Abstract
Proline utilization A from Bradyrhizobium japonicum (BjPutA) is a bifunctional flavoenzyme that catalyzes the oxidation of proline to glutamate using fused proline dehydrogenase (PRODH) and Δ1-pyrroline-5-carboxylate dehydrogenase (P5CDH) domains. Recent crystal structures and kinetic data suggest an intramolecular channel connects the two active sites, promoting substrate channeling of the intermediate Δ1-pyrroline-5-carboxylate/glutamate-γ-semialdehyde (P5C/GSA). In this work, the structure of the channel was explored by inserting large side chain residues at four positions along the channel in BjPutA. Kinetic analysis of the different mutants revealed replacement of D779 with Tyr (D779Y) or Trp (D779W) significantly decreased the overall rate of the PRODH–P5CDH channeling reaction. X-ray crystal structures of D779Y and D779W revealed that the large side chains caused a constriction in the central section of the tunnel, thus likely impeding the travel of P5C/GSA in the channel. The D779Y and D779W mutants have PRODH activity similar to that of wild-type BjPutA but exhibit significantly lower P5CDH activity, suggesting that exogenous P5C/GSA enters the channel upstream of Asp779. Replacement of nearby Asp778 with Tyr (D778Y) did not impact BjPutA channeling activity. Consistent with the kinetic results, the X-ray crystal structure of D778Y shows that the main channel pathway is not impacted; however, an off-cavity pathway is closed off from the channel. These findings provide evidence that the off-cavity pathway is not essential for substrate channeling in BjPutA.
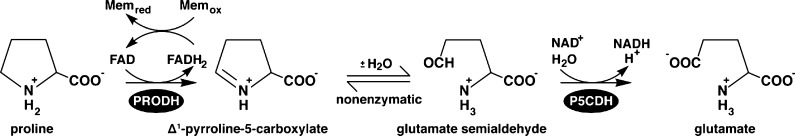
The proline catabolic pathway catalyzes the oxidation of proline to glutamate (Scheme 1). In the first step, proline dehydrogenase (PRODH) uses an FAD cofactor to remove two electrons (as H–) from proline, resulting in Δ1-pyrroline-5-carboxylate (P5C). P5C then undergoes a nonenzymatic hydrolysis, which opens the pyrroline ring to create glutamate-γ-semialdehyde (GSA). Finally, GSA is oxidized to glutamate by the NAD+-dependent P5C dehydrogenase (P5CDH) to complete the overall four-electron oxidation process. Proline and proline metabolism are important for the pathogenicity of Helicobacter pylori and Helicobacter hepaticus,1,2 energy production in procyclic trypanosomes,3,4 and regulation of metabolites linked to pathogenesis in Photorhabdus and Xenorhabdus.5 In humans, inborn errors in proline catabolism lead to hyperprolinemia disorders, and defects in PRODH are linked to schizophrenia.6,7 Also, PRODH is regulated by p53 and has been shown to function in tumor suppression.8
Scheme 1. Overall Reaction Catalyzed by Proline Utilization A (PutA).
Flavin-dependent proline dehydrogenase (PRODH) catalyzes the oxidation of proline to Δ1-pyrroline-5-carboxylate (P5C) and reduction of respiratory quinones in the membrane (Mem). P5C undergoes a nonenzymatic hydrolysis, resulting in glutamate-γ-semialdehyde (GSA). GSA is oxidized to glutamate by P5C dehydrogenase (P5CDH) using an NAD+ cofactor.
PRODH and P5CDH are combined into a single polypeptide chain known as proline utilization A (PutA) in Gram-negative bacteria and Corynebacterium.9 The covalent linking of enzymes catalyzing consecutive reactions in a metabolic pathway affords the possibility of substrate channeling; i.e., the intermediate is transferred between the enzymes without equilibrating with the bulk medium. Several physiological benefits of substrate channeling versus free diffusion have been identified. For example, channeling improves kinetic efficiency by decreasing the transit time between active sites and preventing the loss of intermediates.10,11 Thus, channeling enzymes can operate at maximal rates when cellular substrate concentrations are below saturating levels.12 Also, labile intermediates can be concealed from the bulk environment, preventing decay or interaction with other molecules.13,14 Finally, channeling can influence metabolic flux by segregating intermediates from competing pathways.15
Substrate channeling of P5C/GSA in proline catabolism may be necessary to retain proper metabolic flux and avoid metabolic futile cycling.14 In addition, free P5C/GSA is an inhibitor of three different enzymes in Escherichia coli, including glucosamine-6-phosphate synthase, cytidine-5′-triphosphate synthase, and the amidotransferase domain of carbamoyl phosphate synthetase.16−18 P5C has also been shown to form adducts with other metabolites such as oxaloacetic acid, pyruvic acid, and acetoacetic acid.19 Consistent with the physiological importance of controlling the release of P5C/GSA, kinetic studies have firmly established substrate channeling in PutAs. Early studies of Salmonella typhimurium PutA using 14C-labeled proline are consistent with a channeling mechanism.20 More recent steady-state and rapid reaction transient time measurements of PutAs from Bradyrhizobium japonicum (BjPutA) and Geobacter sulfurreducens (GsPutA) also indicate substrate channeling.21,22 Furthermore, a comprehensive analysis of the complete kinetic mechanism of E. coli PutA showed that substrate channeling is rate-limiting, and the rate constant for the channeling step is slowest during the first enzyme turnover and increases with subsequent turnovers, establishing PutA as a new example of a hysteretic enzyme.23
With the kinetic data firmly demonstrating substrate channeling in PutA, the goal of this study is to gain insight into the structural basis of channeling. The crystal structures of BjPutA and GsPutA revealed that the two active sites are separated by a linear distance of 41–45 Å, implying that substrate channeling involves substantial movement of the P5C/GSA intermediate.21,22 Analysis of potential channeling pathways predicts a curved, 75 Å tunnel that connects the two active sites (Figure 1).
Figure 1.

Tunnel/cavity system of BjPutA. (A) BjPutA protomer with PRODH colored blue, P5CDH pink, and the oligomerization flap green. The FAD and NAD+ are shown as yellow and green sticks, respectively. Catalytic Cys792 of the P5CDH active site is indicated. The gray surface represents the predicted channeling pathway calculated with MOLE. Helices α5a and 770s (residues 773–785) are colored gold and cyan, respectively. We note that in a tetramer of BjPutA, the dimerization flap of one protomer covers the tunnel of the other protomer. (B) Details of the predicted channeling pathway. The predicted path from MOLE is shown as mesh. Models of P5C and GSA in the tunnel are shown for scale (green). (C) Another view of the tunnel/cavity system, with the predicted channeling tunnel calculated from MOLE shown as gray mesh and the off-pathway cavity calculated using VOIDOO shown as red mesh.
Here we use site-directed mutagenesis, kinetics, and X-ray crystallography to gain further insight into the structural features that facilitate substrate channeling in BjPutA. Several residues between the two active sites have been mutated in an effort to obstruct molecular traffic. Kinetic and structural analysis of the mutant enzymes shows that channeling is hindered in some of the variants but not others, which provides information about the pathway traversed by the intermediate. Furthermore, steric considerations suggest that GSA is threaded through the tunnel in a linear conformation, with the aldehyde group facing the P5CDH end of the tunnel. This aspect of substrate channeling in PutA might be considered an example of shape selective catalysis.
Experimental Procedures
Chemicals
All chemicals were purchased from Sigma-Aldrich or Fisher Scientific unless otherwise noted. (DL)-P5C (50/50 mixture) was synthesized according to the method of Williams and Frank and stored in 1 M HCl at 4 °C. The concentration of (DL)-P5C was determined as previously reported.24,25E. coli strain BL21 (DE3) pLysS was purchased from Novagen, and strain DH5α was purchased from Invitrogen. All experiments used Nanopure water.
Site-Directed Mutagenesis
Mutagenic primers (Table 1) were purchased from Integrated DNA Technologies or Eurofins MWG Operon. The GeneTailor Mutagenesis Kit (Invitrogen) was used to generate all mutants except T348Y and D779Y (QuikChange II kit, Agilent Technologies). Mutant plasmids were transformed into DH5α cells, and the resulting plasmids were sequenced by Eurofins MWG Operon to confirm the mutations.
Table 1. Primers Used for Site-Directed Mutagenesis.
| mutant | primers |
|---|---|
| T348Y | Fwd 5′-GCGCCTATTGGGACTACGAGATCAAGCGCGCG-3′ |
| Rev 5′-CGCGCGCTTGATCTCGTAGTCCCAATAGGCGC-3′ | |
| S607Y | Fwd 5′-AGACGCTCGACGATGCGCTCTATGAGCTGCGCG-3′ |
| Rev 5′-GAGCGCATCGTCGAGCGTCTTGCCGCCCTCG-3′ | |
| D778Y | Fwd 5′-GCTGCCGGAGCAGGTCGCCTACGACGTTGTCACC-3′ |
| Rev 5′-GGCGACCTGCTCCGGCAGCGCGGTGGCATCG-3′ | |
| D779A | Fwd 5′-TGCCGGAGCAGGTCGCCGACGCCGTTGTCACCTCC-3′ |
| Rev 5′-GTCGGCGACCTGCTCCGGCAGCGCGGTGGC-3′ | |
| D779W | Fwd 5′-TGCCGGAGCAGGTCGCCGACTGGGTTGTCACCTCC-3′ |
| Rev 5′-GTCGGCGACCTGCTCCGGCAGCGCGGTGGC-3′ | |
| D779Y | Fwd 5′-CCGGAGCAGGTCGCCGACTACGTTGTCACCTCCGC-3′ |
| Rev 5′-GCGGAGGTGACAACGTAGTCGGCGACCTGCTCCGG-3′ |
Expression and Purification of BjPutA Proteins
BjPutA wild-type and mutant proteins were expressed as reported previously, except that induction with isopropyl β-d-1-thiogalactopyranoside was performed at 20 °C for 16 h.26 Cells were harvested by centrifugation and frozen at −80 °C. Frozen cells were resuspended in 50 mL of binding buffer [20 mM Tris base, 0.5 M NaCl, 5 mM imidazole, and 10% glycerol (pH 7.9)] and 100 μM flavin at 4 °C. Protease inhibitors ε-amino-N-caproic acid (3 mM), phenylmethanesulfonyl fluoride (0.3 mM), leupeptin (1.2 μM), tosyl phenylalanyl chloromethyl ketone (48 μM), and tosyllysine chloromethyl ketone hydrochloride (78 μM) were added, and cells were disrupted via sonication. The cell lysate was centrifuged for 1 h at 19000 rpm in a JA-20 rotor (Beckman) and filtered through a 0.2 μm filter (VWR). Cell-free lysate was loaded onto a Ni-NTA Superflow resin (Qiagen) equilibrated with binding buffer. Wash buffer (60 mM imidazole) and then elution buffer (500 mM imidazole) were applied to the column. Elution fractions containing PutA protein were pooled and dialyzed into buffer containing 50 mM Tris (pH 7.5), 10 mM NaCl, 0.5 mM EDTA, and 10% glycerol and loaded onto an anion exchange column (HiTrap Q HP column, GE Life Sciences) equilibrated with dialysis buffer. BjPutA proteins were eluted using a linear 0 to 1 M NaCl gradient (1 L) in dialysis buffer. Purified enzyme was then dialyzed into a final buffer of 50 mM Tris (pH 7.5), 50 mM NaCl, 0.5 mM EDTA, 0.5 mM tris(3-hydroxypropyl)phosphine, and 10% glycerol. The His tag was retained in the subsequent kinetic experiments. The amount of flavin bound in the purified proteins was quantified as described previously (ε451 = 13.62 mM–1 cm–1 for bound flavin).26 The protein concentration was determined from the amount of bound flavin to normalize for differences in flavin content, and the protein was flash-frozen in liquid nitrogen and stored at −80 °C.
Steady-State Kinetic Assays
Steady-state kinetic assays were performed at 23 °C. Kinetic parameters for the PRODH domain were determined for proline and ubiquinone-1 (CoQ1) by following reduction of CoQ1 at 278 nm (ε278 = 14.5 mM–1 cm–1) (Table 2).27 All assays were performed in 50 mM potassium phosphate buffer (pH 7.5) with 0.5 μM PutA enzyme. The Km and kcat values for proline were determined by varying the proline concentration (1–200 mM) while holding the CoQ1 concentration constant (250 μM), and CoQ1 kinetic parameters were determined by varying the CoQ1 concentration (10–350 μM) while holding the proline concentration fixed at 150 mM. Data were collected on a Hi-Tech Scientific SF-61DX2 stopped-flow instrument using a 0.15 cm path length. Initial velocities were fit to the Michaelis–Menten equation using SigmaPlot 12.0.
Table 2. PRODH Kinetic Parameters.
| prolinea |
CoQ1b |
|||||
|---|---|---|---|---|---|---|
| BjPutA | Km (mM) | kcat (s–1) | kcat/Km (M–1 s–1) | Km (μM) | kcat (s–1) | kcat/Km (M–1 s–1) |
| wild-type | 43 ± 5 | 3.1 ± 0.1 | 72 ± 8.6 | 105 ± 6 | 2.9 ± 0.1 | 27619 ± 1713 |
| T348Y | 30 ± 2 | 1.8 ± 0.1 | 60 ± 4.0 | 59 ± 2 | 1.9 ± 0.1 | 32203 ± 1204 |
| S607Y | 46 ± 6 | 1.6 ± 0.1 | 35 ± 4.8 | 131 ± 16 | 2.0 ± 0.1 | 15267 ± 1987 |
| D778Y | 91 ± 38 | 0.36 ± 0.07 | 4.0 ± 1.8 | 82 ± 15 | 0.33 ± 0.02 | 4024 ± 775 |
| D779A | 56 ± 7 | 1.8 ± 0.1 | 32 ± 4.2 | 188 ± 22 | 2.5 ± 0.1 | 13297 ± 1725 |
| D779Y | 43 ± 2 | 2.7 ± 0.1 | 63 ± 3.1 | 56 ± 2 | 3.1 ± 0.1 | 55357 ± 2102 |
| D779W | 30 ± 4 | 1.9 ± 0.1 | 63 ± 8.6 | 109 ± 12 | 2.3 ± 0.1 | 21100 ± 2593 |
Mixture of 1–200 mM proline, 250 μM CoQ1, 0.5 μM enzyme, and 50 mM potassium phosphate (pH 7.5).
Mixture of 150 mM proline, 10–350 μM CoQ1, 0.5 μM enzyme, and 50 mM potassium phosphate (pH 7.5).
Kinetic parameters of P5CDH activity were determined for P5C/GSA (Table 3) using exogenous (DL)-P5C and 0.25 μM PutA enzyme. (DL)-P5C was neutralized with 10 M NaOH immediately prior to assays. The concentration of L-P5C is considered to be half the total (DL)-P5C concentration. To maintain a constant ionic strength, we performed all assays with exogenous (DL)-P5C in 600 mM NaCl as described previously.23Km and kcat for P5C/GSA were determined by varying the L-P5C concentration (0.01–6 mM) while holding the NAD+ concentration constant at 0.2 mM in 50 mM potassium phosphate (pH 7.5, 600 mM NaCl). The effective concentration of GSA was estimated from the pH dependence of the P5C-GSA equilibrium reported previously.16 Initial velocities were determined by following NAD+ reduction at 340 nm (ε340 = 6200 M–1 cm–1). All assays were performed using a Powerwave XS 96-well plate reader (Biotek).
Table 3. P5CDH Kinetic and NAD+ Binding Parameters.
| BjPutA | kcat (s–1)a | Km (mM)a | kcat/Km (M–1 s–1) | Kd (μM, NAD+)b |
|---|---|---|---|---|
| wild-type | 3.4 ± 0.1 | 0.42 ± 0.04 | 8095 ± 822 | 0.60 ± 0.04 |
| T348Y | 4.2 ± 0.2 | 0.42 ± 0.04 | 10000 ± 1017 | 0.75 ± 0.06 |
| S607Y | 4.5 ± 0.2 | 0.48 ± 0.03 | 9375 ± 664 | 1.00 ± 0.04 |
| D778Y | 3.8 ± 0.1 | 0.38 ± 0.02 | 10000 ± 567 | 0.67 ± 0.04 |
| D779A | 5.0 ± 0.1 | 0.38 ± 0.03 | 13157 ± 1102 | 0.64 ± 0.05 |
| D779Y | 0.02 ± 0.01 | 0.20 ± 0.03 | 100 ± 16 | 0.65 ± 0.04 |
| D779W | 0.003 ± 0.001 | 0.35 ± 0.15 | 8.6 ± 4 | 0.78 ± 0.05 |
Mixture of 0.01–6 mM L-P5C, 0.2 mM NAD+, 0.25 μM enzyme, and 50 mM potassium phosphate (pH 7.5, 600 mM NaCl).
From fluorescence quenching with 0.1–25 μM NAD+, 0.25 μM enzyme, and 50 mM potassium phosphate (pH 7.5).
The P5CDH activity of wild-type BjPutA and its D779A, D779Y, and D779W mutants using smaller aldehyde substrates relative to GSA was tested. All assays were performed in 50 mM potassium phosphate (pH 7.5, 25 mM NaCl) containing 0.2 mM NAD+ and variable concentrations of succinate semialdehyde (0.05–20 mM) and propionaldehyde (5–500 mM). For assays with succinate semialdehyde, the concentrations of wild-type BjPutA and mutant D779A were 0.25 μM while those of mutants D779W and D779Y were 1 μM. For propionaldehyde, the concentrations of wild-type BjPutA and mutant D779A were 0.25 μM, that of D779W was 1 μM, and that of D779W was 2 μM. Initial velocities were determined by following NAD+ reduction at 340 nm. All assays were performed using a Powerwave XS 96-well plate reader (Biotek).
The coupled PRODH–P5CDH activity of wild-type BjPutA and its mutants was monitored by following NADH formation at 340 nm or by fluorescence excitation at 340 nm and monitoring fluorescence emission at 460 nm using a Cary Eclipse fluorescence spectrophotometer. Assays were performed at 23 °C in 50 mM potassium phosphate buffer (pH 7.5, 25 mM NaCl, 10 mM MgCl2) containing 0.1 mM CoQ1, 0.2 mM NAD+, 40 mM proline, and the BjPutA enzyme as previously described.21 To determine the kinetic parameters of the overall PRODH–P5CDH reaction for wild-type BjPutA and the D778Y mutant, we performed assays by varying the proline concentration (0.1–550 mM) and holding the CoQ1 (0.1 mM) and NAD+ (0.2 mM) concentrations fixed in 50 mM potassium phosphate buffer (pH 7.5, 25 mM NaCl). NADH formation was followed at 340 nm. Data were fit to a substrate inhibition equation (eq 1) using SigmaPlot 12.0, where [S] is the substrate concentration and Ki is the substrate inhibition constant.
| 1 |
Binding of NAD+ to BjPutA
The binding of NAD+ to BjPutA was assessed by intrinsic tryptophan fluorescence quenching of BjPutA as described previously.23 Tryptophan fluorescence was excited at 295 nm, and fluorescence emission was recorded at 330 nm. Increasing concentrations of NAD+ (0–20 μM) were added to BjPutA (0.25 μM) in 50 mM potassium phosphate (pH 7.5). The inner filter effect caused by the absorption of incident light by NAD+ at 295 nm was corrected using eq 2.27
| 2 |
where Fcorr and Fobs are the corrected and observed fluorescence, respectively, and Aex and Aem are the absorbance values of NAD+ at the excitation and emission wavelengths, respectively. A dissociation constant (Kd) for the BjPutA–NAD+ complex was determined by plotting the fraction of BjPutA bound by NAD+ (θ) versus the free NAD+ concentration using eq 3, where n is the number of binding sites.
| 3 |
The concentration of free NAD+ was determined using eq 4.
| 4 |
The value of θ is obtained from the fluorescence measurements [(F0 – F)/(F0 – Fmax)], where F0 is the fluorescence intensity without NAD+, F is the fluorescence intensity in the presence of NAD+, and Fmax is the maximal fluorescence intensity at saturating NAD+ concentrations.
Binding of NAD to wild-type BjPutA was also estimated by isothermal titration calorimetry (ITC). Titrations were performed at 4 °C using a MicroCal VP-ITC microcalorimeter. Wild-type BjPutA was dialyzed into a buffer composed of 50 mM Tris (pH 7.5), 50 mM NaCl, 0.5 mM EDTA, and 10% glycerol. A NAD+ stock solution of 0.5 mM was made in dialysis buffer. For each titration, 23.4 μM BjPutA was titrated with 2 μL injections (40 total) of 0.5 mM NAD+ at 160 s intervals while the mixture was being stirred at 310 rpm. Data were analyzed using a one-site binding model with Origin ITC Analysis software provided with the instrument.
Prior to the assays described above being performed, the amount of NAD+ bound to purified BjPutA was estimated by high-performance liquid chromatography. BjPutA was denatured with 5% (v/v) trichloroacetic acid and centrifuged at 13000 rpm for 5 min to release bound FAD and NAD+ cofactors. Samples were then filtered with a 0.45 μm filter before being loaded onto the column. FAD and NAD+ were separated on a C18 column using 50 mM potassium phosphate (pH 5.3) and 100% methanol. The cofactors were eluted using a flow rate of 1 mL/min with 5 min of isocratic phosphate buffer, followed by a 25 min linear gradient to 50% methanol, and finally a 5 min linear gradient to 75% methanol. Both cofactors were detected at 280 nm. NAD+ and FAD eluted from the column at 7.9 and 16.6 min, respectively. The concentration of NAD+ was determined using standard solutions of NAD+ (10, 25, 50, 100, and 200 μM). From this analysis, it was estimated that 74% of purified BjPutA contained bound NAD+. Thus, the NAD+ binding experiments report on the remaining 26% of BjPutA that was purified without NAD+ bound.
Single-Turnover Kinetic Experiments
Single-turnover experiments were performed at 21 °C under anaerobic conditions as described previously.21 Briefly, equal volumes of BjPutA enzyme (21.3 μM wild type and 17.9 μM D779Y) were preincubated with 0.1 mM NAD+ in 50 mM potassium phosphate (pH 7.5, 25 mM NaCl) and rapidly mixed with 40 mM proline in 50 mM potassium phosphate buffer (pH 7.5, 25 mM NaCl) (all concentrations reported as final concentrations after mixing).28 Anaerobic conditions were achieved by degassing buffer, substrate, and enzyme solutions by performing repeated vacuum/nitrogen cycles followed by addition of protocatechuate dioxgenase (PCD) (0.05 unit/mL) and protocatechuic acid (PCA) (100 μM), which scrub dissolved oxygen. All enzyme manipulations were performed in an anaerobic glovebox (Belle Technology) prior to the experiments. Rapid-reaction experiments were performed with a Hi-Tech Scientific SF-61DX2 stopped-flow instrument equipped with a photodiode array detector. The stopped-flow mixing cell and tubing were thoroughly washed and incubated overnight with PCA/PCD buffer before stopped-flow syringes were loaded with anaerobic substrate and enzyme solutions. Multiwavelength data (300–700 nm) were recorded, and single-wavelength traces of FAD (451 nm) and NAD+ (340 nm) were extracted and fit to a single-exponential equation to estimate observed rate constants for FAD and NAD+ reduction as previously reported.21
Determination of Crystal Structures and Structural Analysis
Wild-type BjPutA and its mutants were expressed, purified, and crystallized as described previously for wild-type BjPutA.29 Briefly, crystals were grown in sitting drops at room temperature in the presence of ∼2 M ammonium sulfate and cryoprotected with glycerol. For some of the mutants, microseeding was used with a seed stock made initially by crushing crystals of the wild-type enzyme. Seed stocks made from crystals of the mutant enzymes were used in subsequent rounds of crystallization trials. The space group is C2 with a BjPutA dimer in the asymmetric unit. X-ray diffraction data sets were collected at beamline 4.2.2 of the Advanced Light Source using a NOIR-1 detector. The data were integrated with MOSFLM30 and scaled with SCALA.31 Refinements in PHENIX32 were initiated from models derived from the structure of wild-type BjPutA [Protein Data Bank (PDB) entry 3HAZ]. COOT33 was used for model building. The structures were validated with MolProbity34 and the PDB35 validation server. Data collection and refinement statistics are listed in Table 4.
Table 4. X-ray Diffraction Data Collection and Refinementa.
| D779W | D779Y | D778Y | |
|---|---|---|---|
| space group | C2 | C2 | C2 |
| unit cell parameters | a = 166.9 Å | a = 167.1 Å | a = 166.1 Å |
| b = 195.3 Å | b = 196.0 Å | b = 195.1 Å | |
| c = 108.8 Å | c = 108.7 Å | c = 108.4 Å | |
| β = 121.6° | β = 121.4° | β = 121.5° | |
| wavelength (Å) | 1.000 | 1.000 | 1.000 |
| diffraction resolution (Å) | 32.0–2.20 (2.32–2.20) | 32.0–2.30 (2.42–2.30) | 46.9–2.30 (2.42–2.30) |
| no. of observations | 549668 | 490658 | 485882 |
| no. of unique reflections | 149604 | 130815 | 130019 |
| Rmerge(I) | 0.106 (0.464) | 0.103 (0.515) | 0.095 (0.524) |
| Rmeas(I) | 0.124 (0.556) | 0.120 (0.602) | 0.112 (0.612) |
| Rpim(I) | 0.063 (0.302) | 0.061 (0.310) | 0.058 (0.314) |
| mean I/σ | 6.8 (2.1) | 8.1 (2.2) | 10.0 (2.5) |
| completeness (%) | 99.9 (99.3) | 99.3 (98.8) | 99.9 (100) |
| multiplicity | 3.7 (3.3) | 3.8 (3.6) | 3.7 (3.8) |
| no. of protein chains | 2 | 2 | 2 |
| no. of protein residues | 1943 | 1943 | 1941 |
| no. of protein atoms | 14390 | 14386 | 14490 |
| no. of FAD atoms | 106 | 106 | 106 |
| no. of water molecules | 531 | 296 | 419 |
| no. of sulfate ions | 6 | 6 | 8 |
| no. of glycerol molecules | 4 | 3 | 4 |
| Rcryst | 0.208 | 0.216 | 0.195 |
| Rfreeb | 0.241 | 0.251 | 0.235 |
| root-mean-square deviation for bond lengths (Å) | 0.008 | 0.008 | 0.009 |
| root-mean-square deviation for bond angles (deg) | 1.102 | 1.107 | 1.106 |
| Ramachandran plotc | |||
| favored (%) | 98.8 | 98.1 | 98.1 |
| outliers (no. of residues) | 2 | 2 | 0 |
| average B factors (Å2) | |||
| protein | 31.5 | 38.9 | 34.5 |
| FAD | 20.0 | 29.3 | 25.2 |
| water | 28.5 | 31.8 | 30.4 |
| sulfate | 61.4 | 67.6 | 74.3 |
| glycerol | 36.5 | 47.3 | 45.3 |
| coordinate error (Å)d | 0.27 | 0.31 | 0.28 |
| PDB entry | 4Q71 | 4Q72 | 4Q73 |
Values for the outer resolution shell of data are given in parentheses.
A 5% random test set. A common set was used for refinement of all structures.
The Ramachandran plot was generated with RAMPAGE.46
Maximum likelihood-based coordinate error estimate reported by PHENIX.
The substrate-channeling cavity/tunnel system was analyzed and visualized with VOIDOO,36 which characterizes cavities, and MOLE,37,38 which finds tunnels that connect cavities to the bulk medium. Hydrogen atoms were added to the protein with the WHAT IF web services prior to these calculations.39 VOIDOO was run in probe-occupied mode (option O) with a probe radius of 2.9 Å, which approximates P5C/GSA. This radius was chosen on the basis of molecular volume calculations performed with VOIDOO; P5C and GSA have volumes of 104 and 124 Å3, respectively, which correspond to spheres with radii of 2.9 and 3.1 Å, respectively. MOLE was run with default options and using Arg456 of the PRODH active site as the starting point.
Models of P5C and GSA were built into the cavity/tunnel system to understand the steric relationships and estimate the number of intermediates that the system accommodates. The starting models were downloaded from the National Center for Biotechnology Information PubChem database [compound identification numbers 193305 (GSA) and 11966181 (P5C)]. A model of P5C bound in the BjPutA PRODH active site was built using the structure of GsPutA complexed with the proline analogue l-tetrahydro-2-furoic acid (PDB entry 4NMA). A model of GSA bound in the BjPutA P5CDH active site was built using the structure of mouse P5CDH complexed with glutamate (PDB entry 3V9K). Models of GSA were fit manually into the tunnel between the two active sites and the off-pathway cavity.
Results
Rationale for Channel-Blocking Mutagenesis and Purification of BjPutA Mutant Enzymes
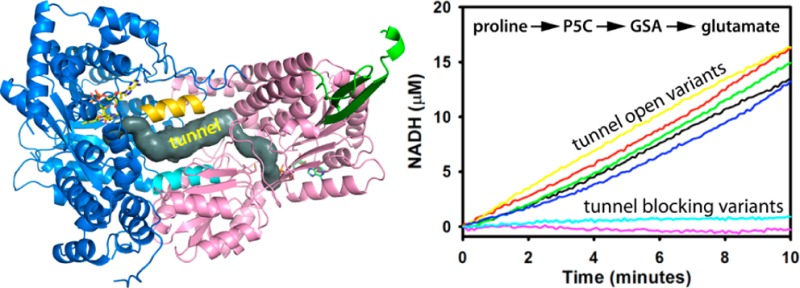
The BjPutA dimer (PDB entry 3HAZ) was analyzed with the PyMOL plug-in CAVER40,41 and MOLE 2.0 to identify residues lining the cavity/tunnel system that, upon mutation to a larger side chain, might eliminate sections of the channeling apparatus. Using starting points in the PRODH site, the programs identified several channels leading to the bulk solvent, including some that connect the two active sites (Figure 1A). (Although the tunnel appears to be open to the bulk medium as shown for the protomer in Figure 1A, we note that it is buried by the dimerization flap of the corresponding protomer in the tetramer that forms in solution.) This tunnel features a prominent central section that runs between and parallel to two helices, helix α5a of the PRODH domain (residues 346–356) and helix 770s of the P5CDH domain (residues 773–785). Side chains of these helices contribute to the walls of the tunnel. The central section is 25 Å in length and 4–8 Å in diameter and can accommodate two to three molecules of GSA (Figure 1B).
Analysis with VOIDOO also identifies a cavity that is connected to the central section of the predicted tunnel (Figure 1C). This “off-pathway” cavity has a volume of ∼700 Å3, which is sufficient to accommodate another two to three molecules of GSA.
Four residues lining the central section of the tunnel were selected for mutagenesis: Thr348, Ser607, Asp778, and Asp779. Thr348 and Ser607 sit near the beginning and end of the central section, respectively, while Asp778 and Asp779 are closer to the middle of the central section, near the off-pathway cavity (Figure 1B). Each of the targeted residues was mutated to Tyr, which retains polarity while increasing steric bulk. Additionally, Asp779 was mutated to Trp and Ala. The Trp mutation further increases side chain bulk, whereas Ala decreases the size and removes the functional property of the side chain carboxylate.
All six BjPutA mutant proteins, T348Y, S607Y, D778Y, D779Y, D779W, and D779A, were purified and shown to have flavin spectra similar to that of wild-type BjPutA with flavin peak absorbances at 380 and 451 nm. From the flavin absorbance spectra, the percent bound flavin was estimated to be 74–99% per monomer for the mutants, which is similar to 79% bound flavin for wild-type BjPutA.
Channeling Assays of BjPutA Mutants
The impact of the mutations on channeling was evaluated by measuring coupled PRODH-P5CDH activity. The assay involves monitoring the progress curve of the production of NADH from proline and determining whether an initial lag phase is apparent in NADH formation.21 As shown in Figure 2, the production of NADH by wild-type BjPutA does not exhibit a perceptible lag time, which is consistent with channeling. The progress curves of NADH formation with BjPutA mutants T348Y, S607Y, D778Y, and D779A likewise show no substantial lag phase, indicating that substrate channeling is unperturbed in these mutants (Figure 2). The linear rate of NADH formation achieved with these mutants is similar to that of the wild type (∼1.4 μM/min) at the same enzyme concentration (0.187 μM). No significant NADH formation, however, was observed with BjPutA mutants D779Y and D779W (Figure 2).
Figure 2.
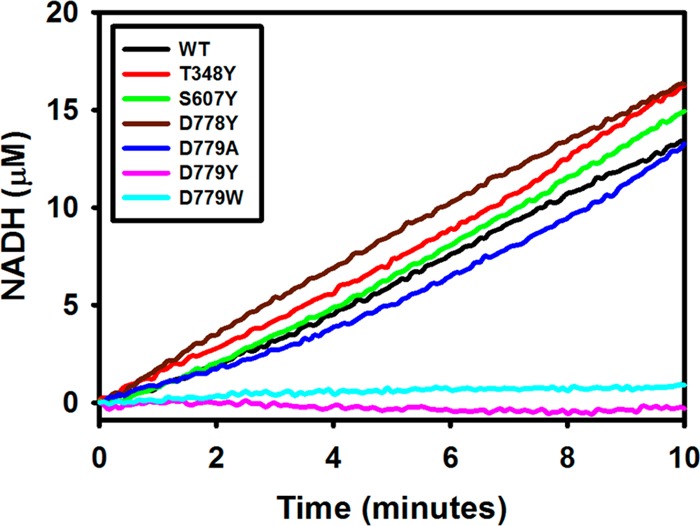
Channeling assays of wild-type BjPutA and its mutants. Assays were performed in 50 mM potassium phosphate (pH 7.5, 25 mM NaCl, 10 mM MgCl2) with 0.187 μM BjPutA enzyme, 40 mM proline, 100 μM CoQ1, and 200 μM NAD+.
Mutants D779Y and D779W were then assayed using an up to 10-fold higher concentration of enzyme (1.87 μM) and fluorescence spectroscopy to detect NADH formation (Figure 3). Increasing the D779Y concentration to 10-fold higher than that of wild-type BjPutA (0.187 μM) resulted in a similar rate of NADH formation, suggesting that the coupled PRODH–P5CDH activity of D779Y is ∼10-fold lower than that of wild-type BjPutA (Figure 3A). At a 10-fold higher D779W concentration, NADH formation remained very slow, indicating that the D779W mutant is severely impaired (Figure 3B).
Figure 3.

Channeling assays with increasing concentrations of D779Y (A) and D779W (B). NADH formation was monitored using fluorescence by exciting at 340 nm and recording the emission at 460 nm. Assays were performed with wild-type BjPutA (0.187 μM) and increasing concentrations of mutants (0.187–1.87 μM) in 50 mM potassium phosphate (pH 7.5, 25 mM NaCl, 10 mM MgCl2) containing 40 mM proline, 100 μM CoQ1, and 200 μM NAD+.
Steady-State Kinetic Properties of Wild-Type BjPutA and Its Mutants
The kinetic parameters of PRODH and P5CDH were then determined for wild-type BjPutA and its mutants. The steady-state kinetic parameters of the PRODH domain were determined using proline and CoQ1 as substrates (Table 2). Similar kcat/Km values (within ∼2-fold) were found for wild-type BjPutA and all the mutants except D778Y. D778Y exhibited comparable Km values for proline (91 mM) and CoQ1 (82 μM), but its kcat value was nearly 9-fold lower than that of wild-type BjPutA, resulting in a significantly lower kcat/Km. This result was unexpected because D778Y exhibited activity similar to that of wild-type BjPutA in the channeling assays (Figure 2).
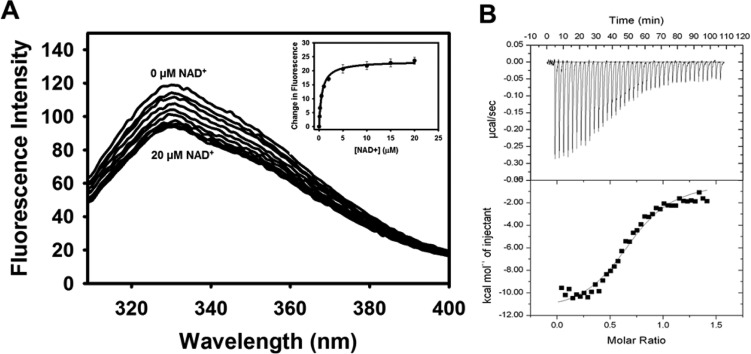
The kinetic parameters of P5CDH were also determined for wild-type BjPutA and its mutants (Table 3). The kcat/Km values for P5CDH activity in the mutants were similar to those of wild-type BjPutA except for mutants D779Y and D779W. The kcat/Km values of D779Y and D779W were 81- and 941-fold lower, respectively, than that of wild-type BjPutA. To determine whether perturbations in NAD+ binding account for the severe loss of P5CDH activity, NAD+ binding was measured for wild-type BjPutA and its mutants (Table 3). For wild-type BjPutA, dissociation constants (Kd) of 0.6 and 1.5 μM were determined by intrinsic tryptophan fluorescence quenching (Figure 4A) and ITC (Figure 4B), respectively. The Kd values of binding of NAD+ to the BjPutA mutants were shown by intrinsic tryptophan fluorescence quenching to be similar to that of wild-type BjPutA (Table 3). Thus, NAD+ binding is unchanged in the mutants, suggesting that the severe decrease in P5CDH activity of D779Y and D779W is not caused by alterations in the Rossmann fold domain.
Figure 4.
Binding of NAD+ to BjPutA. (A) Wild-type BjPutA (0.25 μM) was titrated with increasing concentrations of NAD+ (0–20 μM) in 50 mM potassium phosphate buffer (pH 7.5). The inset is a plot of the change in tryptophan fluorescence vs [NAD+] fit to a single-site binding isotherm. A Kd value of 0.60 ± 0.04 μM was estimated for the NAD+–BjPutA complex. (B) ITC analysis of binding of NAD+ to wild-type BjPutA. The top panel shows the raw data of wild-type BjPutA (23.4 μM) titrated with increasing amounts of NAD+ in 50 mM Tris buffer (pH 7.5). The bottom panel shows the integration of the titration data. The binding of NAD+ to BjPutA is shown to be exothermic, and a best fit of the data to a single-site binding isotherm yielded a Kd of 1.5 ± 0.2 μM.
Because the D778Y mutant exhibited no change in P5CDH activity, we sought to determine whether the 9-fold lower PRODH activity impacts the kinetic parameters of the overall PRODH–P5CDH coupled reaction. Steady-state parameters for the overall reaction were determined for wild-type BjPutA and the D778Y mutant by varying the proline concentration and following NADH formation. The overall reaction shows substrate inhibition at high proline concentrations. A Km of 56 ± 30 mM proline and a kcat of 0.49 ± 0.21 s–1 were determined for wild-type BjPutA with a Ki for proline of 24 ± 12 mM. For D778Y, a Km of 27 ± 9 mM proline and a kcat of 0.25 ± 0.05 s–1 were determined with a Ki for proline of 120 ± 36 mM. The kcat/Km values for the overall reaction are thus similar, 8.8 ± 5.9 and 9.3 ± 3.4 M–1 s–1 for wild-type BjPutA and D778Y, respectively. These results indicate that the 9-fold lower PRODH activity of D778Y does not diminish the overall PRODH–P5CDH reaction rate of this mutant, which is consistent with the channeling assays depicted in Figure 2.
Single-Turnover Rapid-Reaction Kinetics
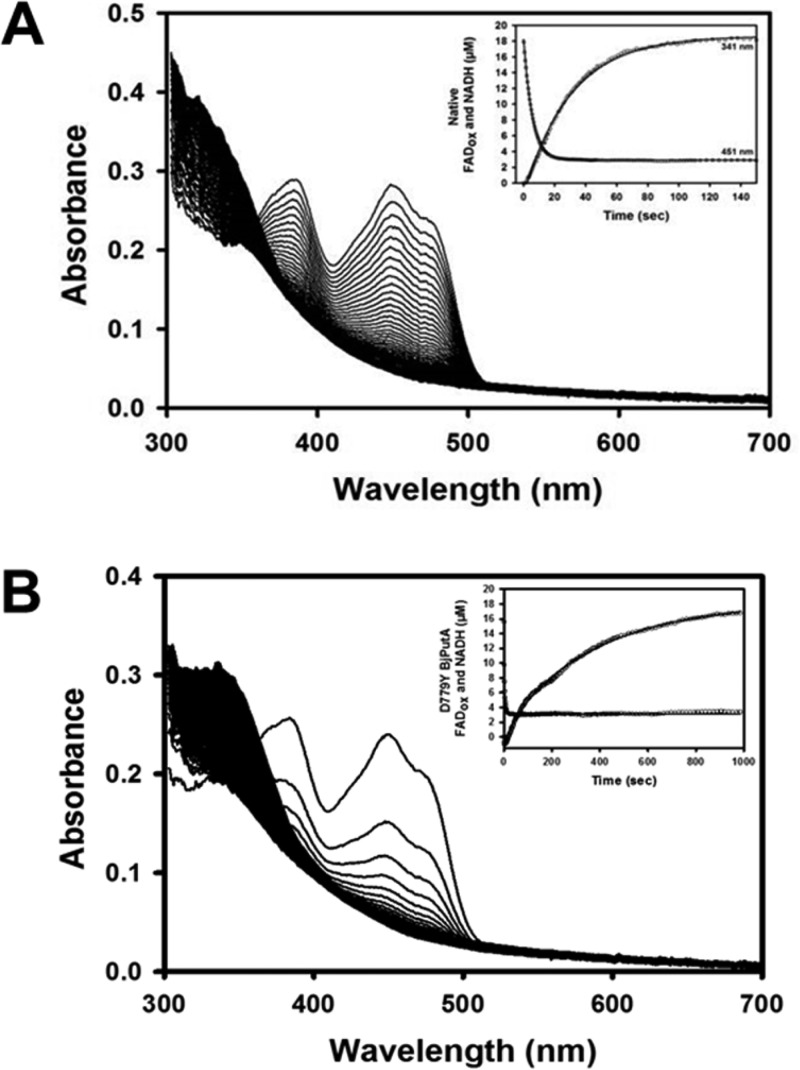
To further corroborate impaired channeling activity in the D779Y mutant, single-turnover experiments were performed anaerobically without an electron acceptor for the flavin cofactor. In this experiment, the PutA enzyme and NAD+ were rapidly mixed with proline and the absorbance spectrum was recorded (Figure 5). Observed rate constants for FAD reduction and NADH formation were estimated by single-exponential fits of absorbance changes at 451 and 340 nm, respectively. The observed rate constant for FAD reduction was faster for BjPutA mutant D779Y (0.46 s–1) than for wild-type BjPutA (0.18 s–1). In contrast, the observed rate constant for NADH formation is 10-fold slower in D779Y (0.003 s–1) than in wild-type BjPutA (0.03 s–1), which is consistent with severely impaired P5CDH activity.
Figure 5.
Single-turnover rapid-reaction kinetic data for wild-type BjPutA and mutant D779Y. (A) Wild-type BjPutA (21.3 μM) and (B) BjPutA mutant D779Y (17.9 μM) were incubated with 100 μM NAD+ and rapidly mixed with 40 mM proline (all concentrations reported as final) and monitored by stopped-flow multiwavelength absorption (300–700 nm). Insets showing FAD (451 nm) and NAD+ (340 nm) reduction vs time fit to a single-exponential equation to obtain the observed rate constant (kobs) of FAD and NAD+ reduction. Note that the inset in panel B is on a longer time scale.
Alternative P5CDH Substrates
The potential tunnel constriction in the D779Y and D779W mutants was explored by measuring P5CDH activity with smaller aldehyde substrates. Table 5 shows the kinetic parameters of wild-type BjPutA and mutants D779A, D779Y, and D779W with exogenous P5C/GSA and smaller substrates succinate semialdehyde and propionaldehyde. Succinate semialdehyde contains one fewer carbon and no amino group, whereas propionaldehyde is a three-carbon aldehyde. The kcat/Km values were significantly lower for each enzyme using the smaller substrates (Table 5). To assess whether succinate semialdehyde and propionaldehyde are more effective substrates in the mutants than P5C/GSA is, the kcat/Km ratio of wild-type BjPutA and each mutant [(kcat/Km)WT/(kcat/Km)mut] was determined for all the substrates. For D779A, the (kcat/Km)WT/(kcat/Km)mut ratio remained ∼1 with each substrate. For the D779Y and D779W mutants, the ratios of (kcat/Km)WT/(kcat/Km)mut ratios were 81 and 941, respectively, with P5C/GSA. The (kcat/Km)WT/(kcat/Km)mut ratios decreased to 30 (D779Y) and 38 (D779W) with succinate semialdehyde, suggesting that relative to P5C/GSA this smaller substrate more readily accesses the P5CDH active site in mutants D779Y and D779W. A further decrease in the (kcat/Km)WT/(kcat/Km)mut ratio, however, was not observed with propionaldehyde.
Table 5. Kinetic Parameters of P5CDH with Alternative Substratesa.
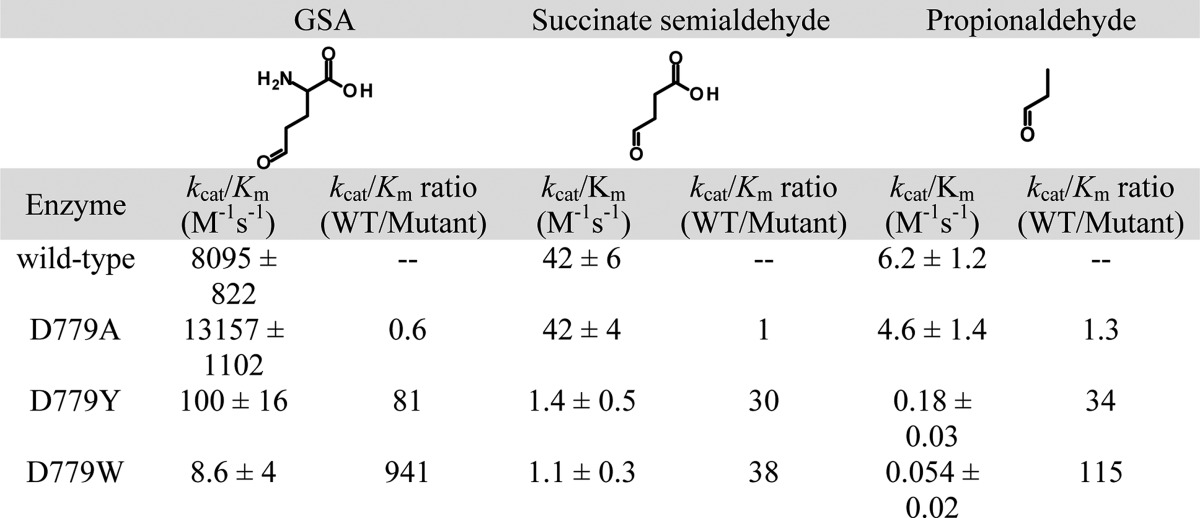
Assays were performed in 50 mM potassium phosphate (pH 7.5, 25 mM NaCl) with 0.2 mM NAD+.
Crystal structures of D778Y, D779Y, and D779W
The structures of D778Y, D779Y, and D779W were determined at 2.2–2.3 Å resolution (Table 4). The electron density features representing the mutated side chains are strong in all three mutant enzymes (Figure 6A–C).
Figure 6.
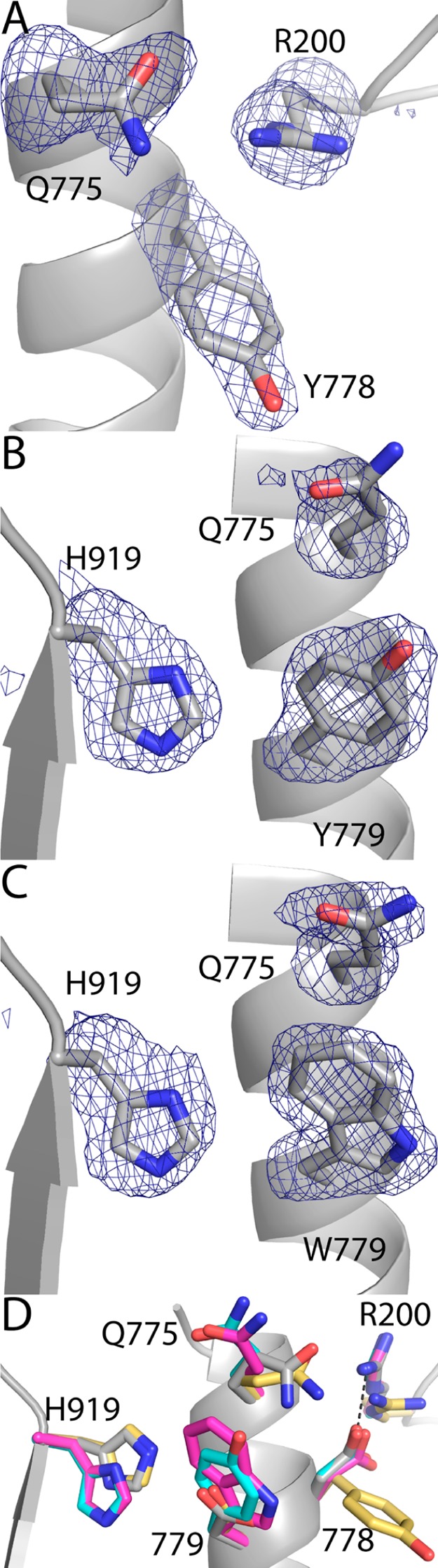
Electron density maps and local conformational changes. (A) Electron density map for D778Y. (B) Electron density map for D779Y. (C) Electron density map for D779W. (D) Superposition of BjPutA (gray), D778Y (gold), D779Y (cyan), and D779W (magenta). The cages in panels A–C represent simulated annealing σA-weighted F0 - Fc omit maps contoured at 2.5σ.
The mutations induce rotations of neighboring side chains but otherwise have minimal impact on the protein structure (Figure 6D). In the wild-type enzyme structure, Asp778 and Arg200 are within 2.8 Å of each other and form an ion pair; the mutation of Asp778 to the larger Tyr would result in steric clash in the absence of conformational changes. Clash is avoided because Tyr778 has rotated by 100° around χ1 relative to Asp778 of the wild-type enzyme. This movement is accompanied by rotation of Arg200 into the space occupied by the carboxylate of Asp778 in the wild-type enzyme. In contrast to D778Y, mutation of Asp779 to Tyr or Trp does not change χ1. Nevertheless, these mutations cause rotations of His919 and Gln775 to prevent steric clash with the new, bulkier side chain at position 779 (Figure 6D). Aside from these local perturbations, no other significant structural changes are evident. In particular, the active site structures are essentially unchanged.
Mutation of Asp778 to Tyr substantially changes the off-pathway cavity located near the central section of the predicted channeling pathway. Asp778 borders this cavity in wild-type BjPutA (Figure 1C). Because of the aforementioned 100° rotation around χ1, the phenol ring of Tyr778 invades the space corresponding to the off-pathway cavity of the wild-type enzyme (Figure 7). The presence of Tyr778 in this region reduces the volume of the cavity by 70% to 200 Å3, so that just a residual cavity remains (Figure 7, blue surface). Furthermore, the close approach of Tyr778 to Arg356 severs the connection between the cavity and the predicted channeling tunnel (using a 2.9 Å probe). Thus, the structure suggests that P5C/GSA molecules that are moving through the tunnel of D778Y cannot enter the off-pathway cavity.
Figure 7.
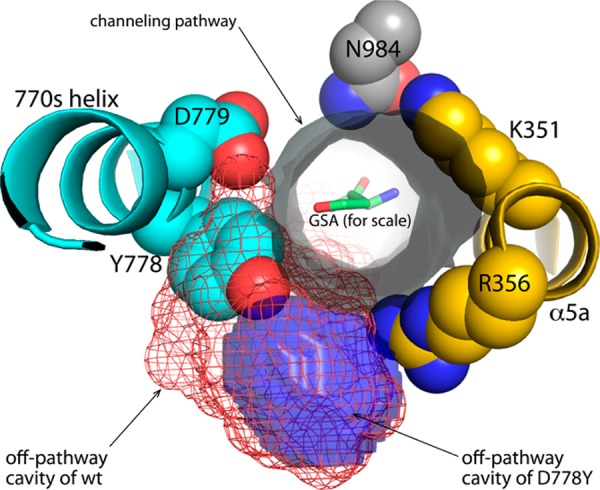
Invasion of the off-pathway cavity by Tyr778 in D778Y. The gray cylinder represents the channeling pathway calculated from the wild-type BjPutA structure (PDB entry 3HAZ) using MOLE, and the view is from the P5CDH active site looking through the tunnel toward the PRODH site. The red mesh represents the off-pathway cavity of wild-type BjPutA calculated using VOIDOO, while the blue surface represents the residual off-pathway cavity of D778Y, also calculated with VOIDOO.
In contrast to the D778Y mutation, the mutation of Asp779 to Tyr constricts the predicted channeling tunnel without affecting the off-cavity pathway (Figure 8). The side chain of Tyr779 pokes into the space corresponding to the central section of the tunnel in the wild-type enzyme (Figure 8A). As a result, the predicted tunnel of D779Y has a 2.0 Å invagination near the phenol hydroxyl (Figure 8B). This narrowing of the tunnel reflects a decrease in distance between helices 770s and α5a. In particular, the distance between the side chains of residue 779 and Lys351 decreases from 9.3 Å in the wild-type enzyme to only 6.8 Å in D779Y. Thus, the gap between these side chains decreases by ∼2.5 Å, which accounts for the invagination of the tunnel near Tyr779.
Figure 8.
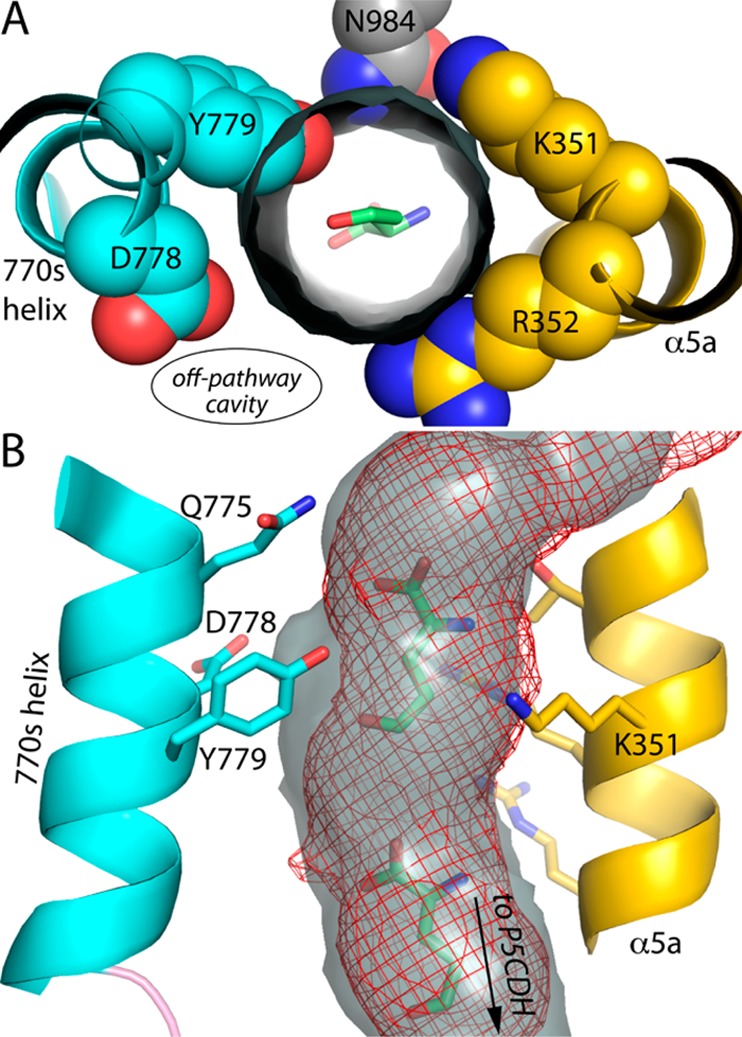
Constriction of the channeling tunnel by Tyr779 in D779Y. (A) The gray cylinder represents the channeling pathway calculated from the wild-type BjPutA structure (PDB entry 3HAZ) using MOLE, and the view is from the P5CDH active site looking through the tunnel toward the PRODH site. (B) Comparison of the predicted channeling pathway of wild-type BjPutA (gray surface) and D779Y (red mesh).
The mutation of Asp779 to Trp similarly reshapes the predicted channeling tunnel (Figure 9). As in D779Y, the bulky side chain of Trp779 penetrates the space corresponding to the tunnel in the wild-type enzyme (Figure 9A). Also, Gln775, which has rotated relative to the wild-type enzyme, protrudes into the tunnel just upstream from Trp779. The invasion of the tunnel by these residues reshapes the predicted channeling pathway, essentially shaving a 2 Å slice off one side of the tunnel (Figure 9B).
Figure 9.
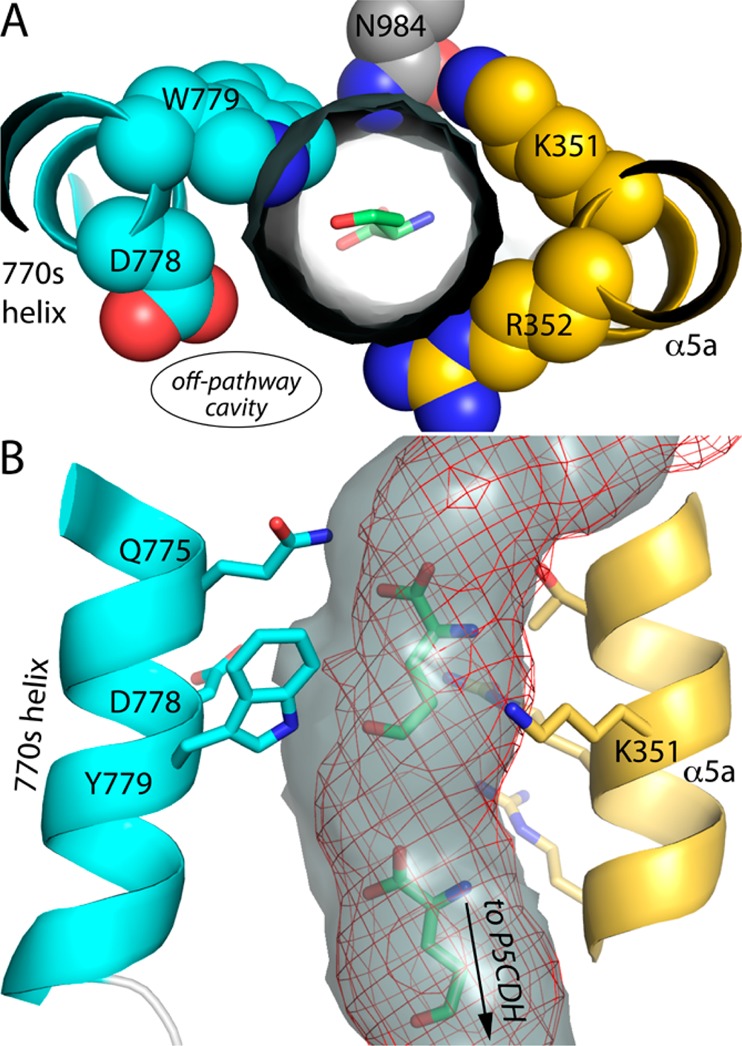
Constriction of the channeling tunnel by Trp779 in D779W. (A) The gray cylinder represents the channeling pathway calculated from the wild-type BjPutA structure (PDB entry 3HAZ) using MOLE, and the view is from the P5CDH active site looking through the tunnel toward the PRODH site. (B) Comparison of the predicted channeling pathway of wild-type BjPutA (gray surface) and D779W (red mesh).
Discussion
Introducing residues with bulkier side chains into a predicted channeling path is a useful approach for validating substrate channeling and exploring the structural architecture of an interconnecting path between active sites. In tryptophan synthase, substitution of βCys170 with Trp in the tunnel pathway significantly hindered passage of the indole intermediate between active sites and also impacted communication between subunits.42 In the bifunctional enzyme dethiobiotin synthetase (DTBS)-diaminopelargonic acid aminotransferase (DAPAT-AT) from Arabidopsis, two mutations were made in a crevice on the surface connecting the two active sites.43 The surface crevice was proposed to be a channel pathway for movement of the intermediate from DAPA-AT to DTBS. Mutation of two crevice residues, Ser360 to Tyr and Ile793 to Trp, resulted in long lag times (∼10–12 min) for product formation, whereas no lag phase was observed with the wild-type enzyme. These results were consistent with the predicted function of the crevice as a channeling path.
Here, we substituted four residues at different points along the predicted channeling path in BjPutA with bulkier side chains. Although Thr348 and Ser607 are located at apparent bottleneck regions and Asp778 points toward the middle of the channel, substitutions of these residues with Tyr did not impact PRODH–P5CDH channeling activity in BjPutA. Only replacement of Asp779 with Tyr or Trp disrupted coupled PRODH–P5CDH activity. Substitution of Asp779 with Ala did not diminish channeling, indicating that the carboxylate group of Asp779 is not critical for channel function. The decrease in the substrate channeling activity of the D779Y and D779W mutants correlates with a significant drop in P5CDH activity, whereas the PRODH activity of the mutants is similar to that of wild-type BjPutA. The X-ray crystal structures of the D779Y and D779W mutants show that the PRODH and P5CDH domains are essentially unchanged from that of wild-type BjPutA. The only structural perturbations are in the side chain conformations of residues near Asp779. Thus, the severely impaired substrate channeling and P5CDH activities of the D779Y and D779W mutants are likely caused by local effects of substituting a larger side chain in the channel. Replacing Asp779 with Tyr decreased the internal width of the predicted channeling path between helices 770s (residues 773–785) and α5a by 2.5 Å or ∼25%. In D779W, the Trp residue carves into the channel by 2.0 Å. These changes result in a narrowing of the tunnel that is sufficient to disrupt substrate channeling and illustrates that the channel structure is finely tuned for transporting P5C/GSA. The results with D779Y and D779W also validate the tunnel in BjPutA identified by X-ray crystallography as the path for channeling the P5C/GSA intermediate.
An outstanding question in PutA enzymes is how P5C/GSA accesses the P5CDH active site. Because the X-ray crystal structures of D779Y and D779W show no changes in the P5CDH active site relative to that of wild-type BjPutA, the significantly lower P5CDH activity of the D779Y and D779W mutants indicates exogenous P5C enters the tunnel upstream of Asp779 possibly via the PRODH active site. If P5C/GSA were able to enter the P5CDH active site from a point downstream of Asp779, the P5CDH activity of the D779Y/W mutants would be expected to be similar to that of the wild-type enzyme. These results indicate that exogenous P5C/GSA must access the P5CDH domain through the channel, a feature that is similar to tryptophan synthase in which the indole intermediate enters the β-subunit active site only via the intramolecular tunnel.44 The kinetic results using smaller aldehydes as exogenous substrates are consistent with this interpretation. Although the activity of D779W with succinate semialdehyde is still lower than that of wild-type BjPutA, the difference in kcat/Km between wild-type BjPutA and D779W is reduced by 25-fold relative to that of GSA.
Even though it neighbors Asp779, replacing Asp778 with Tyr did not diminish the substrate channeling and P5CDH activities of BjPutA. Similar to the D779Y and D779W mutants, the X-ray crystal structure of D778Y shows no changes in the PRODH and P5CDH domains as only perturbations in local residues of the channel were observed. Introducing a bulkier side chain at Asp778 appears to close the off-pathway cavity from the main channeling path. The coupled PRODH–P5CDH activity of the D778Y mutant is similar to that of wild-type BjPutA, demonstrating that the off-pathway cavity is not required for substrate channeling. The function of the off-cavity pathway in substrate channeling thus remains unknown. An interesting finding with the D778Y mutant was its significantly lower PRODH activity. This result may provide additional evidence of a communication link between the PRODH domain and the channel. Recently, we have shown in PutA from E. coli that a substrate channeling step becomes activated during enzyme turnover, thereby increasing the overall PRODH–P5CDH activity by nearly 40-fold.23 PutA also undergoes a conformational change upon flavin reduction, with a conserved ion pair (Arg456–Glu197) proposed to act as a gate between the PRODH domain and the main channeling pathway.21,45 Residues that are critical for communication between the PRODH domain and the channel are unknown, but the findings with D778Y suggest that helix 770s (residues 773–785) may be involved. Despite having 9-fold lower PRODH activity, D778Y exhibited substrate channeling activity similar to that of wild-type BjPutA, consistent with the rate of the coupled PRODH–P5CDH reaction being limited by a channeling step as found previously for E. coli PutA.23
Structural analysis of the channeling path in BjPutA provides new insight into how P5C/GSA is shuttled between the PRODH and P5CDH active sites. Our results suggest that the off-pathway cavity is dispensable for channeling, which implies that the intermediate is constrained to travel through the cylindrical middle section of the tunnel that runs parallel to helices α5a and 770s (residues 773–785) (Figure 1B). The dimensions of this section are consistent with a maximum of two to three intermediates simultaneously occupying the middle section. Furthermore, because the tunnel diameter is similar to the length scales of P5C and GSA, rotational and torsional motions of the intermediates are constrained. In particular, it is unlikely that P5C or GSA can flip orientation while in the tunnel, and torsional motion of GSA is probably restricted. Thus, if the hydrolysis reaction occurs upstream of the P5CDH active site, GSA likely travels though the tunnel with the aldehyde group directed toward the P5CDH active site, as shown in Figure 1B. Potentially, the amino and carboxylic groups of GSA may have a critical role in properly directing its movement and orientation in the tunnel.
Acknowledgments
We thank Dr. Jay Nix of beamline 4.2.2 for help with data collection and processing. Part of this work was conducted at the Advanced Light Source, which is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract DE-AC02-05CH11231.
Glossary
Abbreviations
- CoQ1
ubiquinone-1
- D778Y
site-directed mutant of BjPutA in which Asp778 is replaced with Tyr
- D779A, D779Y, and D779W
site-directed mutants of BjPutA in which Asp779 is replaced with Ala, Tyr, and Trp, respectively
- S607Y
site-directed mutant of BjPutA in which Ser607 is replaced with Tyr
- T348Y
site-directed mutant of BjPutA in which Thr348 is replaced with Tyr
- BjPutA
proline utilization A from B. japonicum
- FAD
flavin adenine dinucleotide
- GSA
glutamate-γ-semialdehyde
- PRODH
proline dehydrogenase
- PCD
protocatechuate dioxygenase
- PCA
protocatechuic acid
- P5C
Δ1-pyrroline-5-carboxylate
- P5CDH
Δ1-pyrroline-5-carboxylate dehydrogenase
- PutA
proline utilization A
- ITC
isothermal titration calorimetry.
Accession Codes
Atomic coordinates and structure factors have been deposited in the Protein Data Bank as entries 4Q71 (D779W), 4Q72 (D779Y), and 4Q73 (D778Y).
Research reported here was supported by National Institutes of Health Grants GM065546 and P30GM103335 and is a contribution of the University of Nebraska Agricultural Research Division, supported in part by funds provided by the Hatch Act.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Nakajima K.; Inatsu S.; Mizote T.; Nagata Y.; Aoyama K.; Fukuda Y.; Nagata K. (2008) Possible involvement of putA gene in Helicobacter pylori colonization in the stomach and motility. Biomed. Res. 29, 9–18. [DOI] [PubMed] [Google Scholar]
- Krishnan N.; Doster A. R.; Duhamel G. E.; Becker D. F. (2008) Characterization of a Helicobacter hepaticus putA mutant strain in host colonization and oxidative stress. Infect. Immun. 76, 3037–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weelden S. W.; Fast B.; Vogt A.; van der Meer P.; Saas J.; van Hellemond J. J.; Tielens A. G.; Boshart M. (2003) Procyclic Trypanosoma brucei do not use Krebs cycle activity for energy generation. J. Biol. Chem. 278, 12854–12863. [DOI] [PubMed] [Google Scholar]
- Bringaud F.; Riviere L.; Coustou V. (2006) Energy metabolism of trypanosomatids: Adaptation to available carbon sources. Mol. Biochem. Parasitol. 149, 1–9. [DOI] [PubMed] [Google Scholar]
- Crawford J. M.; Kontnik R.; Clardy J. (2010) Regulating alternative lifestyles in entomopathogenic bacteria. Curr. Biol. 20, 69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis A.; Bender H. U.; Steel G.; Valle D. (2008) PRODH variants and risk for schizophrenia. Amino Acids 35, 673–679. [DOI] [PubMed] [Google Scholar]
- Chakravarti A. (2002) A compelling genetic hypothesis for a complex disease: PRODH2/DGCR6 variation leads to schizophrenia susceptibility. Proc. Natl. Acad. Sci. U.S.A. 99, 4755–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phang J. M.; Donald S. P.; Pandhare J.; Liu Y. (2008) The metabolism of proline, a stress substrate, modulates carcinogenic pathways. Amino Acids 35, 681–690. [DOI] [PubMed] [Google Scholar]
- Tanner J. J., and Becker D. F. (2013) PutA and proline metabolism. In Handbook of Flavoproteins (Hille R., Miller S. M., and Palfey B., Eds.) pp 31–56, Walter de Gruyter, Boston. [Google Scholar]
- Ovadi J. (1991) Physiological significance of metabolic channelling. J. Theor. Biol. 152, 1–22. [PubMed] [Google Scholar]
- Easterby J. S. (1981) A generalized theory of the transition time for sequential enzyme reactions. Biochem. J. 199, 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph J.; Stubbe J. (1995) Investigation of the mechanism of phosphoribosylamine transfer from glutamine phosphoribosylpyrophosphate amidotransferase to glycinamide ribonucleotide synthetase. Biochemistry 34, 2241–2250. [DOI] [PubMed] [Google Scholar]
- Huang X.; Holden H. M.; Raushel F. M. (2001) Channeling of substrates and intermediates in enzyme-catalyzed reactions. Annu. Rev. Biochem. 70, 149–180. [DOI] [PubMed] [Google Scholar]
- Arentson B. W.; Sanyal N.; Becker D. F. (2012) Substrate channeling in proline metabolism. Front. Biosci. 17, 375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K. S. (1999) Fundamental mechanisms of substrate channeling. Methods Enzymol. 308, 111–145. [DOI] [PubMed] [Google Scholar]
- Bearne S. L.; Wolfenden R. (1995) Glutamate γ-semialdehyde as a natural transition state analogue inhibitor of Escherichia coli glucosamine-6-phosphate synthase. Biochemistry 34, 11515–11520. [DOI] [PubMed] [Google Scholar]
- Bearne S. L.; Hekmat O.; Macdonnell J. E. (2001) Inhibition of Escherichia coli CTP synthase by glutamate γ-semialdehyde and the role of the allosteric effector GTP in glutamine hydrolysis. Biochem. J. 356, 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoden J. B.; Huang X.; Raushel F. M.; Holden H. M. (1999) The small subunit of carbamoyl phosphate synthetase: Snapshots along the reaction pathway. Biochemistry 38, 16158–16166. [DOI] [PubMed] [Google Scholar]
- Farrant R. D.; Walker V.; Mills G. A.; Mellor J. M.; Langley G. J. (2001) Pyridoxal phosphate de-activation by pyrroline-5-carboxylic acid. Increased risk of vitamin B6 deficiency and seizures in hyperprolinemia type II. J. Biol. Chem. 276, 15107–15116. [DOI] [PubMed] [Google Scholar]
- Surber M. W.; Maloy S. (1998) The PutA protein of Salmonella typhimurium catalyzes the two steps of proline degradation via a leaky channel. Arch. Biochem. Biophys. 354, 281–287. [DOI] [PubMed] [Google Scholar]
- Srivastava D.; Schuermann J. P.; White T. A.; Krishnan N.; Sanyal N.; Hura G. L.; Tan A.; Henzl M. T.; Becker D. F.; Tanner J. J. (2010) Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. U.S.A. 107, 2878–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H.; Arentson B. W.; Becker D. F.; Tanner J. J. (2014) Structures of the PutA peripheral membrane flavoenzyme reveal a dynamic substrate-channeling tunnel and the quinone binding site. Proc. Natl. Acad. Sci. U.S.A. 111, 3389–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxley M. A.; Sanyal N.; Krishnan N.; Tanner J. J.; Becker D. F. (2014) Evidence for hysteretic substrate channeling in the proline dehydrogenase and Δ1-pyrroline-5-carboxylate dehydrogenase coupled reaction of proline utilization A (PutA). J. Biol. Chem. 289, 3639–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams I.; Frank L. (1975) Improved chemical synthesis and enzymatic assay of Δ1-pyrroline-5-carboxylic acid. Anal. Biochem. 64, 85–97. [DOI] [PubMed] [Google Scholar]
- Moxley M. A.; Becker D. F. (2012) Rapid reaction kinetics of proline dehydrogenase in the multifunctional proline utilization A protein. Biochemistry 51, 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N.; Becker D. F. (2005) Characterization of a bifunctional PutA homologue from Bradyrhizobium japonicum and identification of an active site residue that modulates proline reduction of the flavin adenine dinucleotide cofactor. Biochemistry 44, 9130–9139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxley M. A.; Tanner J. J.; Becker D. F. (2011) Steady-state kinetic mechanism of the proline:ubiquinone oxidoreductase activity of proline utilization A (PutA) from Escherichia coli. Arch. Biochem. Biophys. 516, 113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil P. V.; Ballou D. P. (2000) The use of protocatechuate dioxygenase for maintaining anaerobic conditions in biochemical experiments. Anal. Biochem. 286, 187–192. [DOI] [PubMed] [Google Scholar]
- Schuermann J. P.; White T. A.; Srivastava D.; Karr D. B.; Tanner J. J. (2008) Three crystal forms of the bifunctional enzyme proline utilization A (PutA) from Bradyrhizobium japonicum. Acta Crystallogr. F64, 949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A. G. (2006) The integration of macromolecular diffraction data. Acta Crystallogr. D62, 48–57. [DOI] [PubMed] [Google Scholar]
- Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D62, 72–82. [DOI] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkoczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L. W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V. B.; Arendall W. B. III; Headd J. J.; Keedy D. A.; Immormino R. M.; Kapral G. J.; Murray L. W.; Richardson J. S.; Richardson D. C. (2010) MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M.; Westbrook J.; Feng Z.; Gilliland G.; Bhat T. N.; Weissig H.; Shindyalov I. N.; Bourne P. E. (2000) The Protein Data Bank. Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt G. J.; Jones T. A. (1994) Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D50, 178–185. [DOI] [PubMed] [Google Scholar]
- Berka K.; Hanak O.; Sehnal D.; Banas P.; Navratilova V.; Jaiswal D.; Ionescu C. M.; Svobodova Varekova R.; Koca J.; Otyepka M. (2012) MOLEonline 2.0: Interactive web-based analysis of biomacromolecular channels. Nucleic Acids Res. 40, W222–W227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrek M.; Kosinova P.; Koca J.; Otyepka M. (2007) MOLE: A Voronoi diagram-based explorer of molecular channels, pores, and tunnels. Structure 15, 1357–1363. [DOI] [PubMed] [Google Scholar]
- Hekkelman M. L.; Te Beek T. A.; Pettifer S. R.; Thorne D.; Attwood T. K.; Vriend G. (2010) WIWS: A protein structure bioinformatics Web service collection. Nucleic Acids Res. 38, W719–W723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chovancova E.; Pavelka A.; Benes P.; Strnad O.; Brezovsky J.; Kozlikova B.; Gora A.; Sustr V.; Klvana M.; Medek P.; Biedermannova L.; Sochor J.; Damborsky J. (2012) CAVER 3.0: A tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput. Biol. 8, e1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, version 1.3r1 (2010) Schrodinger, LLC, Portland, OR. [Google Scholar]
- Anderson K. S.; Kim A. Y.; Quillen J. M.; Sayers E.; Yang X. J.; Miles E. W. (1995) Kinetic characterization of channel impaired mutants of tryptophan synthase. J. Biol. Chem. 270, 29936–29944. [DOI] [PubMed] [Google Scholar]
- Cobessi D.; Dumas R.; Pautre V.; Meinguet C.; Ferrer J. L.; Alban C. (2012) Biochemical and structural characterization of the Arabidopsis bifunctional enzyme dethiobiotin synthetase-diaminopelargonic acid aminotransferase: Evidence for substrate channeling in biotin synthesis. Plant Cell 24, 1608–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn M. F.; Aguilar V.; Brzovic P.; Drewe W. F. Jr.; Houben K. F.; Leja C. A.; Roy M. (1990) The tryptophan synthase bienzyme complex transfers indole between the α- and β-sites via a 25–30 Å long tunnel. Biochemistry 29, 8598–8607. [DOI] [PubMed] [Google Scholar]
- Brown E. D.; Wood J. M. (1993) Conformational change and membrane association of the PutA protein are coincident with reduction of its FAD cofactor by proline. J. Biol. Chem. 268, 8972–8979. [PubMed] [Google Scholar]
- Lovell S. C.; Davis I. W.; Arendall W. B. III; de Bakker P. I.; Word J. M.; Prisant M. G.; Richardson J. S.; Richardson D. C. (2003) Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins 50, 437–450. [DOI] [PubMed] [Google Scholar]