Abstract
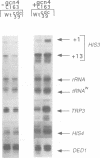
The yeast HIS3 promoter region contains two functionally distinct TATA elements, TC and TR, that are responsible respectively for initiation from the +1 and +13 sites. Both TC and TR support basal HIS3 transcription and require the TATA binding protein TFIID, but only TR responds to transcriptional activation by GCN4 and GAL4. By selecting for yeast strains that increase transcription by a GCN4 derivative with a defective activation domain, we have isolated a temperature-sensitive mutation in CDC39, a previously defined gene implicated in cell-cycle control and the pheromone response. This cdc39-2 mutation causes increased basal transcription of many, but not all genes, as well as increased transcriptional activation by GCN4 and GAL4. Surprisingly, basal HIS3 transcription from the +1 initiation site is strongly increased, while initiation from the +13 site is barely affected. Thus, unlike acidic activator proteins that function through TR, CDC39 preferentially affects transcription mediated by TC. CDC39 is an essential gene that encodes a very large nuclear protein (2108 amino acids) containing two glutamine-rich regions. These observations suggest that CDC39 negatively regulates transcription either by affecting the general RNA polymerase II machinery or by altering chromatin structure.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chen W., Struhl K. Saturation mutagenesis of a yeast his3 "TATA element": genetic evidence for a specific TATA-binding protein. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2691–2695. doi: 10.1073/pnas.85.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Tabor S., Struhl K. Distinguishing between mechanisms of eukaryotic transcriptional activation with bacteriophage T7 RNA polymerase. Cell. 1987 Sep 25;50(7):1047–1055. doi: 10.1016/0092-8674(87)90171-1. [DOI] [PubMed] [Google Scholar]
- Clark-Adams C. D., Norris D., Osley M. A., Fassler J. S., Winston F. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 1988 Feb;2(2):150–159. doi: 10.1101/gad.2.2.150. [DOI] [PubMed] [Google Scholar]
- Clark K. L., Sprague G. F., Jr Yeast pheromone response pathway: characterization of a suppressor that restores mating to receptorless mutants. Mol Cell Biol. 1989 Jun;9(6):2682–2694. doi: 10.1128/mcb.9.6.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L., Tanese N., Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992 Mar 6;68(5):965–976. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- Cormack B. P., Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992 May 15;69(4):685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- Davis J. L., Kunisawa R., Thorner J. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992 Apr;12(4):1879–1892. doi: 10.1128/mcb.12.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan J. W., Kirkman C., Fields S. The yeast STE12 protein binds to the DNA sequence mediating pheromone induction. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5703–5707. doi: 10.1073/pnas.86.15.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrin L. K., Mann R. K., Grunstein M. Nucleosome loss activates CUP1 and HIS3 promoters to fully induced levels in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992 Apr;12(4):1621–1629. doi: 10.1128/mcb.12.4.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynlacht B. D., Hoey T., Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991 Aug 9;66(3):563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- Eisenmann D. M., Arndt K. M., Ricupero S. L., Rooney J. W., Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 1992 Jul;6(7):1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- Errede B., Ammerer G. STE12, a protein involved in cell-type-specific transcription and signal transduction in yeast, is part of protein-DNA complexes. Genes Dev. 1989 Sep;3(9):1349–1361. doi: 10.1101/gad.3.9.1349. [DOI] [PubMed] [Google Scholar]
- Han M., Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988 Dec 23;55(6):1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- Han M., Kim U. J., Kayne P., Grunstein M. Depletion of histone H4 and nucleosomes activates the PHO5 gene in Saccharomyces cerevisiae. EMBO J. 1988 Jul;7(7):2221–2228. doi: 10.1002/j.1460-2075.1988.tb03061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbury P. A., Struhl K. Functional distinctions between yeast TATA elements. Mol Cell Biol. 1989 Dec;9(12):5298–5304. doi: 10.1128/mcb.9.12.5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I., Jensen R. E. Putting the HO gene to work: practical uses for mating-type switching. Methods Enzymol. 1991;194:132–146. doi: 10.1016/0076-6879(91)94011-z. [DOI] [PubMed] [Google Scholar]
- Hill D. E., Hope I. A., Macke J. P., Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986 Oct 24;234(4775):451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Mahadevan S., Struhl K. Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature. 1988 Jun 16;333(6174):635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Inostroza J. A., Mermelstein F. H., Ha I., Lane W. S., Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992 Aug 7;70(3):477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- Kanazawa S., Driscoll M., Struhl K. ATR1, a Saccharomyces cerevisiae gene encoding a transmembrane protein required for aminotriazole resistance. Mol Cell Biol. 1988 Feb;8(2):664–673. doi: 10.1128/mcb.8.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keleher C. A., Redd M. J., Schultz J., Carlson M., Johnson A. D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992 Feb 21;68(4):709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Kruger W., Herskowitz I. A negative regulator of HO transcription, SIN1 (SPT2), is a nonspecific DNA-binding protein related to HMG1. Mol Cell Biol. 1991 Aug;11(8):4135–4146. doi: 10.1128/mcb.11.8.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. S., Kao C. C., Bryant G. O., Liu X., Berk A. J. Adenovirus E1A activation domain binds the basic repeat in the TATA box transcription factor. Cell. 1991 Oct 18;67(2):365–376. doi: 10.1016/0092-8674(91)90188-5. [DOI] [PubMed] [Google Scholar]
- Lieberman P. M., Berk A. J. The Zta trans-activator protein stabilizes TFIID association with promoter DNA by direct protein-protein interaction. Genes Dev. 1991 Dec;5(12B):2441–2454. doi: 10.1101/gad.5.12b.2441. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Mahadevan S., Struhl K. Tc, an unusual promoter element required for constitutive transcription of the yeast HIS3 gene. Mol Cell Biol. 1990 Sep;10(9):4447–4455. doi: 10.1128/mcb.10.9.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisterernst M., Roeder R. G. Family of proteins that interact with TFIID and regulate promoter activity. Cell. 1991 Nov 1;67(3):557–567. doi: 10.1016/0092-8674(91)90530-c. [DOI] [PubMed] [Google Scholar]
- Nicolet C. M., Craig E. A. Inducing and assaying heat-shock response in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:710–717. doi: 10.1016/0076-6879(91)94052-e. [DOI] [PubMed] [Google Scholar]
- Oettinger M. A., Struhl K. Suppressors of Saccharomyces cerevisiae his3 promoter mutations lacking the upstream element. Mol Cell Biol. 1985 Aug;5(8):1901–1909. doi: 10.1128/mcb.5.8.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S. G., van der Aart Q. J., Agostoni-Carbone M. L., Aigle M., Alberghina L., Alexandraki D., Antoine G., Anwar R., Ballesta J. P., Benit P. The complete DNA sequence of yeast chromosome III. Nature. 1992 May 7;357(6373):38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- Peterson C. L., Kruger W., Herskowitz I. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell. 1991 Mar 22;64(6):1135–1143. doi: 10.1016/0092-8674(91)90268-4. [DOI] [PubMed] [Google Scholar]
- Ponticelli A. S., Struhl K. Analysis of Saccharomyces cerevisiae his3 transcription in vitro: biochemical support for multiple mechanisms of transcription. Mol Cell Biol. 1990 Jun;10(6):2832–2839. doi: 10.1128/mcb.10.6.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S. I., Ferguson J., Jahng K. Y. Isolation and characterization of two genes encoding yeast mating pheromone signaling elements: CDC72 and CDC73. Cold Spring Harb Symp Quant Biol. 1988;53(Pt 2):621–627. doi: 10.1101/sqb.1988.053.01.071. [DOI] [PubMed] [Google Scholar]
- Reed S. I. The selection of S. cerevisiae mutants defective in the start event of cell division. Genetics. 1980 Jul;95(3):561–577. doi: 10.1093/genetics/95.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Schultz J., Marshall-Carlson L., Carlson M. The N-terminal TPR region is the functional domain of SSN6, a nuclear phosphoprotein of Saccharomyces cerevisiae. Mol Cell Biol. 1990 Sep;10(9):4744–4756. doi: 10.1128/mcb.10.9.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster E. O., Byers B. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics. 1989 Sep;123(1):29–43. doi: 10.1093/genetics/123.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer V. L., Wobbe C. R., Struhl K. A wide variety of DNA sequences can functionally replace a yeast TATA element for transcriptional activation. Genes Dev. 1990 Apr;4(4):636–645. doi: 10.1101/gad.4.4.636. [DOI] [PubMed] [Google Scholar]
- Strubin M., Struhl K. Yeast and human TFIID with altered DNA-binding specificity for TATA elements. Cell. 1992 Feb 21;68(4):721–730. doi: 10.1016/0092-8674(92)90147-5. [DOI] [PubMed] [Google Scholar]
- Struhl K. Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol Cell Biol. 1986 Nov;6(11):3847–3853. doi: 10.1128/mcb.6.11.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7865–7869. doi: 10.1073/pnas.81.24.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Hill D. E. Two related regulatory sequences are required for maximal induction of Saccharomyces cerevisiae his3 transcription. Mol Cell Biol. 1987 Jan;7(1):104–110. doi: 10.1128/mcb.7.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. The DNA-binding domains of the jun oncoprotein and the yeast GCN4 transcriptional activator protein are functionally homologous. Cell. 1987 Sep 11;50(6):841–846. doi: 10.1016/0092-8674(87)90511-3. [DOI] [PubMed] [Google Scholar]
- Timmers H. T., Sharp P. A. The mammalian TFIID protein is present in two functionally distinct complexes. Genes Dev. 1991 Nov;5(11):1946–1956. doi: 10.1101/gad.5.11.1946. [DOI] [PubMed] [Google Scholar]
- Vidal M., Strich R., Esposito R. E., Gaber R. F. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol Cell Biol. 1991 Dec;11(12):6306–6316. doi: 10.1128/mcb.11.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Clark I., Nicholson P. R., Herskowitz I., Stillman D. J. The Saccharomyces cerevisiae SIN3 gene, a negative regulator of HO, contains four paired amphipathic helix motifs. Mol Cell Biol. 1990 Nov;10(11):5927–5936. doi: 10.1128/mcb.10.11.5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteway M., Hougan L., Dignard D., Thomas D. Y., Bell L., Saari G. C., Grant F. J., O'Hara P., MacKay V. L. The STE4 and STE18 genes of yeast encode potential beta and gamma subunits of the mating factor receptor-coupled G protein. Cell. 1989 Feb 10;56(3):467–477. doi: 10.1016/0092-8674(89)90249-3. [DOI] [PubMed] [Google Scholar]
- Williams F. E., Trumbly R. J. Characterization of TUP1, a mediator of glucose repression in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Dec;10(12):6500–6511. doi: 10.1128/mcb.10.12.6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams F. E., Varanasi U., Trumbly R. J. The CYC8 and TUP1 proteins involved in glucose repression in Saccharomyces cerevisiae are associated in a protein complex. Mol Cell Biol. 1991 Jun;11(6):3307–3316. doi: 10.1128/mcb.11.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobbe C. R., Struhl K. Yeast and human TATA-binding proteins have nearly identical DNA sequence requirements for transcription in vitro. Mol Cell Biol. 1990 Aug;10(8):3859–3867. doi: 10.1128/mcb.10.8.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Barros Lopes M., Ho J. Y., Reed S. I. Mutations in cell division cycle genes CDC36 and CDC39 activate the Saccharomyces cerevisiae mating pheromone response pathway. Mol Cell Biol. 1990 Jun;10(6):2966–2972. doi: 10.1128/mcb.10.6.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]