Abstract
Alpha-satellite DNA (AS) is a main DNA component of primate centromeres, consisting of tandemly repeated units of ∼170 bp. The AS of humans contains sequences organized into higher-order repeat (HOR) structures, in which a block of multiple repeat units forms a larger repeat unit and the larger units are repeated tandemly. The presence of HOR in AS is widely thought to be unique to hominids (family Hominidae; humans and great apes). Recently, we have identified an HOR-containing AS in the siamang, which is a small ape species belonging to the genus Symphalangus in the family Hylobatidae. This result supports the view that HOR in AS is an attribute of hominoids (superfamily Hominoidea) rather than hominids. A single example is, however, not sufficient for discussion of the evolutionary origin of HOR-containing AS. In the present study, we developed an efficient method for detecting signs of large-scale HOR and demonstrated HOR of AS in all the three other genera. Thus, AS organized into HOR occurs widely in hominoids. Our results indicate that (i) HOR-containing AS was present in the last common ancestor of hominoids or (ii) HOR-containing AS emerged independently in most or all basal branches of hominoids. We have also confirmed HOR occurrence in centromeric AS in the Hylobatidae family, which remained unclear in our previous study because of the existence of AS in subtelomeric regions, in addition to centromeres, of siamang chromosomes.
Keywords: alpha-satellite DNA, higher-order repeat, centromere, hominoids, evolutionary origin
1. Introduction
The centromere is the part of the chromosome to which spindle fibres attach via the kinetochore, and it serves as the forefront of chromosome migration during cell division. The main DNA components of centromeres are usually tandemly repeated sequences, and the most abundant sequence in primate centromeres is alpha-satellite DNA (AS), which has basic repeat units of ∼170 bp in length.1,2 Human AS has been classified into two types according to the organization and nucleotide sequence properties of its repeat units.3,4 One is monomeric AS, which is a simple repetition of the basic repeat units. The other is higher-order AS, whose sequence is organized into a higher-order repeat (HOR) structure, in which a block of multiple basic repeat units forms a larger repeat unit. In a typical HOR structure, any combination of two basic repeat units within the larger repeat unit shows nucleotide sequence identities of 70–90%, whereas the identity often exceeds 95% between a basic repeat unit and its counterpart (located at the same position) in another larger unit. Higher-order AS is known to be present in all normal human centromeres,4 to occupy central portions of centromeres (whereas monomeric AS is found mostly in peripheral regions),5 and to be more important than monomeric AS in centromere function.3,4,6 It is unclear, however, what aspects of higher-order AS are associated with these facts. One approach to identifying these associations would be to clarify the history of higher-order AS, including its evolutionary origin and expansion. The present study addresses the evolutionary origin of higher-order AS.
Humans (genus Homo) are included in hominids (family Hominidae), along with chimpanzees (genus Pan), gorillas (genus Gorilla), and other great apes. Small apes (gibbons) are not included in hominids and form another taxon (family Hylobatidae). Collectively, organisms included in these two families are called hominoids (superfamily Hominoidea). All hominids examined to date carry higher-order AS.7,8 In small apes, however, AS of this type had not been found until we recently reported one in the siamang (Sympha-langus syndactylus).9
Although the HOR structure observed in siamang AS was clear (HOR units consisting of four or six basic repeat units), there remained two unresolved questions: (i) whether HOR structures are common among small apes or are limited to the siamang and (ii) whether AS residing in the centromere (without migrating to another chromosomal position) exhibits HOR structures or if HOR is limited to non-centromeric AS in small apes. The second question arose because of the fact that the siamang carries AS in terminal regions of chromosomes as large blocks of constitutive heterochromatin as well as centromeres.10 The function and formation processes of these heterochromatin blocks are not yet clear. Similar structures have also been observed in chromosomes of chimpanzees and gorillas. However, their DNA component is not AS, but is an unrelated tandem repeat sequence (called the StSat sequence) comprising 32-bp repeat units.11,12 In the present study, we obtained results that can be used to answer the two questions described above. Our results indicate that higher-order AS widely occurs in hominoids.
Structures of tandem repeat sequences are difficult to accurately determine using data from whole-genome shotgun sequencing or next-generation sequencing, because of difficulty in assembling sequence reads into contigs. Primer walking on a cloned DNA fragment is also inadequate because of the presence of multiple primer annealing sites in the clone. In fact, centromere regions are left as large gaps even in the human genome database.13,14 To overcome this problem, we used a traditional method consisting of cloning long genomic DNA fragments, preparation of deletion clones, and sequencing end regions of these clones. There was, however, another problem to be solved before sequencing analyses: which clones should be sequenced? Our idea was to detect signs of HOR structures by comparing sequences of two different regions within a clone. For this purpose, we used the transposition reaction of a bacterial transposon, which provided AS sequences with unique sites for annealing by sequencing primers. Below, we describe our methods, results, and their significance with regard to the evolutionary origin of higher-order AS.
2. Materials and methods
2.1. Animals for collection of genomic DNA
In the latest taxonomy of small apes, the family Hylobatidae consists of four genera: Hoolock, Hylobates, Nomascus, and Symphalangus.15,16 A species of the genus Symphalangus (S. syndactylus) has been examined in a previous study.10 In the present study, we focused on the other three genera. The species and the origin of the individuals that served as DNA sources were as follows: the Western hoolock gibbon (Hoolock hoolock), an adult female (named Faisal) bred at Bangabandhu Sheikh Mujib Safari Park, Bangladesh; the agile gibbon (Hylobates agilis), an adult male (named Raja) bred at the Primate Research Institute of Kyoto University; and the Southern white-cheeked gibbon (Nomascus siki), an adult female (identification number 001064376) bred at Khao Kheow Forest and Wildlife Park, Thailand.
2.2. Preparation of genomic libraries
We collected genomic DNA from white blood cells by following a standard method. A genomic library of each species was constructed, following a previously described protocol.10,17 The vector was an 8.1-kb fosmid pCC1FOS, and the inserts were 40- to 44-kb genomic DNA fragments that were generated by mechanical shearing and isolated by gel electrophoresis and subsequent recovery from a gel piece. Identification of AS-containing sequences from these libraries is described in the Results section together with strategies and results.
2.3. DNA sequencing
We performed subcloning of a part of the fosmid clones, preparation of a series of deletion clones from the subclone plasmids and sequenced the deletion clones by using the M13 universal primer. The details of our strategy are described in the Results section along with the results. The sequencing method used was Sanger's method, which involved using an Applied Biosystems 3730xl DNA Analyzer.
2.4. Fluorescent in situ hybridization analysis of chromosomes
We performed fluorescent in situ hybridization (FISH) analysis following the procedures described previously.10 Specific conditions are explained in each case.
3. Results
3.1. Collection of genomic DNA clones containing repetitive sequences
We screened the genomic libraries of the three gibbon species for repetitive sequences by the genomic hybridization method described previously.10,17 In this screening, the target of the hybridization was fosmid clones randomly selected from the library and fixed on nylon membranes. The probe was genomic DNA (fragmented by mechanical shearing in advance) of the same species as that of the target DNA. For example, fragmented genomic DNA of Hoolock hoolock was chemically labelled and used as probe in the hybridization assay for Hoolock hoolock fosmid clones. If a fosmid carries repetitive DNA (such as AS) as its insert, the same sequence occupies a large fraction of the probe DNA. A large number of labelled DNA fragments anneal to this fosmid, and the fosmid produces an intense signal. If a fosmid clone carries a single-copy region, the same sequence is present at a low frequency in the labelled DNA, and the fosmid displays a weak (or even undetectable) signal. A big advantage of this method is that any repetitive sequences are identified irrespective of their nucleotide sequences (even if the sequences are unknown), with more chances of identification (stronger signals and larger numbers of signals) for sequences of higher copy numbers.18 For each species, we screened 384 fosmid clones (four membranes corresponding to four 96-well plates) and picked up 12 clones exhibiting the highest level of signal intensities.
3.2. Selection of AS-containing clones
We sequenced one end of the clones obtained by genomic hybridization and selected clones whose sequence reads showed tandem repeat structures with unit sizes between 100 and 200 bp (the size of the basic repeat units of AS is uniform among hominoids at ∼170 bp and is included in this range). Of the 12 Hoolock, 12 Hylobates, and 12 Nomascus clones, 4, 9, and 6 clones, respectively, met this criterion. Sequence reads from all of these clones exhibited >70% nucleotide identity with the consensus sequence of human AS and with that of siamang AS.10
3.3. Detection of signs of HOR
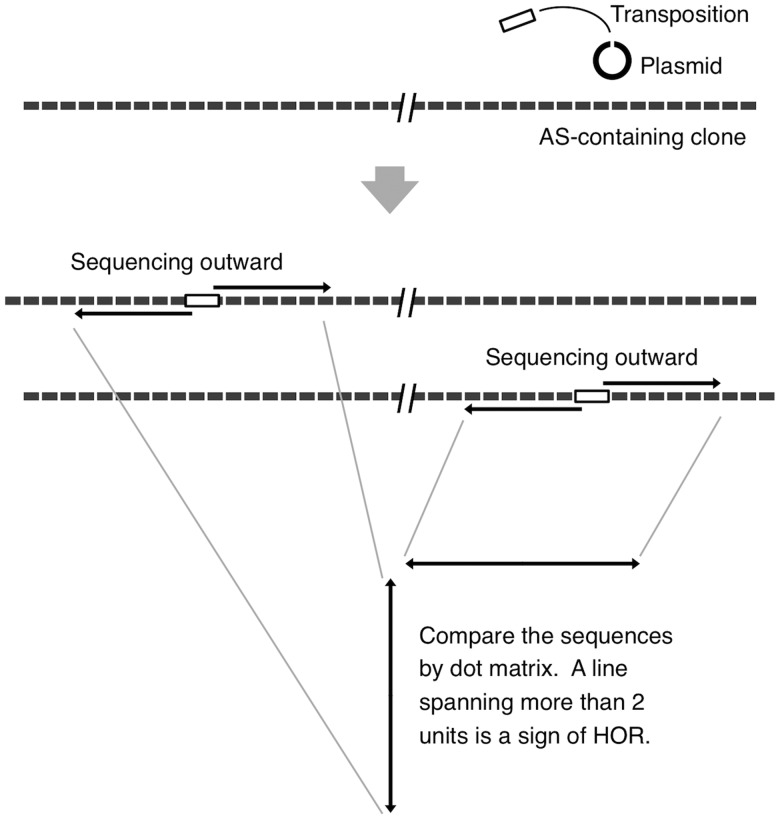
We first tried to detect HOR signs by comparing the sequences of two terminal regions (∼900 bp each), which we had successfully carried out in the case of Symphalangus in our previous study.9 This test resulted in an HOR sign in one (HylFos15) of the clones from Hylobates (data not shown). The test, however, did not give a positive result in any of the four clones from Hoolock or six clones from Nomascus, suggesting that, even if these clones contain an HOR structure, its scale (the number of basic repeat units constituting an HOR unit) is larger than that found in Symphalangus. We developed another method to obtain longer sequences, with the expectation that longer sequences would increase the possibility of detecting signs. The strategy used for this purpose is shown in Fig. 1. We prepared a clone of the Tn5 transposon that carried the kanamycin-resistance gene. We mixed it with an AS-containing genomic DNA clone and induced the transposition reaction by supplying the Tn5 transposase enzyme. The fosmid vector (pCC1FOS) for the genomic DNA clone carried the chloramphenicol-resistance gene. After the transposition reaction, the reaction mixture was introduced into bacterial cells by electroporation, and single colonies that survived on LB/agar plates containing chloramphenicol (5 µg/ml) and kanamycin (5 µg/ml) were selected. Fosmid DNA extracted from these colonies mostly carried a single Tn5 insertion at different positions. We sequenced two of these clones by using sequencing primers that annealed to the left or right terminal regions of Tn5 and were oriented outward.
Figure 1.
Strategy for the detection of HOR signs. Transposition of the Tn5 transposon to DNA molecules of an AS-containing fosmid clone was induced, and two clones carrying Tn5 at different positions were selected. Each clone was sequenced using two primers that annealed to Tn5 and oriented outward. The sequence reads (∼900 bp, including the tail region of a low reliability) were combined, and a longer sequence (∼1800 bp) was obtained. These sequences were compared between the two clones by the dot-matrix analysis. When a line spanning more than two basic units was found, it was regarded as a sign of HOR.
Each sequence read contained data for >855 (5 × 171) nucleotides. Although the tail region of the sequence read (distant by >700 nucleotides from the sequencing primer) had quite a high possibility of containing sequencing errors, this was not a problem for our purpose because the data were used for detecting signs of HOR, but sequencing errors masked such signs. By combining the sequence reads for both sides of a single Tn5 insertion, we could obtain nucleotide sequence information for a region of >1710 (10 × 171) nucleotides, and it contained sequence information for at least nine consecutive repeat units.
In each clone, we compared two sets of such sequences by dot-matrix analysis. When a line spanning more than two repeat units was found in the dot matrix, we regarded it as a sign of an HOR. The detection efficiency of this method depends on the scale of the HOR to be revealed, namely, how many (designated as n hereafter) basic repeat units constitute the HOR. For HORs with n ≤ 16, our method is expected to be powerful enough to detect a sign with only two sets of sequence data. For n ≥ 17, the probability of detecting a sign with two sets of sequence data is 15/n. The value of the probability is as large as 0.5 for n = 30 and 0.375 for n = 40. If we use sequence data of additional insertion points, we can increase the probability.
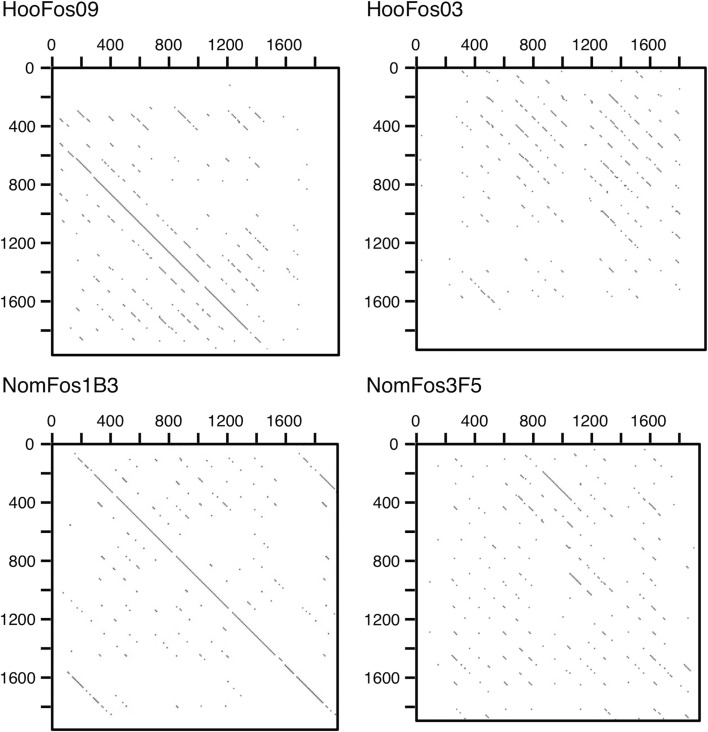
We examined the AS-containing fosmid clones by the transposition assay and subsequent sequencing analysis. Of the four Hoolock and six Nomascus clones, one and three clones, respectively, exhibited signs of HOR. Figure 2 shows examples of the results of this dot-matrix analysis. We selected one clone each (clones HooFos09 of Hoolock and NomFos1B3 of Nomascus) and moved on to determining the long sequences of the AS arrays contained in these two clones, and also the clone HylFos15 from Hylobates.
Figure 2.
Dot-matrix analysis of clone partial sequences for signs of HOR. The criterion in the dot-matrix analysis was that a 19-nucleotide match should exist over a window of 20 nucleotides. Examples of the results are shown. HooFos09 and NomFos1B3 exhibited a line spanning more than two basic repeat units, and HooFos03 and NomFos3F5 did not.
3.4. Transfer of fragments to plasmid
As explained earlier, sequencing strategies relying on shotgun segmentation or primer walking are not adequate for tandemly repeated sequences, such as AS. The strategy we employed was preparation of a series of deletion clones by using exonuclease III and Mung bean nuclease.19 By digesting the DNA fragment with an appropriate combination of restriction endonucleases (KpnI whose 3′-protruding cutting site is resistant to the exonuclease, and BamHI or SalI whose 5′-protruding cutting sites are sensitive), the deletion starts at one end of the fragment and continues to various degrees. The maximum fragment size for which these treatments work smoothly is ∼16 kb. The size of the fosmid clones was 40–44 kb. Therefore, it was necessary to transfer part of the fosmid clones into a plasmid vector. For delimiting the part to be analysed, PCR was not adequate because of the repetitive nature of the object sequence.
Restriction endonuclease NotI recognizes an eight-nucleotide segment (GCGGCCGC), which rarely appears even in the insert size of the fosmid vector. The vector carried two NotI recognition sites located closely to its cloning site, and we manipulated the Tn5 transposon in advance to have an NotI site in it. These NotI sites enabled us to determine the position of the Tn5 insertion by cutting Tn5-carrying fosmid DNAs and running an electrophoresis gel. We checked 12 Tn5-carrying clones originating from the HooFos09, HylFos15, and NomFos1B3 clones, and selected one in which Tn5 had been inserted at a position 10–16 kb distant from one end of its AS array. We then subcloned the NotI-delimited DNA fragment into the NotI site of an ampicillin-resistant plasmid (pBluescript II SK+), and subsequently transferred the fragment to a chloramphenicol-resistant plasmid (originating from pHSG399) to which we had, in advance, added some restriction enzyme sites necessary for the preparation of deletion clones. We used the ampicillin-resistant plasmid because transfer of a long fragment from a chloramphenicol-resistant vector to another chloramphenicol-resistant vector becomes easier by using an intermediate vector carrying a resistance gene to a different antibiotic (ampicillin in this case). The plasmid clones carrying the AS fragments from the HooFos09, HylFos15, and NomFos1B3 clones are designated HooCam09, HylCam15, and NomCam1B3, respectively.
3.5. Sequencing analysis
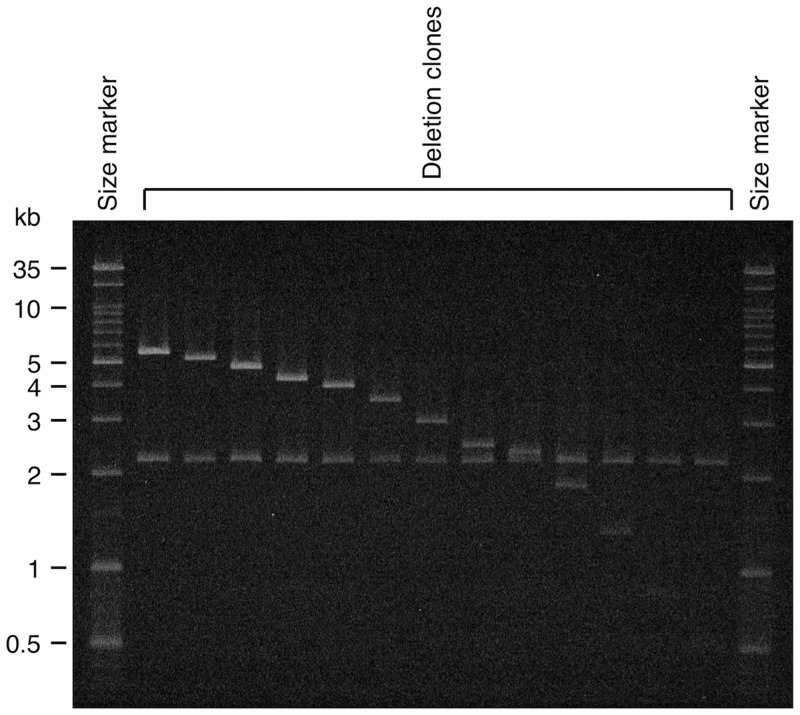
We prepared deletion clones of various lengths by treating HooCam09, HylCam15, and NomCam1B3 with exonuclease III and then with Mung bean nuclease. We checked the insert size of these deletion clones, arranged them in the order of longest to shortest, and selected 55, 28, and 41 clones, respectively, to be used as sources of partial sequences. The criterion for this selection was any two adjacent clones that did not differ in insert size by >0.5 kb. Figure 3 shows an example of gel electrophoresis in which clones of 5.3–0.5 kb were placed according to their sizes. We sequenced the selected clones by using the M13 universal primer. We then manually assembled the obtained sequence reads into a contig sequence.
Figure 3.
Confirmation of deletion clone insert size. Deletion clones of various sizes were prepared from the HooCam09 and NomCam1B3 clones. Out of these pools, clones of sizes adequate for sequencing analysis were selected. This photograph is an example of gel electrophoresis in which deletion clones of insert sizes of 5.3–0.5 kb, originating from HooCam09, are contained. The 2.1-kb vector plasmid carried two cutting sites for restriction endonuclease XhoI just outside of the cloning site, and the insert fragments in the original clones did not have a site for this enzyme. Each deletion clone was digested with XhoI and divided into two fragments: the 2.1-kb plasmid vector and its insert fragment.
3.6. Evidence for HORs
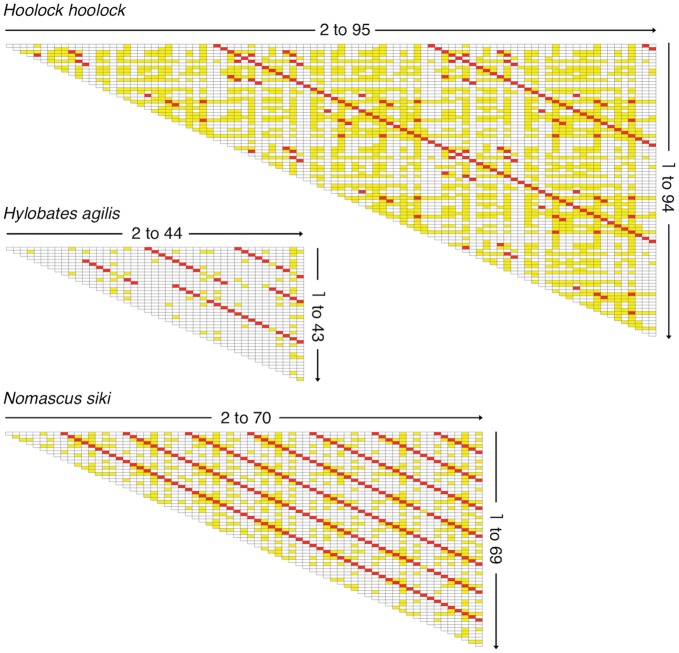
We obtained a 16 429-bp contig sequence for the HooCam09 clone, a 7828-bp contig sequence for the HylCam15 clone, and a 12 093-bp contig sequence for the NomCam1B3 clone. These were deposited in DDBJ with accession numbers AB861929, AB900117, and AB846956, respectively. We defined the boundaries and orientation of the basic repeat unit, so that they would be the same as those used in our previous studies.10,20 The three clones contained 95, 44, and 70 basic repeat units, respectively.
We made pairwise comparisons between basic repeat units, and the essence of the obtained results is shown as matrices in Fig. 4. In the matrices, red and yellow cells indicate identities of >95 and 90–95%, respectively. Matrices containing the nucleotide identity of each cell are shown in Supplementary Figs S1–S3. In HooCam09 and NomCam1B3, clear step-like lines consisting of red cells appeared with a constant interval throughout the sequence. The interval was 31 basic repeat units for HooCam09 and 9 basic repeat units for NomCam1B3. HylCam15 showed step-like lines exhibiting two types of intervals (8-unit interval for units 5–12 and 13-unit interval for units 13–44). Thus, it is evident that the AS sequences contained in these clones are organized into HOR structures.
Figure 4.
Pairwise comparisons of basic repeat unit sequences. The comparisons were made using the MEGA5 program21 under default settings. The horizontal and vertical axes of each matrix represent consecutive basic repeat units contained in the AS contig sequence. Cells showing nucleotide identities of 90–95 and >95% are indicated by yellow and red, respectively. The same matrices containing the identity values in the cells are shown in Supplementary Figs S1–S3.
From the matrices shown in Fig. 4, we picked up cells that represented pairs of basic units located at identical positions in different HOR units (e.g. the second basic repeat unit in the third HOR unit and the second basic repeat unit in the fourth HOR unit). In the HooCam09 clone, the average pairwise nucleotide identity among these pairs was 99.8%. The average value among the pairs of the other relationships was 89.2%. In the NomCam1B3 clone, the values were 98.6 and 88.0%, respectively. In the analysis of the HylCam15 clone, in which the 13th–44th basic repeat units (exhibiting a 13-unit interval) were used, we obtained the values of 97.0 and 86.5%, respectively. These situations are similar to those observed in human AS sequences: 95–99 and 70–90%, respectively.22
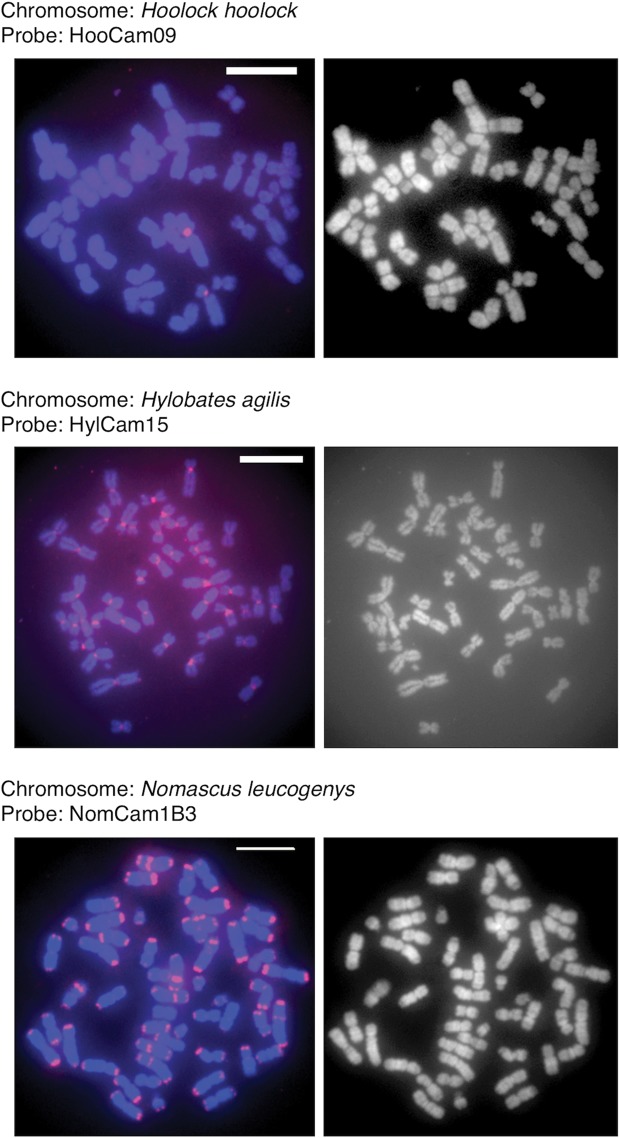
3.7. Chromosomal locations
We conducted FISH analysis of chromosomes, by using HooCam09, HylCam15, and NomCam1B3 as probes, to identify chromosomal locations (Fig. 5). HooCam09 exhibited strong signals on the centromere region of chromosome 4 of Hoolock hoolock chromosome spreads. HylCam15 produced signals in the centromere regions of all Hylobates agilis chromosomes. NomCam1B3 showed clear signals on the centromere regions and terminal regions of most chromosomes of Nomascus siki. These results are consistent with the FISH results we obtained previously by using a clone of siamang AS as a probe.20
Figure 5.
FISH analysis of chromosomes for locations of AS. The left panels show FISH images obtained using a fluorescence scanner, and the right panels are images of the same chromosome spreads stained with 4′,6-diamidino-2-phenylindole (DAPI). In the left panels, red signals indicate hybridization of the probe DNA to chromosome DNA. The combination of chromosome spread and probe is shown on each left panel. The bar in the panels represents 10 µm. All experimental procedures were the same as those described in our previous work.10
3.8. Comparison of AS sequences among hominoid species
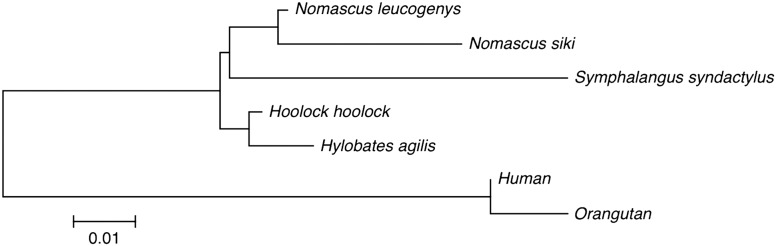
We made within-species consensus sequences of AS repeat units of Hoolock hoolock, Hylobates agilis, and Nomascus siki. Furthermore, from references, we obtained consensus sequence for Nomascus leucogenys, S. syndactylus, human, and orangutan. We then made a neighbour-joining phylogenetic tree of these consensus sequences, which is shown in Fig. 6. The five gibbon species representing four genera formed a cluster distinct from that of the two hominid species, and the two Nomascus species formed an internal cluster within the gibbon cluster.
Figure 6.
Neighbour-joining phylogenetic tree of within-species consensus sequences. The consensus sequences of AS of Hoolock hoolock, Hylobates agilis and Nomascus siki were obtained by sending the contig sequences, which we determined in the present study, to the Consensus Maker program (version 2.0.0; http://www.hiv.lanl.gov/content/sequence/CONSENSUS/consensus.html). The consensus sequences obtained are included in the respective DDBJ files. The consensus sequences for Nomascus leucogenys,23 Symphalangus syndactylus,9 human22, and orangutan7 AS were obtained from the respective references. The neighbour-joining phylogenetic tree of these within-species consensus sequences was made using the ClustalW program contained in MEGA.21.
4. Discussion
In addition to humans, higher-order AS has also been observed in all great ape species examined to date, although it had not been found in small apes (except for our recent report) or other primate taxa more distant from humans. Based on this distribution in primates, it is widely postulated that higher-order AS arose in the common ancestor of hominids.
We have demonstrated the presence of high-order AS in all of the four genera of small apes. Thus, higher-order AS occurs widely in hominoids. The species of the Symphalangus genus that we previously examined was shown to have higher-order AS containing four or six basic repeat units in its HOR units. In the present study, we found clear HOR structures in the AS of species of Hoolock and Nomascus, in which 31 and 9 basic repeat units formed HOR units, respectively. HOR structures were also found in a species of Hylobates, although two types of HORs (8 and 13 basic repeat units) coexisted in a single clone.
Nomascus leucogenys (Northern white-cheeked gibbon) has been extensively examined in the past for HOR structures by computational analysis of its genome sequence database and additional FISH experiments, but no positive evidence was obtained.23 In the present study, we have identified HOR in AS of N. siki (Southern white-cheeked gibbon). These two species are so close phylogenetically that they have been regarded as subspecies of one species until about a decade ago (N. leucogenys leucogenys and N. leucogenys siki, respectively). The most probable reason for this difference of results would be the difference in the method used; we relied on traditional sequencing of long clones preceded by detection of HOR signs. Another possible factor is the difference in the gibbon species. Satellite DNA is known to undergo rapid fluctuation in copy number. The HOR-containing AS we found may have been amplified in a small number of generations in the lineage leading to N. siki (or the individual we used), or its copy number may have been reduced rapidly in the lineage for N. leucogenys (or the individual used by those authors).
Our strategy included a screening step of AS-containing fosmid clones for those exhibiting HOR signs, in which small parts of candidate clones were sequenced. We examined four Hoolock and six Nomascus clones by the method newly developed in the present study, and one and three clones, respectively, exhibited an HOR sign. Considering that our library screening did not involve a factor to favour HOR-containing AS clones, the HOR structure does not appear to be rare in AS sequences contained in their genomes. The lack of a sign does not necessarily mean that the AS clones examined have no HOR structure. In our method, as we described in the Results section, the probability of finding a sign in a single fosmid clone is 15/n, where n is the number of basic repeat units constituting the HOR unit. If n is as large as 100, it would be required to increase the number of fosmid clones to be examined or the number of Tn5 insertions in individual clones. We did not attempt this in the present study because we wanted to develop another, more efficient method.
Another significance of our results is that we found HOR structures in AS residing in the centromere. This question remained from our previous study,9 because Symphalangus syndactylus carries AS in terminal regions of chromosomes in addition to in centromeres. The species of genus Nomascus examined so far also contain AS in terminal regions.20,23 In contrast, such structures are not found in Hoolock hoolock and Hylobates agilis,20 and, therefore, the possibility that the fosmid clones we sequenced originate from chromosomal portions other than the centromere would be negligible.
The currently dominant view about the evolutionary origin of HOR-containing AS is that it arose in the common ancestor of hominids (family Hominidae). The evidence of its widespread occurrence in the other family (Hylobatidae) of hominoids requires reconsideration of the currently dominant view. Likely explanations for the history of HOR-containing AS are as follows: (i) HOR-containing AS was present in the last common ancestor of hominoids and (ii) HOR-containing AS emerged in most or all basal branches of hominoids. As a more parsimonious explanation, (i) would be preferred.
The neighbour-joining phylogenetic tree of AS sequences is consistent with the widely accepted phylogenetic relationship of the host species. This neighbour-joining phylogenetic tree is, however, not powerful enough to extract information concerning the history of HOR structures, because the source sequences of the gibbon species (except for the case of Nomascus leucogenys) are limited to higher-order AS, while the consensus sequences of the hominid species represent both higher-order AS and monomeric AS.
The purpose of the present study was to show evidence, if any, for HOR-containing AS in multiple gibbon genera, which was attained with lines clearly appearing in constant intervals in the pairwise comparison tables. It is now of interest whether AS sequences of gibbons exhibit within-species variation in the HOR structure, and, if so, how large the variation is. In hominids, the within-species HOR structure variation is larger in humans than in great ape species, and the reason for the higher level of variation in humans is not known. Further studies by identifying different types of HOR structures in gibbons are expected to contribute to the understanding of factors leading to the high level of variation of AS structure in humans, and factors that are associated with the more important role of higher-order AS than monomeric AS in centromere function.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This study was supported by Grants-in-Aid (23657165 and 23470098 to A.K. and 22247037 to H.H.) from the Japan Society for the Promotion of Science.
Supplementary Material
Acknowledgements
We are grateful to Hiroshi Masumoto and Masanaru Takai for helpful discussions.
References
- 1.Willard H.F. Evolution of alpha satellite. Curr. Opin. Genet. Dev. 1991;1:509–14. doi: 10.1016/s0959-437x(05)80200-x. [DOI] [PubMed] [Google Scholar]
- 2.Rudd M.K., Wray G.A., Willard H.F. The evolutionary dynamics of alpha-satellite. Genome Res. 2006;16:88–96. doi: 10.1101/gr.3810906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Willard H.F. Chromosome-specific organization of human alpha satellite DNA. Am. J. Hum. Genet. 1985;37:524–32. [PMC free article] [PubMed] [Google Scholar]
- 4.Alexandrov I., Kazakov A., Tumeneva I., Shepelev V., Yurov Y. Alpha-satellite DNA of primates: old and new families. Chromosoma. 2001;110:253–66. doi: 10.1007/s004120100146. [DOI] [PubMed] [Google Scholar]
- 5.Rudd M.K., Willard H.F. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 2004;20:529–33. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 6.Spence J.M., Critcher R., Ebersole T.A., et al. Co-localization of centromere activity, proteins and topoisomerase II within a subdomain of the major human X α-satellite array. EMBO J. 2002;21:5269–80. doi: 10.1093/emboj/cdf511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haaf T., Willard H.F. Orangutan alpha-satellite monomers are closely related to the human consensus sequence. Mamm. Genome. 1998;9:440–7. doi: 10.1007/s003359900793. [DOI] [PubMed] [Google Scholar]
- 8.Waye J.S., Willard H.F. Concerted evolution of alpha satellite DNA: evidence for species specificity and a general lack of sequence conservation among alphoid sequences of higher primates. Chromosoma. 1989;98:273–9. doi: 10.1007/BF00327313. [DOI] [PubMed] [Google Scholar]
- 9.Terada S., Hirai Y., Hirai H., Koga A. Higher-order repeat structure in alpha satellite DNA is an attribute of hominoids rather than hominids. J. Hum. Genet. 2013 doi: 10.1038/jhg.2013.87. doi:10.1038/jhg.2013.87. [DOI] [PubMed] [Google Scholar]
- 10.Koga A., Hirai Y., Hara T., Hirai H. Repetitive sequences originating from the centromere constitute large-scale heterochromatin in the telomere region in the siamang, a small ape. Heredity. 2012;109:180–7. doi: 10.1038/hdy.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Royle N.J., Baird D.M., Jeffreys A.J. A subterminal satellite located adjacent to telomeres in chimpanzees is absent from the human genome. Nat. Genet. 1994;6:52–6. doi: 10.1038/ng0194-52. [DOI] [PubMed] [Google Scholar]
- 12.Koga A., Notohara M., Hirai H. Evolution of subterminal satellite (StSat) repeats in hominids. Genetica. 2011;139:167–75. doi: 10.1007/s10709-010-9534-0. [DOI] [PubMed] [Google Scholar]
- 13.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 14.Eichler E.E., Clark R.A., She X. An assessment of the sequence gaps: unfinished business in a finished human genome. Nat. Rev. Genet. 2004;5:345–54. doi: 10.1038/nrg1322. [DOI] [PubMed] [Google Scholar]
- 15.Brandon-Jones D., Eudey A.A., Geissmann T., et al. Asian primate classification. Int. J. Primatol. 2004;25:97–164. [Google Scholar]
- 16.Thinh V.N., Mootnick A.R., Geissmann T., et al. Mitochondrial evidence for multiple radiations in the evolutionary history of small apes. BMC Evol. Biol. 2010;10:74. doi: 10.1186/1471-2148-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prakhongcheep O., Hirai Y., Hara T., Srikulnath K., Hirai H., Koga A. Two types of alpha satellite DNA in distinct chromosomal locations in Azara's owl monkey. DNA Res. 2013;20:235–40. doi: 10.1093/dnares/dst004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prakhongcheep O., Chaiprasertsri N., Terada S., et al. Heterochromatin blocks constituting the entire short arms of acrocentric chromosomes of Azara's owl monkey: formation processes inferred from chromosomal locations. DNA Res. 2013;20:461–70. doi: 10.1093/dnares/dst023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–9. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 20.Baicharoen S., Arsaithamkul V., Hirai Y., Hara T., Koga A., Hirai H. In situ hybridization analysis of gibbon chromosomes suggests that amplification of alpha satellite DNA in the telomere region is confined to two of the four genera. Genome. 2012;55:809–12. doi: 10.1139/gen-2012-0123. [DOI] [PubMed] [Google Scholar]
- 21.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov I.A., Medvedev L.I., Mashkova T.D., Kisselev L.L., Romanova L.Y., Yurov Y.B. Definition of a new alpha satellite suprachromosomal family characterized by monomeric organization. Nucleic Acids Res. 1993;21:2209–15. doi: 10.1093/nar/21.9.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cellamare A., Catacchio C.R., Alkan C., et al. New insights into centromere organization and evolution from the white-cheeked gibbon and marmoset. Mol. Biol. Evol. 2009;26:1889–900. doi: 10.1093/molbev/msp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.