Abstract
Plasmodium falciparum malaria imposes a serious public health concern throughout the tropics. Although genetic tools are principally important to fully investigate malaria parasites, currently available forward and reverse tools are fairly limited. It is expected that parasites with a high mutation rate can readily acquire novel phenotypes/traits; however, they remain an untapped tool for malaria biology. Here, we generated a mutator malaria parasite (hereinafter called a ‘malaria mutator’), using site-directed mutagenesis and gene transfection techniques. A mutator Plasmodium berghei line with a defective proofreading 3′ → 5′ exonuclease activity in DNA polymerase δ (referred to as PbMut) and a control P. berghei line with wild-type DNA polymerase δ (referred to as PbCtl) were maintained by weekly passage in ddY mice for 122 weeks. High-throughput genome sequencing analysis revealed that two PbMut lines had 175–178 mutations and a 86- to 90-fold higher mutation rate than that of a PbCtl line. PbMut, PbCtl, and their parent strain, PbWT, showed similar course of infection. Interestingly, PbMut lost the ability to form gametocytes during serial passages. We believe that the malaria mutator system could provide a novel and useful tool to investigate malaria biology.
Keywords: mutator, Plasmodium, DNA polymerase δ, genome sequencing
1. Introduction
Malaria imposes a serious public health concern throughout the tropics with estimated 1.2 million deaths worldwide in 2010, due mostly to infections of Plasmodium falciparum, the most virulent human malaria parasite.1 This parasite has developed drug resistance to almost all conventional antimalarial drugs.2 Development of malaria vaccines targeting both insect and blood stage parasites is still ongoing.3,4 These circumstances urge us to further pursue effective intervention strategies to combat malaria. However, our knowledge of the parasite biology to identify new drug and vaccine targets is still inadequate. Current genetic tools, although limited, provided opportunities to specifically alter the parasite genome to explore its biology and gain new insights into gene function and expression.5–11 A combination approach that exploits the advantages of different genetic tools may provide the best strategy to study malaria parasite biology.
Important biological phenotypes/traits of malaria parasites like drug resistance are often caused by a point mutation(s).12–14 If the mutation rate of malaria parasites is artificially increased, it is expected that parasites can rapidly acquire novel phenotypes/traits that can be selected for under appropriate selectable conditions: e.g. drug resistance would be readily produced under the pressure of a drug of interest. To date, little attempts have been made to utilize such parasites with an accelerated mutation rate to study malaria parasite biology.
In eukaryotic organisms, the spontaneous mutation rate is very low, e.g. 3.3 × 10−10/base pair (bp)/cell division in yeast,15 2.7 × 10−9/bp/generation in Caenorhabditis elegans,16 and 5.8 × 10−9/bp/generation in Drosophila melanogaster.17 The mutation rate of in vitro P. falciparum culture was recently reported as 1.0–9.7 × 10−9/bp/generation.18 These low mutation rates are ensured by the high fidelity of DNA replication due to the strict selectivity of correct bases and the proofreading function of DNA polymerases, as well as the mismatch repair system. DNA polymerases α, δ, and ε are involved in nuclear genome DNA synthesis, of which polymerases δ and ε possess a proofreading 3′ → 5′ exonuclease activity for lagging and leading DNA strand replication, respectively. The proofreading activity domain of DNA polymerases δ and ε is widely conserved from prokaryotes to eukaryotes,19,20 including Plasmodium.21 Disruption of the proofreading activity by site-directed mutagenesis significantly increased a mutation rate by as much as 10-fold to >100-fold,20,22,23 and the mutator effect is particularly strong in DNA polymerase δ.20,24 Furthermore, in microorganisms such as yeast and fungi, the mutator system has been employed to generate various mutated phenotypes including drug resistance.25–28
In the present study, we generated a mutator malaria parasite (hereinafter called a ‘malaria mutator’) using a rodent malaria parasite, Plasmodium berghei ANKA by eliminating the proofreading activity of its DNA polymerase δ and performed genome-wide analysis of mutations in order to see whether mutation rate in the mutator increased as expected. The mutation rate of the malaria mutator was 86- to 90-fold higher than the control parasite.
2. Materials and methods
2.1. Generation of mutator parasites and its phenotypic analyses
The rodent malaria parasite, P. berghei ANKA strain (clone 2.34), was used. The plasmid and primer sequences used for generation of mutator parasite are described in Supplementary Data (Supplementary Table S1). In brief, the two amino acids critical for proofreading in DNA polymerase δ were replaced with alanine in the mutator parasites. The phenotype analyses (growth rate, gametocytogenesis, male gametogenesis, and ookinete formation) of the mutator parasites were performed as described in Supplementary Data.
2.2. Maintenance of parasites in mice
Mutator (referred to PbMut) and control parasites (PbCtl) were maintained by weekly passages in mice. We injected intraperitoneally 100 or 1,000 infected erythrocytes into two female ddY mice (6- to 10-week-old; Japan SLC), respectively. Infected blood of either mouse reaching adequate parasitaemia (several percent) was collected 1 week later by cardiac puncture and used for the next passage. Cloning of parasites by limiting dilution was done in the 53rd and 119th week of passage for PbMut, and in the 43rd and 119th week of passage for PbCtl.
2.3. High-throughput genome sequencing
Genomic DNA from the cloned lines and PbWT (Supplementary Data) was sequenced genome wide using an Illumina HiSeq 2000 or an Illumina Genome Analyzer II. Paired-end read sequencing was performed using read lengths of 90 or 100 bases. Low quality reads with an N-rate of >10% and/or reads for >50% all bases with a PHRED quality score of ≤5 were removed. To exclude mouse DNA-derived reads, the sequence reads were aligned to the Mus musculus (C57BL/6J) genome sequence from the NCBI Genome database using SOAP2,29 and matched reads were removed.
2.4. Read mapping and mutation identification
The sequence data were further processed using two pipelines, i.e. one by read alignment using Bowtie version 0.12.730 and variant calling with VarScan version 2.2.8,31 and the other with the CLC Genomics Workbench software (CLC Bio). We used the genome sequence (18.2 Mb) of the P. berghei ANKA strain32 as a reference sequence, which was downloaded from the Wellcome Trust Sanger Institute website (http://www.sanger.ac.uk/). Further details are provided in Supplementary Data. Mutations detected by Illumina genome sequencing were validated preliminarily by Sanger sequencing. For comparison, all insertions and deletions (indels), all base substitutions causing non-sense mutations, and all base substitutions detected using either of two pipelines were validated preliminarily by Sanger sequencing (Supplementary Table S2). The Sanger sequencing confirmed all the mutations (data not shown). Thus, we inferred that the sequences produced by Illumina sequencing were consistent.
2.5. Detection of mutation clusters in a chromosome
The r-scan statistics was used to detect significant clustering of mutations along each chromosome as described previously.33 We used two probability cut-offs: 1 and 5%. Overlapping clustering regions were considered as a single mutation cluster region.
2.6. Estimation of mutation rates
We estimated the mutation rate using the base substitution data generated by the Bowtie and VarScan pipeline. The mutation rate was calculated using the equation µbs = m/(nT), where µbs is the base substitution rate (per nucleotide site per day), m is the number of mutations detected, n is the number of nucleotide sites, and T corresponds to the passage days.34 The mutation rate was calculated for the following specific regions: synonymous sites (Syn) and non-synonymous sites (NonSyn) in protein-coding regions; introns; intergenic regions; and the overall sequences. Syn and NonSyn sites were estimated as described by Nei and Gojobori.35 The mutation rate per replication was also calculated. In the ddY mice used here, P. berghei multiplied ∼10-fold per day during the logarithmic growth phase, and hence, we assumed log210 (≈3.3) replications per day. We considered that the number of base substitutions were the same as that of point mutations because no complicated mutations were found at a given substituted site. Fisher's exact test for the 2 × 2 contingency table was performed to assess statistical significance of difference in variant proportion between specific regions (i.e. Syn vs. NonSyn, NonSyn vs. intron, and intron vs. Syn). For this test, we rounded the numbers of Syn and NonSyn sites to unit.
2.7. Data access
The raw sequence reads used in this study are available from the DDBJ Sequence Read Archive (DRA) under accession No. DDBJ: DRA000656. The information for P. berghei genes discussed in this publication is available from PlasmoDB (www.plasmodb.org).
3. Results
3.1. Generation of malaria mutator
Using site-directed mutagenesis and gene transfection techniques, we established a mutator P. berghei line where DNA polymerase δ had a defective proofreading activity (referred to as PbMut) and a control P. berghei line with wild-type (non-mutated) DNA polymerase δ (referred to as PbCtl) (Supplementary Fig. S1). These parasite lines were then subjected to serial passages and cloning, and two PbMut lines (M122A and M122B) and one PbCtl line (C122), as well as one PbWT line, were obtained (Supplementary Fig. S2).
3.2. Mutations generated in the malaria mutator
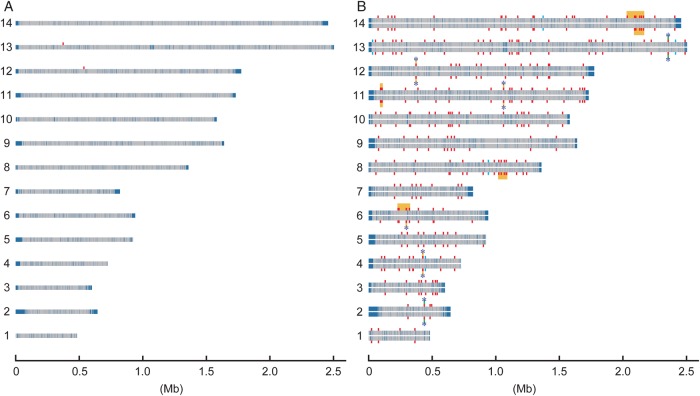
High-throughput genome sequencing of PbMut lines (M122A and M122B), one PbCtl line (C122), and one PbWT line produced each 0.9–1.2 Gb, which corresponded to ∼50-fold redundancy relative to the P. berghei ANKA genome sequence from the Wellcome Trust Sanger Institute (18.2 Mb) (Supplementary Table S3). We reconstructed ∼86–95% sequence regions that encompassed the 14 chromosomes and analysed them for mutations generated in PbMut and PbCtl. Preliminary analysis revealed that PbWT (clone 2.34) used in this study had 71 substitutions compared with the Wellcome Trust Sanger ANKA reference sequence (Supplementary Table S4). It is not certain whether these differences are true substitutions or due to artefacts such as mapping errors and/or sequencing/assembly errors from the lower coverage (×4) reference genome sequence.32 We therefore compared the sequence of PbWT (clone 2.34) with those of PbMut and PbCtl lines, and different sites were regarded as variants that occurred in each line. The number of point mutations in PbMut and PbCtl differed dramatically (Table 1, Fig. 1, and Supplementary Table S5). We only detected two base substitutions in C122, whereas 178 and 175 base substitutions were detected in M122A and M122B, respectively (Supplementary Table S5). A large proportion of the substitutions (n = 162) were shared by the two PbMut lines. This high rate of shared mutations between the two PbMut clones was probably due to a severe bottleneck effect during each mouse passage, where 100–1,000 infected erythrocytes were inoculated intraperitoneally, corresponding to intravenous injection of 10–100 infected erythrocytes. There were 107 and 110 base substitutions in the protein-coding gene regions of M122A and M122B, respectively. Most of these substitutions, 89 and 92 substitutions in M122A and M122B, respectively, were NonSyn substitutions (missense mutations).
Table 1.
Summary of substitutions in the P. berghei control and mutator clonesa
| Control | Mutator |
||
|---|---|---|---|
| C122b | M122Ab | M122Bb | |
| Total number of sites | 16.5M | 16.1M | 16.6M |
| Number of base substitutions | 2 | 176 | 173 |
| Total number of sites in protein-coding regions | 9.7M | 9.6M | 9.7M |
| Number of substitutions in protein-coding regions | 2 | 105 | 108 |
| dSc | 0 | 1.1 × 10−8 | 1.1 × 10−8 |
| Total number of Syn sites | 1.9M | 1.9M | 1.9M |
| Number of Syn substitutions | 0 | 18 | 18 |
| dNc | 2.9 × 10−10 | 1.3 × 10−8 | 1.3 × 10−8 |
| Total number of NonSyn sites | 7.8M | 7.7M | 7.8M |
| Number of NonSyn substitutions | 2(0)d | 87(3)d | 90(3)d |
| Total number of sites in introns | 1.1M | 1.1M | 1.1M |
| Number of substitutions in introns | 0 | 8 | 7 |
| Total number of sites in intergenic regions | 5.6M | 5.4M | 5.7M |
| Number of substitutions in intergenic regions | 0 | 63 | 58 |
aBase substitutions detected from alignments by the Bowtie software are listed.
bNumbers followed by ‘C’ (control) and ‘M’ (mutator) indicate the number of weeks of mouse passaging for each clone.
cdS and dN are the number of Syn substitutions per Syn sites and the number of NonSyn substitutions per NonSyn sites, respectively.
dIn parenthesis, the number of substitutions resulting in non-sense mutation is shown.
Figure 1.
Distribution of mutations on the 14 chromosomes of P. berghei control and mutator lines. (A) Mutations in a P. berghei control line (C122). (B) Mutations in two P. berghei mutator lines (the upper and lower horizontal bars represent lines M122A and M122B, respectively). The 14 chromosomes are represented by horizontal bars with grey and dark blue blocks. The grey blocks indicate the sequence regions used for mapping to detect mutations, and the dark blue blocks were those regions not used for mapping because of their lower coverage (see Materials and Methods for details). The red vertical bars indicate point mutations detected using the Bowtie program. The cyan vertical bars indicate short indels detected by the CLC Genomics Workbench software. The green and yellow blocks indicate clusters of mutations with the significance levels of P < 0.01 and <0.05, respectively. Asterisks show closely spaced vertical bars that could not be separated at the resolution scale used for this figure.
There were five non-sense mutations: in the serine/threonine protein kinase gene (PBANKA_090380), the secreted ookinete protein gene (PBANKA_123360), and three genes with unknown functions (Table 2). PbMut lines also had seven to eight base substitutions in introns and 58–63 base substitutions in intergenic regions. PbCtl had two NonSyn substitutions in genes with unknown function (Supplementary Table S5). Five single-base indel mutations were detected in single-nucleotide repeat regions in PbMut lines (Table 3), but none in the PbCtl line. Two were found in protein-coding genes, causing frameshifts in the phosphatidylinositol transfer protein gene (PBANKA_136380) and a gene encoding for ApiAP2 (PBANBKA_143750).
Table 2.
Non-sense mutations in P. berghei mutator clones
| Gene ID | Product (total amino acid length) | AA position | Clone |
|---|---|---|---|
| PBANKA_083220 | Conserved Plasmodium protein, unknown function (300) | E87 | M122B |
| PBANKA_090380 | Serine/threonine protein kinase (965) | E677 | M122Aa, M122Ba |
| PBANKA_113490 | Conserved Plasmodium protein, unknown function (870) | E336 | M122A, M122B |
| PBANKA_123360 | Secreted ookinete protein (205) | E144 | M122A |
| PBANKA_123730 | Conserved Plasmodium protein, unknown function (486) | S465 | M122A, M122B |
aBase substitutions were detected using CLC Genomics Workbench.
Table 3.
Indel mutations in P. berghei mutator clones
| Chr | Position | Change | Region | Gene ID | Product | Clone |
|---|---|---|---|---|---|---|
| 4 | 446,169 | (A)4→(A)3 | Intergenic | M122A, M122B | ||
| 8 | 941,899 | (T)10→ (T)11 | Intergenic | M122A, M122B | ||
| 13 | 30,917 | (A)2→ (A)1 | Intergenic | M122A, M122B | ||
| 13 | 2,414,137 | (T)3→ (T)2 | Exon | PBANKA_136380 | Phosphatidylinositol transfer protein | M122A, M122B |
| 14 | 1,372,440 | (C)3→ (C)2 | Exon | PBANKA_143750 | Transcription factor with AP2 domain(s) (ApiAP2) | M122A, M122B |
Indels were detected using CLC Genomics Workbench.
Analysis of chromosomal distribution of the mutations by r-scan statistics33 revealed that there were nine chromosomal regions, where mutations were significantly clustered (Fig. 1) (P < 0.05). This might suggest the presence of mutation hot spots in the P. berghei genome.
3.3. Mutation rate of the mutator
Overall substitution rate (mean ± SE) was 0.14 × 10−9/bp/day (0.42 × 10−10/bp/cell division) for C122, 12.7 × 10−9/bp/day (3.8 × 10−9/bp/cell division) for M122A, and 12.2 × 10−9/bp/day (3.7 × 10−9/bp/cell division) for M122B, respectively. Thus, the mutation rate was 86- to 90-fold higher in PbMut than in PbCtl. Since no mutation was detected in the intergenic regions in PbCtl, we were unable to calculate the mutation rate in the intergenic regions, but it should be <0.21 × 10−9/bp/day (0.62 × 10−10/bp/cell division), the rate estimated by assuming one mutation in the regions. The mutation rates (M122A: 13.6 × 10−9/bp/day and M122B: 11.9 × 10−9/bp/day) in the intergenic regions in PbMut were similar to those in overall regions (Fig. 2). In intragenic regions, the mutation rate in NonSyn sites was comparable with that of overall sites. Although the NonSyn mutation rates (M122A: 13.2 × 10−9/bp/day and M122B: 13.4 × 10−9/bp/day) appeared to be higher than the rates in Syn sites (M122A: 11.3 × 10−9/bp/day and M122B: 11.2 × 10−9/bp/day) and in introns (M122A: 8.5 × 10−9/bp/day and M122B: 7.3 × 10−9/bp/day), these differences were not significant (P > 0.05 by Fisher's exact test) (Fig. 2).
Figure 2.
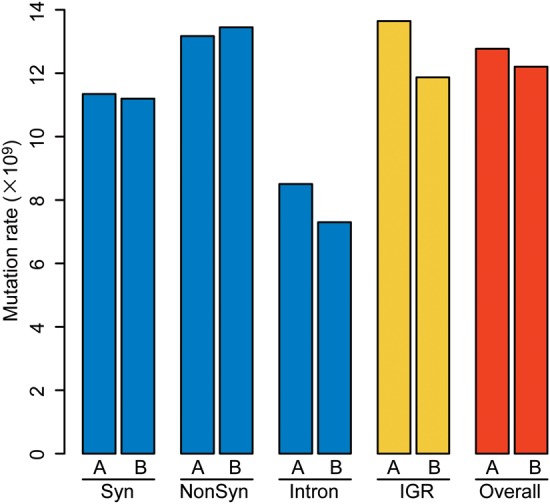
Mutation rate of P. berghei mutator lines. The mutation rate (per nucleotide site per day) was calculated for the following specific sequence regions: Syn and NonSyn sites in protein-coding regions; introns; IGR; and the overall sequences. A and B indicate M122A and M122B, respectively. This figure appears in colour in the online version of DNA Research.
3.4. Growth rate and sexual differentiation of the mutator
The growth rate analysis revealed that two PbMut lines (M122A and B) proliferated in the mouse with a similar rate to those of PbCtl (C122) and PbWT (Supplementary Fig. S3). This result suggests that elimination of the proofreading activity of DNA polymerase δ and the generated mutations did not affect asexual blood stage parasite growth. It seems likely that parasites with mutations deleterious to parasite survival were eliminated during the propagation of parasites in mice, and thus parasites that may have showed slow growth rate were masked under the present experimental conditions. Interestingly, M122A and M122B did not produce gametocytes, while C122 did (Supplementary Table S5). Consistently, neither exflagellation nor ookinete formation was observed in PbMut lines (Supplementary Table S5).
4. Discussion
The present study demonstrated that elimination of the proofreading activity of the P. berghei DNA polymerase δ produced malaria parasites (malaria mutator) with a high mutation rate (86- to 90-fold higher mutation rate in PbMut vs. control PbCtl). The increased mutation rate in PbMut is comparable with a ≥60 time increase observed in Saccharomyces cerevisiae.23 We consider this 86- to 90-fold higher mutation rate as a tentative one that needs to be refined by further investigation. In this study, only one parasite lineage each for PbCtl and PbMut was used to calculate the relative mutation rate. To obtain more accurate mutation rates in both PbCtl and PbMut, mutation rates from several lineages would be required.
PbMut lines grew with a similar rate to those of PbCtl and PbWT lines (Supplementary Fig. S3). Because of this, it is unlikely that the 89–92 NonSyn mutations (and other mutations) in the PbMut lines were deleterious to parasite survival. Apart from genes with unknown function, most of the genes with NonSyn changes code for housekeeping functions but not proteins involved in immune evasion (Supplementary Table S5). These genes may well have had relaxed constraints, not seriously affecting parasite survival during the asexual blood stage. We identified five non-sense mutations in PbMut lines (Table 2), causing truncations in each gene. The truncation of these genes, likewise, appeared not to affect parasite growth, suggesting that expression of these genes may not be essential for the asexual blood stage development. This is the case for the ookinete protein gene (PBANKA_123360), which is not expressed at the blood stage (PlasmoDB). Previously, it was shown that knockout of the serine/threonine protein kinase gene (PBANKA_090380) was lethal, which suggested the importance of this gene during the asexual blood stage.36 However, the mutation detected in PbMut lines was located downstream of the protein's kinase domain, thus the kinase activity was probably not impaired in the truncated protein.
Two indel mutations causing frameshifts were also detected: one in the phosphatidylinositol transfer protein gene (PBANKA_136380) and the other in the transcription factor gene with AP2 domain(s) (ApiAP2) (PBANKA_143750). The frameshift mutation at amino acid position 506 caused a truncation (1513 amino acids) in the Pleckstrin homology domain of the phosphatidylinositol transfer protein gene. This domain binds to phosphatidylinositol lipids, suggesting that the truncated protein lost its function. BLASTP search did not identify any homologue of PBANKA_136380 in PlasmoDB. Thus, the expression of the phosphatidylinositol transfer protein gene may not be essential during the asexual blood stage. The ApiAP2 gene is a member of the gene family of DNA binding transcription factor proteins and is involved in the regulation of gene expression at various stages of parasite development.37 The frameshift in the ApiAP2 gene caused an extension of the gene product (from 2339 to 2377 amino acids in non-mutated gene and frameshift-mutated gene, respectively). Recently, it was reported that two clones of gametocyte-deficient P. falciparum produced by piggyBac random mutagenesis were attributed to insertions in ApiAP2 gene.38 Notably, we did detect no gametocyte in PbMut lines at the 122nd mouse passage. The ApiAP2 gene which we identified is not orthologous to one that Ikadai et al. reported, but the APiAP2 family may function as a key molecule for sexual differentiation of Plasmodium parasites.
Random mutagenesis using artificial mutagens such as alkylating agents or irradiation remains viable tools to generate malaria mutants. However, artificial mutagen-induced mutations are highly biased, e.g. G/C to A/T transition by ethyl methanesulfonate,25,39 A to T and T to A transversions by N-ethyl-N-nitrosourea,39 or pyrimidine dinucleotide-associated C to T transitions by UV or neutron irradiation.40 These biases may influence studies such as drug resistance and vaccine resistance, which should reproduce the actual mutants in the field. The mutations generated in the malaria mutator were probably generated during natural processes of DNA replication as the principle of the mutator relies on a defective proofreading activity of DNA polymerase δ. Malaria parasites with identified DNA mismatch repair activities have been linked to drug-resistant parasites.41 Because of this, we believe that the malaria mutator system could be useful for reproducing drug resistance observed in the field. We have, in fact, generated mutator parasites resistant to sulfadoxine and could identify a NonSyn mutation in dihydropteroate synthase gene, a known target of the drug (data not shown). It would be interesting if we could also use the malaria mutator to demonstrate emergence of vaccine-resistant parasites similar to the mutator phenotype of the bacterial pathogen, Neisseria meningitidis, which rapidly became resistant to bactericidal antibody B5 and had a competitive advantage over a non-mutator strain.42
Constructing the mutant library including various mutants will be useful to readily acquire novel phenotypes/traits. The analysed PbMut lines, however, shared a large proportion of variants, suggesting that the diverse population was not constructed after long-term passages in this study. Mouse passage by inoculating a large number of mutator parasites (e.g. 106 infected erythrocytes) and increasing passage lineages will serve to construct the mutant library. If this attempt improved the diversity of mutant library, more precise mutation hot spots might be identified by analysing the several number of mutant parasite lines.
In the malaria mutator system, there is one limitation to overcome; unrelated mutations may be inevitably accumulated in the parasite during the propagation of parasites through mouse passages. Once the mutator parasite with a novel phenotype is isolated, accumulation of further unrelated mutations in the parasite must be stopped. Genetic crossing with the wild-type parasites can be used to replace the mutated DNA polymerase δ with the wild-type enzyme. Also, genetic crossing will be useful to reduce extra mutations uninvolved in the phenotypes. Although PbMut lines at the 122nd mouse passage lost the ability of gametocyte formation, accumulation of deleterious mutations in genes essential for mosquito or liver stages will be suppressed by conducting a long-term mouse passage in combination with mosquito passage in the future. The candidate mutation(s) involved in novel phenotypes could be inferred with whole genome sequencing and/or linkage group selection.6 Furthermore, screening test with recent molecular technology like an artificial chromosome library could be useful to accelerate the identification of genes involved in novel phenotypes of interest.7,8 We believe that the mutator system could be useful for rapid generation of novel mutants, as rodent malaria parasites are ideally suited for ease with which the whole life cycle of Plasmodium asexual stage could be achieved. The characterization of the malaria mutator will also allow rapid correlation of sequence data with biological function.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (18073013, 24112705, and 24117504; http://www.mext.go.jp/english/) and from the Japan Society for the Promotion of Sciences (22406012, 23659211, 24590501, and 24790401; http://www.jsps.go.jp/english/), and a grant for research into emerging and re-emerging infectious diseases from the Ministry of Health, Labour and Welfare of Japan (H23-Shinkosaiko-ippan-014; http://www.mhlw.go.jp/english/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr Yoshiya Matsubara for useful suggestions on statistical analysis, Dr Kenji Hikosaka for assistance with animal experiments, and MR4 for providing pL0007 plasmid. We would like to dedicate this paper to the late Professor Tanabe who was one of the corresponding authors of this paper.
References
- 1.Murray C.J.L., Rosenfeld L.C., Lim S.S., et al. Global malaria mortality between 1980 and 2010: a systematic analysis. Lancet. 2012;379:413–31. doi: 10.1016/S0140-6736(12)60034-8. [DOI] [PubMed] [Google Scholar]
- 2.Dondorp A.M., Nosten F., Yi P., et al. Artemisinin resistance in Plasmodium falciparum malaria. New Engl. J. Med. 2009;361:455–67. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The RTS, S Clinical Trials Partnership. A phase 3 trial of RTS,S/AS01 malaria vaccine in African infants. New Engl. J. Med. 2012;367:2284–95. doi: 10.1056/NEJMoa1208394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz L., Brown G.V., Genton B., Moorthy V.S. A review of malaria vaccine clinical projects based on the WHO rainbow table. Malar. J. 2012;11:11. doi: 10.1186/1475-2875-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Koning-Ward T.F., Janse C.J., Waters A.P. The development of genetic tools for dissecting the biology of malaria parasites. Annu. Rev. Microbiol. 2000;54:157–85. doi: 10.1146/annurev.micro.54.1.157. [DOI] [PubMed] [Google Scholar]
- 6.Culleton R., Martinelli A., Hunt P., Carter R. Linkage group selection: rapid gene discovery in malaria parasites. Genome Res. 2004;15:92–7. doi: 10.1101/gr.2866205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwanaga S., Kaneko I., Yuda M. A high-coverage artificial chromosome library for the genome-wide screening of drug-resistance genes in malaria parasites. Genome Res. 2012;22:985–92. doi: 10.1101/gr.124164.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iwanaga S., Khan S.M., Kaneko I., et al. Functional identification of the Plasmodium centromere and generation of a Plasmodium artificial chromosome. Cell Host Microbe. 2010;7:245–55. doi: 10.1016/j.chom.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins C.R., Das S., Wong E.H., et al. Robust inducible Cre recombinase activity in the human malaria parasite Plasmodium falciparum enables efficient gene deletion within a single asexual erythrocytic growth cycle. Mol. Microbiol. 2013;88:687–701. doi: 10.1111/mmi.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Straimer J., Lee M.C.S., Lee A.H., et al. Site-specific genome editing in Plasmodium falciparum using engineered zinc-finger nucleases. Nat. Methods. 2012;9:993–8. doi: 10.1038/nmeth.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pino P. From technology to biology: a malaria genetic toolbox for the functional dissection of essential genes. Mol. Microbiol. 2013;88:650–4. doi: 10.1111/mmi.12232. [DOI] [PubMed] [Google Scholar]
- 12.Rottmann M., McNamara C., Yeung B.K.S., et al. Spiroindolones, a potent compound class for the treatment of malaria. Science. 2010;329:1175–80. doi: 10.1126/science.1193225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wongsrichanalai C., Pickard A.L., Wernsdorfer W.H., Meshnick S.R. Epidemiology of drug-resistant malaria. Lancet Infect. Dis. 2002;2:209–18. doi: 10.1016/s1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- 14.Hoepfner D., McNamara C.W., Lim C.S., et al. Selective and specific inhibition of the Plasmodium falciparum lysyl-tRNA synthetase by the fungal secondary metabolite cladosporin. Cell Host Microbe. 2012;11:654–63. doi: 10.1016/j.chom.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch M. The cellular, developmental and population-genetic determinants of mutation-rate evolution. Genetics. 2008;180:933–43. doi: 10.1534/genetics.108.090456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denver D.R., Dolan P.C., Wilhelm L.J., et al. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc. Natl. Acad. Sci. USA. 2009;106:16310–14. doi: 10.1073/pnas.0904895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haag-Liautard C., Dorris M., Maside X., et al. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature. 2007;445:82–5. doi: 10.1038/nature05388. [DOI] [PubMed] [Google Scholar]
- 18.Bopp S.E.R., Manary M.J., Bright A.T., et al. Mitotic evolution of Plasmodium falciparum shows a stable core genome but recombination in antigen families. PLoS Genet. 2013;9:e1003293. doi: 10.1371/journal.pgen.1003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shevelev I.V., Hubscher U. The 3′–5′ exonucleases. Nat. Rev. Mol. Cell Biol. 2002;3:364–76. doi: 10.1038/nrm804. [DOI] [PubMed] [Google Scholar]
- 20.Morrison A., Sugino A. The 3′→5′ exonucleases of both DNA polymerases δ and ε participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol. Gen. Genet. 1994;242:289–96. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- 21.Ridley R.G., White J.H., McAleese S.M., et al. DNA polymerase δ: gene sequences from Plasmodium falciparum indicate that this enzyme is more highly conserved than DNA polymerase α. Nucleic Acids Res. 1991;19:6731–6. doi: 10.1093/nar/19.24.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon M., Giot L., Faye G. The 3′ to 5′ exonuclease activity located in the DNA polymerase delta subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991;10:2165–70. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fortune J.M., Pavlov Y.I., Welch C.M., Johansson E., Burgers P.M.J., Kunkel T.A. Saccharomyces cerevisiae DNA polymerase δ: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005;280:29980–87. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 24.Karthikeyan R., Vonarx E.J., Straffon A.F., Simon M., Faye G., Kunz B.A. Evidence from mutational specificity studies that yeast DNA polymerases delta and epsilon replicate different DNA strands at an intracellular replication fork. J. Mol. Biol. 2000;299:405–19. doi: 10.1006/jmbi.2000.3744. [DOI] [PubMed] [Google Scholar]
- 25.Shiwa Y., Fukushima-Tanaka S., Kasahara K., Horiuchi T., Yoshikawa H. Whole-genome profiling of a novel mutagenesis technique using proofreading-deficient DNA polymerase δ. Int. J. Evol. Biol. 2012;2012:860797. doi: 10.1155/2012/860797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park E.Y., Ito Y., Nariyama M., Sugimoto T., Lies D., Kato T. The improvement of riboflavin production in Ashbya gossypii via disparity mutagenesis and DNA microarray analysis. Appl. Microbiol. Biotechnol. 2011;91:1315–26. doi: 10.1007/s00253-011-3325-0. [DOI] [PubMed] [Google Scholar]
- 27.Shimoda C., Itadani A., Sugino A., Furusawa M. Isolation of thermotolerant mutants by using proofreading-deficient DNA polymerase δ as an effective mutator in Saccharomyces cerevisiae. Genes Genet. Syst. 2006;81:391–7. doi: 10.1266/ggs.81.391. [DOI] [PubMed] [Google Scholar]
- 28.Furusawa M. Implications of fidelity difference between the leading and the lagging strand of DNA for the acceleration of evolution. Front. Oncol. 2012;2:144. doi: 10.3389/fonc.2012.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li R., Yu C., Li Y., et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–7. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 30.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koboldt D.C., Zhang Q., Larson D.E., et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall N., Karras M., Raine J.D., et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–6. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 33.Karlin S., Macken C. Some statistical problems in the assessment of inhomogeneities of DNA sequence data. J. Am. Stat. Assoc. 1991;86:27–35. [Google Scholar]
- 34.Denver D.R., Morris K., Lynch M., Thomas W.K. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature. 2004;430:679–82. doi: 10.1038/nature02697. [DOI] [PubMed] [Google Scholar]
- 35.Nei M., Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986;3:418–26. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 36.Tewari R., Straschil U., Bateman A., et al. The systematic functional analysis of Plasmodium protein kinases identifies essential regulators of mosquito transmission. Cell Host Microbe. 2010;8:377–87. doi: 10.1016/j.chom.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell T.L., De Silva E.K., Olszewski K.L., Elemento O., Llinás M. Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite. PLoS Path. 2010;6:e1001165. doi: 10.1371/journal.ppat.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikadai H., Shaw Saliba K., Kanzok S.M., et al. Transposon mutagenesis identifies genes essential for Plasmodium falciparum gametocytogenesis. Proc. Natl. Acad. Sci. USA. 2013;110:E1676–1684. doi: 10.1073/pnas.1217712110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flibotte S., Edgley M.L., Chaudhry I., et al. Whole-genome profiling of mutagenesis in Caenorhabditis elegans. Genetics. 2010;185:431–41. doi: 10.1534/genetics.110.116616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belfield E.J., Gan X., Mithani A., et al. Genome-wide analysis of mutations in mutant lineages selected following fast-neutron irradiation mutagenesis of Arabidopsis thaliana. Genome Res. 2012;22:1306–15. doi: 10.1101/gr.131474.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castellini M.A., Buguliskis J.S., Casta L.J., et al. Malaria drug resistance is associated with defective DNA mismatch repair. Mol. Biochem. Parasitol. 2011;177:143–7. doi: 10.1016/j.molbiopara.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bayliss C.D., Hoe J.C., Makepeace K., Martin P., Hood D.W., Moxon E.R. Neisseria meningitidis escape from the bactericidal activity of a monoclonal antibody is mediated by phase variation of lgtG and enhanced by a mutator phenotype. Infect. Immun. 2008;76:5038–48. doi: 10.1128/IAI.00395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.