mixed group), winter OSR, spring OSR and semi-winter OSR were assigned to three major clusters. The analysis of relative kinship showed that the average relative kinship between any two lines was 0.0856. A total of 51.4% of kinship coefficients between lines were equal to 0, and 32.6% kinship coefficients ranged from 0 to 0.2 (Supplementary Fig. S5). This pattern of genetic relatedness revealed that most lines have no or weak kinship in this OSR panel. To assess the extent of LD, we calculated the mean pairwise r2 for all SNPs as 0.0176.
Figure 1.
Analysis of the population structure of 472 rapeseed accessions by STRUCTURE. (a) Estimated LnP(D) of possible clusters (k) from 1 to 10; (b) Δk based on the rate of change of LnP(D) between successive k; (c) population structure based on k = 3. Each individual is represented by a vertical bar, partitioned into coloured segments with the length of each segment representing the proportion of the individual's genome. A given group is represented: Red, Group 1; Green, Group 2; Purple, Group 3.
3.5. Association mapping
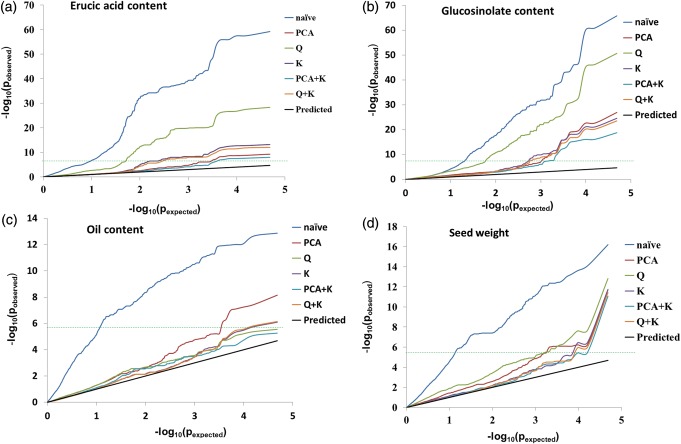
We assessed the utility of six statistical models for controlling type I and II errors in this B. napus panel. As can be seen in the QQ plots (Fig. 2), the distribution of observed −log10(p) values from the naive model departed quite far from the expected distribution leading to a high level of false-positive signals. For all four traits, any model controlling population structure or relative kinship performed significantly better than the naive model. The models controlling relative kinship, i.e. K, Q + K and P + K, had similar effects in reducing the false positives, while P + K showed slightly greater effect than K and Q + K models. For erucic acid content and glucosinolate content, the PCA, K, Q + K and P + K models were better than the Q model. All of the models except the naive model performed well for oil content and seed weight, and no significant difference was observed among these five models for the seed weight. There were no significant associations between SNPs and oil content in the Q, P + K models based on threshold −log10(p) = 5.7, which might indicate false negatives in this analysis.
Figure 2.
Quantile–quantile plots of estimated −log10(p) from association analysis using six methods for four traits: (a) Erucic acid content; (b) glucosinolate content; (c) oil content and (d) seed weight. The black line is the expected line under the null distribution, and the deviations from expectation indicate that the statistical analysis may cause spurious associations. The horizontal dashed green lines indicates genome-wide significance threshold −log10(p) = 5.7.
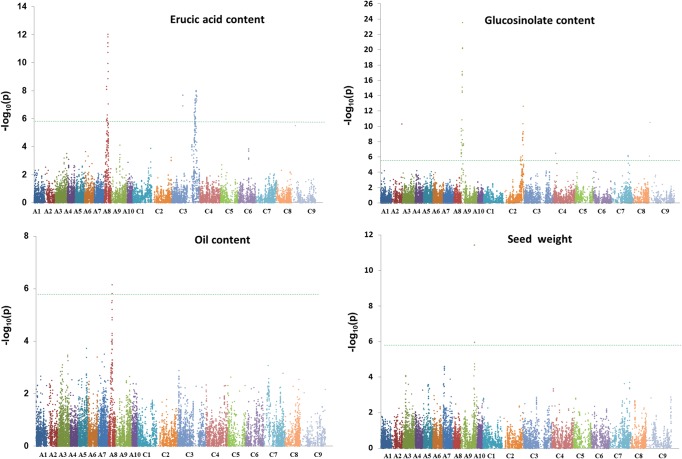
According to the Q−Q plots of the six models, we used PCA and Q + K models to identify association signals. First, in order to evaluate the power of association analysis in this panel, the erucic acid content, for which genetic control was well known, was first used to perform association analysis. Two significant regions located at 9.5 and 63.7 Mb of A8 and C3, respectively, in the ‘pseudomolecules’ of B. napus were detected by both models (Fig. 3a, Supplementary Fig. S6a and Table 3). The FDRs for associations detected in Q + K and PCA models were 0.046 and 0.312%, respectively. The two GWAS peaks, at Bn-A08-p12599446 and Bn-scaff_15794_3-p29807, contributed to 12.2 and 7.9% of phenotypic variance, respectively, based on R2 values. These two peak SNPs were found to be 233 and 128 kb away from the key genes BnaA.FAE1 and BnaC.FAE1, respectively, indicating that our association genetics approach was successful.
Figure 3.
Manhattan plots of association analysis using the Q + K model for four traits: (a) erucic acid content; (b) glucosinolate content; (c) oil content and (d) seed weight. Each dot represents a SNP. The horizontal dashed green line represents the Bonferroni-corrected significance threshold −log10(p) = 5.7.
Table 3.
Genome-wide significant association signals of seed quality and seed weight
| Trait | SNP | Linkage group | Position in ‘pseudomolecules’ (kb) | Major allele | Minor allele | Minor allele frequency | −log10 (p)a | Contributiona (%) | Candidate gene | Position of candidate genes in ‘pseudomolecules' (kb) |
|---|---|---|---|---|---|---|---|---|---|---|
| Erucic acid content | Bn-A08-p12599446 | A8 | 9514 | T | C | 0.432 | 12.0 | 12.27 | BnaA.FAE1 | A8: 9281 |
| Bn-scaff_15794_3-p29807 | C3 | 63 672 | A | C | 0.446 | 8.0 | 7.9 | BnaC.FAE1 | C1: 63 800 | |
| Glucosinolate content | Bn-A01-p9004629 | A9 | 3210 | T | C | 0.314 | 23.6 | 25.17 | BnaA.HAG1c | A9: 3426 |
| Bn-scaff_26086_1-p11779 | C2 | 50 098 | G | A | 0.292 | 12.7 | 13.19 | BnaC.HAG1a | C2: 49 620 | |
| Bn-scaff_16534_1-p2070156b | C4 | 6327 | T | C | 0.084 | 10.1 | 5 | |||
| Bn-scaff_15705_1-p1628802 | C7 | 39 871 | G | A | 0.284 | 6.2 | 6.06 | BnaC.HAG1b | C7: 40 676 | |
| Bn-scaff_17526_1-p1140588 | C9 | 2823 | C | T | 0.335 | 10.5 | 12.26 | BnaC.HAG1c | C9: 4725 | |
| Oil content | Bn-A08-p12599446 | A8 | 9514 | T | C | 0.432 | 6.1 | 6.22 | BnaA.FAE1 | A8: 9281 |
| Seed weight | Bn-A10-p12639538b | A7 | 2674 | T | G | 0.265 | 6.2 | 4.9 | ||
| Bn-A09-p30654305 | A9 | 34 653 | A | C | 0.275 | 11.4 | 13.87 |
aOnly the values estimated by the Q + K model were shown, if the SNPs were detected by both Q + K and PCA models.
bAssociation signals identified only by the PCA model.
Four and five significant associations for glucosinolate content were detected by Q + K and PCA models, respectively, with the FDR of 0.083 and 0.106%. Both models detected four common regions at 3.2, 50.0, 39.9 and 2.8 Mb of A9, C2, C7 and C9, respectively, in the ‘pseudomolecules’ of B. napus (Fig. 3b, Supplementary Fig. S6b and Table 3). The cumulative phenotypic variance explained by all significant SNPs was 56.7%. As the deletions of the A. thaliana orthologous gene HAG1 (At5g61420) in C2 and A9 were regarded to lead to low glucosinolate content,34 we searched the HAG1 paralogues in the ‘pseudomolecules’ of B. napus and found four paralogues at 3.4 Mb of A9, 49.6 Mb of C2, 40.7 Mb of C7 and 4.7 Mb of C9, which were very similar to the four GWAS peaks. The candidate gene BnaC.HAG1c was 1.9 Mb away from the peak SNP Bn-scaff_17526_1-p1140588, which might be due to the low density of SNPs around the candidate gene. In addition, the PCA model also detected a novel locus at 6.33 Mb of C4, which contributed 5.0% of the phenotypic variation (Supplementary Fig. S6b and Table 3). For oil content, only one significantly associated region in A8 was detected by Q + K and PCA models with FDRs of 2.5 and 0.714%, respectively (Fig. 3c, Supplementary Fig. S6c and Table S3). The GWAS peaks explained 6.1% of the total phenotypic variance. The GWAS peak for oil content was similar to that for erucic acid. For seed weight, Q + K and PCA models detected one association in A9 and A7 with the FDR of 1.667 and 0.384%, respectively. The GWAS peak detected by Q + K, i.e. Bn-A09-p30654305, at 34.65 Mb of A9, explained 11.4% of the phenotypic variance (Fig. 3d, Supplementary Fig. S6d and Table 3). The SNP Bn-A09-p30654305 was also detected by the PCA model as a single associated SNP in A9. In addition, using the PCA model, we also detected an associated SNP Bn-A10-p12639538 at 2.67 Mb of A7 (Supplementary Fig. S5d and Table 3), which explained 4.9% of the total seed weight variation.
4. Discussion
High-quality molecular markers and reliable genotype data are the precondition of genetic diversity analysis and association mapping. The amphidiploid nature of the B. napus genome might make automated SNP detection challenging and confounding, with homologue, paralogue and also homoeologue variation.30,51 Trick et al.30 termed three types of SNPs, i.e. inter-homoeologue polymorphisms, hemi-SNPs and simple SNPs in B. napus. The inter-homoeologue polymorphisms do not represent allelic variation per se, and as hemi-SNPs are the allelic polymorphisms in the homoeologous sequences, the short flanking sequences of the hemi-SNPs could not be specifically mapped in the ‘pseudomolecules’ representative of the genome of B. napus. By scanning the rapeseed association panel using the 60K SNP array, the clustering results showed that ∼10 000 SNPs had zero call frequency of AA or BB, which could, in part, be caused by inter-homoeologue polymorphisms. Meanwhile, ∼10 000 SNPs showed more than three clusters, which are indicative of two loci in a duplicated sequence as suggested by Ganal et al.,52 and are possibly hemi-SNPs. By strictly filtering, the genotypes of the finally selected SNPs of all DH lines were completely homozygous, and the DNA pedigree consistency and technical reproducibility were nearly perfect, suggesting the high quality of these selected SNPs.
On the whole, over 76% of the SNPs have the PIC of 0.25–0.5, which was regarded as the PIC range of medium polymorphism of the DNA marker.53 On average, the polymorphism level was slightly higher for the A than for the C linkage groups in our population, contrary to the results of Delourme et al.32 In our panel, nearly half of the genotypes were Asiatic, while those of Delourme et al. were European.32 Asiatic cultivars were selected for improved adaptation to the local environment by introgression of B. rapa into B. napus germplasm,54,55 increasing the genetic diversity of the A genome. Our finding suggests that genetic diversity of B. napus germplasm should be broadened by introgression of B. oleracea, as described by previous studies.24,56
Understanding the population structure of the association mapping population contributes to reducing both Type I and II errors between molecular markers and traits of interest.46,57 In our study, the association population was classified into three groups, semi-winter, winter and spring OSR were the main components of these three groups. Distinct clustering of winter and spring types has been reported by previous studies.24,32,58,59 In our study, although Groups 2 and 3 mainly distinguished winter and spring types, respectively, some winter and spring types were found in the mixed group, located at an intermediate position between Groups 2 and 3. Delourme et al.32 also observed that some spring OSR lines did not show distinct clustering. These intermediate lines might be derived from hybrids of the lines from different gene pools.60,61 The kinship analysis revealed that most lines in the panel have no or weak kinship, together with significant differences in phenotype performance, indicating that this panel is suitable for association analysis. The mean pairwise r2 values is close to previous estimates of 0.011726 or 0.024734 and lower than 0.037 estimated for the population used by Delourme et al.,32 confirming the low overall level of LD in B. napus. At the genome level, mean LD decay was estimated to be 0.5–1.2 cM in rapeseed germplasm.24,26,32 Given the length of the genetic map of 2500 cM, >2100–5000 evenly spaced markers would be necessary to perform GWASs in rapeseed. Therefore, by selecting in excess of 20 000 SNPs, our study should have more than sufficient markers to perform a good association analysis.
In the previous association analysis study in rapeseed, generally only population structure estimated by STRUCTURE40 was considered.27–29,62 However, spurious associations cannot be controlled completely by population structure since the Q matrix only gives a rough dissection of population differentiation. Furthermore, the programme STRUCTURE needs intensive computational cost on large data sets. The current PCA software such as EIGENSTRAT63 and GCTA39 could analyse tens of thousands of samples with millions of SNPs with high performance and infer continuous axes of genetic variation; therefore, the PCA based on genome-wide SNPs was broadly used to detect and correct for population stratification in GWAS;18,19 however, a few residual false-positive associations still existed in the PCA model.64 Yu et al.46 suggested that integration of the pairwise kinship into a mixed model to correct for relatedness could reduce spurious association, which was subsequently supported by the study in maize,48 sorghum65 and barley.66 In our study, six models, i.e. naive, Q, PCA, K, Q + K and P + K models, were first compared for controlling the false positives in B. napus association mapping. The models considering population structure or relatedness, especially PCA, K, Q + K and P + K models, could effectively eliminate the excess of low p-values for all traits; however, the P + K model also likely eliminated true positives, which is a common problem seen in other systems as well.16,67 Therefore, in order to reduce the false positives and false negatives, both Q + K and PCA models were considered to identify association signals.
The position of the DNA markers relative to the genome sequence or linkage map is also preferable for successful association mapping. In this study, the SNPs of the array were in silico mapped in B. napus ‘pseudomolecules’ to establish their hypothetical order. The positions of the significant association signals detected in this study are consistent with previous studies. It has been shown that two homoeologues, namely BnaA.FAE1 and BnaC.FAE1 on linkage groups A8 and C3, respectively, are responsible for controlling erucic acid content.68,69 In the present study, the peaks of the two association signals for erucic acid content (represented by the markers Bn-A08-p12599446 and Bn-scaff_15794_3-p29807, which have the highest significance of association on A8 and C3, respectively) were within 233 kb of these two genes, indicating that our association approach was successful.
For glucosinolate content, we identified four association signals on A9, C2, C7 and C9, which were detected independently in different studies.7,70,71 Howell et al.70 regarded that the QTLs on A9, C2 and C9 were homoeologous loci, whereas the underlying control genes were not uncovered until Harper et al.34 identified genomic deletions that underlie the QTLs on A9 and C2. In the low-glucosinolate rapeseed accessions, the deleted segments contained orthologs of the transcription factor HAG1 (At5g61420),34 which controls aliphatic glucosinolate biosynthesis in A. thaliana.72 In Brassica juncea, silencing of the orthologs of HAG1 was reported to cause low glucosinolate content.73 Interestingly, in the present study, four peak SNPs associated with glucosinolate content were identified to be close to the paralogues of HAG1. Further studies are necessary to reveal the DNA variation of the other two paralogues of HAG1 on C7 and C9.
Control of seed oil content is complex and previous studies have detected at least 10 QTLs, which were environmentally sensitive and with minor effects.74–76 These studies commonly regarded that two QTLs on A8 and C3 overlapped with the QTLs for erucic acid content. Since the BnaA.FAE1 and BnaC.FAE1 were cloned and confirmed as the key factors controlling the synthesis of erucic acid in rapeseed,68,69 it was regarded that the pleiotropy of FAE1 was responsible for the decrease in seed oil content along with the reduction of seed erucic acid content in the modern cultivars. Our study showed that the peak SNP on A8 was only 233 kb away from the BnaA.FAE1, indicating the role of fine mapping of GWAS.
Seed weight of rapeseed is also a complex trait, which has a relatively high heritability and may primarily be controlled by genes with additive effects.10,11 So far, >80 QTLs across the 19 linkage groups have been identified and individual QTL contributed to 2.0–28.2% of the phenotypic variation.10–12,77–80 The seed weight QTLs on A7 were repeatedly detected in many studies with diverse genetic materials,10–12,78,80 and the QTL on A9 was identified as a major QTL with the most contribution of 28.2% for the total seed weight variation.12 Interestingly, our association mapping also detected a major QTL on A9 and a minor QTL on A7, indicating our association panel might be suitable for dissecting complex traits in B. napus. Since associated markers and causal polymorphisms underlying traits of interest are in a narrow interval equivalent to the distance of LD decay,57 the genes flanking the peak SNPs and falling within the window of LD decay were always used to predict the candidate genes.81 As the average LD across our panel is very low (mean pairwise r2 = 0.0176), it is reasonable to suppose that the detected loci in our study will be located in close proximity to the candidate genes controlling seed weight, and that these tightly associated SNPs would be of significant benefit in a molecular design breeding approach to improve seed weight.
Supplementary Data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by the Chinese National Basic Research and Development Program (2011CB109302), the National Natural Science Foundation of China (31301360), Strategic Japanese-Chinese Cooperative Program on ‘Climate Change’ (2012DFG90290), National Science and Technology Pillar Program during the Twelfth Five-year Plan Period (2011BAD35B09) and Chengguang Program for Young Scientists of Wuhan Municipal Government (2013070104010031).
Supplementary Material
References
- 1.Rana D., van den Boogaart T., O'Neill C.M., et al. Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 2004;40:725–33. doi: 10.1111/j.1365-313X.2004.02244.x. [DOI] [PubMed] [Google Scholar]
- 2.Nagaharu U. Genome-analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japan J. Bot. 1935;7:389–452. [Google Scholar]
- 3.Gomez-Campo C., Prakash S. Origin and domestication. In: Gomez-Campo C., editor. Biology of Brassica coenospecies. Amsterdam: Elsevier; 1999. pp. 33–58. [Google Scholar]
- 4.Prakash S., Wu X.-M., Bhat S.R. History, evolution, and domestication of Brassica crops. In: Janick J., editor. Plant Breeding Reviews. vol. 35. New Jersey: John Wiley & Sons, Inc.; 2011. pp. 19–84. [Google Scholar]
- 5.Toxopeus H. In: The domestication of Brassica crops. Van Marrewijk N.P.A., Toxopeus H., editors. Wageningen: Proc Eucarpia Conference on the breeding of Cruciferous crops; 1979. pp. 47–56. [Google Scholar]
- 6.Zhao J., Becker H., Zhang D., Zhang Y., Ecke W. Conditional QTL mapping of oil content in rapeseed with respect to protein content and traits related to plant development and grain yield. Theor. Appl. Genet. 2006;113:33–8. doi: 10.1007/s00122-006-0267-5. [DOI] [PubMed] [Google Scholar]
- 7.Uzunova M., Ecke W., Weissleder K., Röbbelen G. Mapping the genome of rapeseed (Brassica napus L.). I. Construction of an RFLP linkage map and localization of QTLs for seed glucosinolate content. Theor. Appl. Genet. 1995;90:194–204. doi: 10.1007/BF00222202. [DOI] [PubMed] [Google Scholar]
- 8.Zhao J., Dimov Z., Becker H., Ecke W., Möllers C. Mapping QTL controlling fatty acid composition in a doubled haploid rapeseed population segregating for oil content. Mol. Breed. 2008;21:115–25. [Google Scholar]
- 9.Long Y., Shi J., Qiu D., et al. Flowering time quantitative trait loci analysis of oilseed Brassica in multiple environments and genomewide alignment with Arabidopsis. Genetics. 2007;177:2433–44. doi: 10.1534/genetics.107.080705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi J., Li R., Qiu D., et al. Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics. 2009;182:851–61. doi: 10.1534/genetics.109.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Radoev M., Becker H., Ecke W. Genetic analysis of heterosis for yield and yield components in rapeseed (Brassica napus L.) by quantitative trait locus mapping. Genetics. 2008;179:1547–58. doi: 10.1534/genetics.108.089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang P., Shu C., Chen L., Xu J., Wu J., Liu K. Identification of a major QTL for silique length and seed weight in oilseed rape (Brassica napus L.) Theor. Appl. Genet. 2012;125:285–96. doi: 10.1007/s00122-012-1833-7. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L., Li S., Chen L., Yang G. Identification and mapping of a major dominant quantitative trait locus controlling seeds per silique as a single Mendelian factor in Brassica napus L. Theor. Appl. Genet. 2012;125:695–705. doi: 10.1007/s00122-012-1861-3. [DOI] [PubMed] [Google Scholar]
- 14.Nordborg M., Weigel D. Next-generation genetics in plants. Nature. 2008;456:720–3. doi: 10.1038/nature07629. [DOI] [PubMed] [Google Scholar]
- 15.Nordborg M., Tavare S. Linkage disequilibrium: what history has to tell us. Trends Genet. 2002;18:83–90. doi: 10.1016/s0168-9525(02)02557-x. [DOI] [PubMed] [Google Scholar]
- 16.Zhao K., Tung C.W., Eizenga G.C., et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011;2:467. doi: 10.1038/ncomms1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian F., Bradbury P.J., Brown P.J., et al. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 2011;43:159–62. doi: 10.1038/ng.746. [DOI] [PubMed] [Google Scholar]
- 18.Huang X., Wei X., Sang T., et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010;42:961–7. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 19.Huang X., Zhao Y., Wei X., et al. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 2012;44:32–9. doi: 10.1038/ng.1018. [DOI] [PubMed] [Google Scholar]
- 20.Li H., Peng Z., Yang X., et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 2013;45:43–50. doi: 10.1038/ng.2484. [DOI] [PubMed] [Google Scholar]
- 21.Atwell S., Huang Y.S., Vilhjalmsson B.J., et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–31. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandakova T., Lysak M.A. Chromosomal phylogeny and karyotype evolution in x=7 crucifer species (Brassicaceae) Plant Cell. 2008;20:2559–70. doi: 10.1105/tpc.108.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parkin I., Gulden S., Sharpe A., et al. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics. 2005;171:765–81. doi: 10.1534/genetics.105.042093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bus A., Korber N., Snowdon R.J., Stich B. Patterns of molecular variation in a species-wide germplasm set of Brassica napus. Theor. Appl. Genet. 2011;123:1413–23. doi: 10.1007/s00122-011-1676-7. [DOI] [PubMed] [Google Scholar]
- 25.Ecke W., Clemens R., Honsdorf N., Becker H.C. Extent and structure of linkage disequilibrium in canola quality winter rapeseed (Brassica napus L.) Theor. Appl. Genet. 2010;120:921–31. doi: 10.1007/s00122-009-1221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao Y., Cai D., Yang W., et al. Genetic structure and linkage disequilibrium pattern of a rapeseed (Brassica napus L.) association mapping panel revealed by microsatellites. Theor. Appl. Genet. 2012;125:437–47. doi: 10.1007/s00122-012-1843-5. [DOI] [PubMed] [Google Scholar]
- 27.Zou J., Jiang C., Cao Z., et al. Association mapping of seed oil content in Brassica napus and comparison with quantitative trait loci identified from linkage mapping. Genome. 2010;53:908–16. doi: 10.1139/G10-075. [DOI] [PubMed] [Google Scholar]
- 28.Honsdorf N., Becker H.C., Ecke W. Association mapping for phenological, morphological, and quality traits in canola quality winter rapeseed (Brassica napus L.) Genome. 2010;53:899–907. doi: 10.1139/G10-049. [DOI] [PubMed] [Google Scholar]
- 29.Snowdon R.J., Wittkop B., Rezaidad A., et al. Regional association analysis delineates a sequenced chromosome region influencing antinutritive seed meal compounds in oilseed rape. Genome. 2010;53:917–28. doi: 10.1139/G10-052. [DOI] [PubMed] [Google Scholar]
- 30.Trick M., Long Y., Meng J., Bancroft I. Single nucleotide polymorphism (SNP) discovery in the polyploid Brassica napus using Solexa transcriptome sequencing. Plant Biotechnol. J. 2009;7:334–46. doi: 10.1111/j.1467-7652.2008.00396.x. [DOI] [PubMed] [Google Scholar]
- 31.Bancroft I., Morgan C., Fraser F., et al. Dissecting the genome of the polyploid crop oilseed rape by transcriptome sequencing. Nat. Biotechnol. 2011;29:762–8. doi: 10.1038/nbt.1926. [DOI] [PubMed] [Google Scholar]
- 32.Delourme R., Falentin C., Fomeju B., et al. High-density SNP-based genetic map development and linkage disequilibrium assessment in Brassica napus L. BMC Genomics. 2013;14:120. doi: 10.1186/1471-2164-14-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X., Wang H., Wang J., et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011;43:1035–9. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 34.Harper A.L., Trick M., Higgins J., et al. Associative transcriptomics of traits in the polyploid crop species Brassica napus. Nat. Biotechnol. 2012;30:798–802. doi: 10.1038/nbt.2302. [DOI] [PubMed] [Google Scholar]
- 35.Thies W. Schnelle und einfache Analysen der Fettsäurezusammensetzung in einzelnen Rapskotyledonen I. Gaschromatographische und papierchromatographische Methoden. Z Pflanzenzücht. 1971;65:181–202. [Google Scholar]
- 36.Merk H.L., Yarnes S.C., Van Deynze A., et al. Trait diversity and potential for selection indices based on variation among regionally adapted processing tomato germplasm. J. Am. Soc. Hortic. Sci. 2012;137:427–37. [Google Scholar]
- 37.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Liu K., Muse S. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128–9. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- 39.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evanno G., Regnaut S., Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 2005;14:2611–20. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 42.Jakobsson M., Rosenberg N.A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–6. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 43.Rosenberg N. DISTRUCT: a program for the graphical display of population structure. Mol. Ecol. Notes. 2004;4:137–8. [Google Scholar]
- 44.Nei M., Takezaki N. Estimation of genetic distances and phylogenetic trees from DNA analysis. Proceedings of the Fifth World Congress on Genetics Applied to Livestock Production; Guelph, Ontario, Canada: University of Guelph; 1983. pp. 405–12. [Google Scholar]
- 45.Hardy O., Vekemans X. SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol. Ecol. Notes. 2002;2:618–20. [Google Scholar]
- 46.Yu J., Pressoir G., Briggs W.H., et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006;38:203–8. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 47.Bradbury P.J., Zhang Z., Kroon D.E., Casstevens T.M., Ramdoss Y., Buckler E.S. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–5. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 48.Yang X., Yan J., Shah T., et al. Genetic analysis and characterization of a new maize association mapping panel for quantitative trait loci dissection. Theor. Appl. Genet. 2010;121:417–31. doi: 10.1007/s00122-010-1320-y. [DOI] [PubMed] [Google Scholar]
- 49.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 50.Olsen H.G., Hayes B.J., Kent M.P., et al. Genome-wide association mapping in Norwegian Red cattle identifies quantitative trait loci for fertility and milk production on BTA12. Anim. Genet. 2011;42:466–74. doi: 10.1111/j.1365-2052.2011.02179.x. [DOI] [PubMed] [Google Scholar]
- 51.Kaur S., Francki M.G., Forster J.W. Identification, characterization and interpretation of single-nucleotide sequence variation in allopolyploid crop species. Plant Biotechnol. J. 2012;10:125–38. doi: 10.1111/j.1467-7652.2011.00644.x. [DOI] [PubMed] [Google Scholar]
- 52.Ganal M.W., Durstewitz G., Polley A., et al. A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE. 2011;6:e28334. doi: 10.1371/journal.pone.0028334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Botstein D., Wlllte R.L., Skolinck M. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980;32:314–31. [PMC free article] [PubMed] [Google Scholar]
- 54.Qian W., Meng J., Li M., et al. Introgression of genomic components from Chinese Brassica rapa contributes to widening the genetic diversity in rapeseed (B. napus L.), with emphasis on the evolution of Chinese rapeseed. Theor. Appl. Genet. 2006;113:49–54. doi: 10.1007/s00122-006-0269-3. [DOI] [PubMed] [Google Scholar]
- 55.Shiga T. Rape breeding by interspecific crossing between Brassica napus and Brassica campestris in Japan. Japan Agric. Res. Quart. 1970;5:5–10. [Google Scholar]
- 56.Rahman M.H., Bennett R., Yang R.-C., Kebede B., Thiagarajah M. Exploitation of the late flowering species Brassica oleracea L. for the improvement of earliness in B. napus L.: an untraditional approach. Euphytica. 2011;177:365–74. [Google Scholar]
- 57.Flint-Garcia S.A., Thornsberry J.M., Buckler E.S., IV Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003;54:357–74. doi: 10.1146/annurev.arplant.54.031902.134907. [DOI] [PubMed] [Google Scholar]
- 58.Diers B., Osborn T. Genetic diversity of oilseed Brassica napus germplasm based on restriction fragment length polymorphisms. Theor. Appl. Genet. 1994;88:662–8. doi: 10.1007/BF01253968. [DOI] [PubMed] [Google Scholar]
- 59.Hasan M., Seyis F., Badani A., et al. Analysis of genetic diversity in the Brassica napus L. Gene pool using SSR markers. Genet. Res. Crop Evol. 2006;53:793–802. [Google Scholar]
- 60.Qian W., Chen X., Fu D., Zou J., Meng J. Intersubgenomic heterosis in seed yield potential observed in a new type of Brassica napus introgressed with partial Brassica rapa genome. Theor. Appl. Genet. 2005;110:1187–94. doi: 10.1007/s00122-005-1932-9. [DOI] [PubMed] [Google Scholar]
- 61.Hammerli A., Reusch T.B. Genetic neighbourhood of clone structures in eelgrass meadows quantified by spatial autocorrelation of microsatellite markers. Heredity (Edinb) 2003;91:448–55. doi: 10.1038/sj.hdy.6800310. [DOI] [PubMed] [Google Scholar]
- 62.Hasan M., Friedt W., Pons-Kuhnemann J., Freitag N.M., Link K., Snowdon R.J. Association of gene-linked SSR markers to seed glucosinolate content in oilseed rape (Brassica napus ssp. napus) Theor. Appl. Genet. 2008;116:1035–49. doi: 10.1007/s00122-008-0733-3. [DOI] [PubMed] [Google Scholar]
- 63.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 64.Yang X., Gao S., Xu S., et al. Characterization of a global germplasm collection and its potential utilization for analysis of complex quantitative traits in maize. Mol. Breed. 2011;28:511–26. [Google Scholar]
- 65.Upadhyaya H., Wang Y.-H., Gowda C.L.L., Sharma S. Association mapping of maturity and plant height using SNP markers with the sorghum mini core collection. Theor. Appl. Genet. 2013;126:2003–15. doi: 10.1007/s00122-013-2113-x. [DOI] [PubMed] [Google Scholar]
- 66.Pasam R.K., Sharma R., Malosetti M., et al. Genome-wide association studies for agronomical traits in a world wide spring barley collection. BMC Plant Biol. 2012;12:16. doi: 10.1186/1471-2229-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao K., Aranzana M.J., Kim S., et al. An Arabidopsis example of association mapping in structured samples. PLoS Genet. 2007;3:e4. doi: 10.1371/journal.pgen.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang N., Wang Y., Tian F., et al. A functional genomics resource for Brassica napus: development of an EMS mutagenized population and discovery of FAE1 point mutations by TILLING. New Phytol. 2008;180:751–65. doi: 10.1111/j.1469-8137.2008.02619.x. [DOI] [PubMed] [Google Scholar]
- 69.Wu G., Wu Y., Xiao L., Li X., Lu C. Zero erucic acid trait of rapeseed (Brassica napus L.) results from a deletion of four base pairs in the fatty acid elongase 1 gene. Theor. Appl. Genet. 2008;116:491–9. doi: 10.1007/s00122-007-0685-z. [DOI] [PubMed] [Google Scholar]
- 70.Howell P., Sharpe A., Lydiate D. Homoeologous loci control the accumulation of seed glucosinolates in oilseed rape (Brassica napus) Genome. 2003;46:454–60. doi: 10.1139/g03-028. [DOI] [PubMed] [Google Scholar]
- 71.Zhao J., Meng J. Detection of loci controlling seed glucosinolate content and their association with Sclerotinia resistance in Brassica napus. Plant Breed. 2003;122:19–23. [Google Scholar]
- 72.Hirai M.Y., Sugiyama K., Sawada Y., et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA. 2007;104:6478–83. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Augustine R., Mukhopadhyay A., Bisht N. C. Targeted silencing of BjMYB28 transcription factor gene directs development of low glucosinolate lines in oilseed Brassica juncea. Plant Biotechnol. J. 2013;11:855–66. doi: 10.1111/pbi.12078. [DOI] [PubMed] [Google Scholar]
- 74.Burns M.J., Barnes S.R., Bowman J.G., Clarke M.H., Werner C.P., Kearsey M.J. QTL analysis of an intervarietal set of substitution lines in Brassica napus: (i) seed oil content and fatty acid composition. Heredity (Edinb) 2003;90:39–48. doi: 10.1038/sj.hdy.6800176. [DOI] [PubMed] [Google Scholar]
- 75.Ecke W., Uzunova M., Weißleder K. Mapping the genome of rapeseed (Brassica napus L.). II. Localization of genes controlling erucic acid synthesis and seed oil content. Theor. Appl. Genet. 1995;91:972–7. doi: 10.1007/BF00223908. [DOI] [PubMed] [Google Scholar]
- 76.Qiu D., Morgan C., Shi J., et al. A comparative linkage map of oilseed rape and its use for QTL analysis of seed oil and erucic acid content. Theor. Appl. Genet. 2006;114:67–80. doi: 10.1007/s00122-006-0411-2. [DOI] [PubMed] [Google Scholar]
- 77.Basunanda P., Radoev M., Ecke W., Friedt W., Becker H., Snowdon R. Comparative mapping of quantitative trait loci involved in heterosis for seedling and yield traits in oilseed rape (Brassica napus L.) Theor. Appl. Genet. 2010;120:271–81. doi: 10.1007/s00122-009-1133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fan C., Cai G., Qin J., et al. Mapping of quantitative trait loci and development of allele-specific markers for seed weight in Brassica napus. Theor. Appl. Genet. 2010;121:1289–301. doi: 10.1007/s00122-010-1388-4. [DOI] [PubMed] [Google Scholar]
- 79.Quijada P., Udall J., Lambert B., Osborn T. Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 1. Identification of genomic regions from winter germplasm. Theor. Appl. Genet. 2006;113:549–61. doi: 10.1007/s00122-006-0323-1. [DOI] [PubMed] [Google Scholar]
- 80.Zhang L., Yang G., Liu P., Hong D., Li S., He Q. Genetic and correlation analysis of silique-traits in Brassica napus L. by quantitative trait locus mapping. Theor. Appl. Genet. 2011;122:21–31. doi: 10.1007/s00122-010-1419-1. [DOI] [PubMed] [Google Scholar]
- 81.Weng J., Xie C., Hao Z., et al. Genome-wide association study identifies candidate genes that affect plant height in Chinese elite maize (Zea mays L.) inbred lines. PLoS ONE. 2011;6:e29229. doi: 10.1371/journal.pone.0029229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.