Abstract
Background:
Germline alterations in the proapoptotic protein Bcl-2–like 11 (BIM) can have a crucial role in tumor response to treatment. To determine the clinical utility of detecting BIM deletion polymorphism in non–small-cell lung cancer positive for epidermal growth factor receptor (EGFR) mutation, we examined outcomes of patients with and without BIM alterations.
Methods:
We studied 70 patients with EGFR mutation-positive non–small-cell lung cancer who were treated with an EGFR tyrosine kinase inhibitor between January 2008 and January 2013. BIM deletion was analyzed by polymerase chain reaction in 58 samples of peripheral blood and 24 formalin-fixed paraffin-embedded slides of surgical specimens (20 of lung tissue and four of brain tissue); both blood and tissue specimens were available for 12 patients. We retrospectively analyzed clinical characteristics, response rate, toxicity, and outcomes among patients with and without BIM deletion.
Results:
BIM deletion was present in 13 of 70 patients (18.6%). There were no significant differences between patients with and without BIM deletion in clinical characteristics, rate of response to EGFR tyrosine kinase inhibitor, or incidence of adverse events. Patients with BIM deletion had significantly shorter progression-free survival (PFS) than those without BIM deletion (median, 227 versus 533 days; p < 0.001). Multivariate Cox regression analysis showed that BIM deletion was an independent indicator of shorter PFS (hazard ratio, 3.99; 95% confidence interval, 1.864–8.547; p < 0.001).
Conclusions:
Polymerase chain reaction successfully detected BIM deletion in samples of peripheral blood and formalin-fixed paraffin-embedded slides of surgical specimens. BIM deletion was the most important independent prognostic factor in shorter PFS.
Keywords: BIM, Non–small-cell lung cancer, Epidermal growth factor receptor tyrosine kinase inhibitor
An activating mutation of the epidermal growth factor receptor (EGFR) gene is a promising target in the treatment of non–small-cell lung cancer (NSCLC).1,2 The frequency of EGFR mutations depends on the population studied. In North America and Western Europe, approximately 10% of patients with NSCLC harbor mutations, whereas in East Asia approximately 30% have EGFR mutations.3,4 EGFR tyrosine kinase inhibitors (EGFR-TKI) such as gefitinib and erlotinib are recommended for treating EGFR mutation-positive NSCLC.5,6 NSCLC patients with such mutations who were treated with EGFR-TKI as first-line therapy had longer progression-free survival (PFS) than did those who received platinum-based chemotherapy.7–10 Therefore, detection of EGFR mutations in patients with metastatic NSCLC is important in selecting individualized therapies.
However, most patients develop a recurrence within 10 to 16 months after initial EGFR-TKI treatment.11 Approximately 50% of patients with acquired resistance to EGFR-TKI were found to have the EGFR T790M mutation.12,13 Other reported mechanisms responsible for acquired resistance are MET amplification, in 5% to 10% of cases,14,15 and small-cell cancer transformation, in less than 5% of cases.16 However, the mechanisms responsible for acquired EGFR-TKI resistance are not known in approximately 30% to 40% of patients.11
Bcl-2–like 11 (BIM) is a proapoptotic member of the B-cell CLL/Lymphoma 2 (Bcl-2) family of proteins17,18 and has emerged as a key modulator of apoptosis triggered by EGFR-TKI.19,20 Low expression levels of BIM in primary tumors are reported to be associated with shorter PFS in patients treated with EGFR-TKI.21 Recently, Ng et al.22 reported a common intronic deletion polymorphism in the gene encoding BIM. This polymorphism switched BIM splicing from exon 4 to exon 3, which resulted in increased expression of BIM isoforms lacking the proapoptotic Bcl-2-homology domain 3 (BH3). After TKI exposure, cells with the BIM deletion polymorphism showed decreased induction of exon-4-containing transcripts and, consequently, impaired BH3-domain–dependent apoptosis. This germline alteration could have a crucial role in determining how a tumor responds to treatment. However, few studies have examined the clinical usefulness of detecting BIM deletion polymorphism or the clinical characteristics of EGFR mutation-positive NSCLC.
To determine the clinical utility of detecting BIM deletion polymorphism in patients with EGFR mutation-positive NSCLC, we examined the outcomes of patients with and without BIM alterations.
PATIENTS AND METHODS
Polymerase Chain Reaction
To detect BIM deletion polymorphism, we performed two types of polymerase chain reaction (PCR) analysis, according to the method of Ng et al.22 In brief, we used a single primer set that contains the deletion area in intron 2 and two separate primer sets designed for wild-type and deletion alleles. The DNA was subjected to PCR amplification using primers designed to detect deletion site (2903 bp) in intron 2 of the BCL2L11 gene. The resulting PCR products from the deletion (1285 bp) and wild-type (4188 bp) alleles were analyzed on agarose gels. In addition, the PCR products for the deletion (177 bp) and wild-type (174 bp) alleles were analyzed on agarose gel. We analyzed 20 cell lines, including the KCL-22 cell (which was reported to have the BIM deletion),22 and 30 DNA samples from healthy Japanese volunteers.
Clinical Samples
We studied 70 patients with EGFR mutation-positive NSCLC who were treated with EGFR-TKI during the period from January 2008 to January 2013. BIM deletion polymorphism was analyzed by PCR in 58 samples of peripheral blood (cell-free DNA in 34, leukocyte DNA in 35) and on 24 formalin-fixed paraffin-embedded (FFPE) slides of surgical specimens (20 specimens of lung tissue and four of brain tissue); both blood and tissue specimens were available for 12 patients. To confirm the validity of PCR analysis of two types of samples, we compared the results for BIM deletion polymorphism identified in lung tissue on FFPE slides with those from peripheral blood (cell-free DNA or leukocyte DNA) from the same patients (n = 12). DNA was extracted from FFPE slides using the QIAamp FFPE Tissue Kit (QIAGEN KK, Tokyo, Japan). DNA extraction blood samples were diluted in lysis solution to lyse the red cells and the white cell fraction was pelleted and washed once in phosphate-buffered saline. DNA was extracted from the white cell pellets using the QIAamp DNA mini Kit (QIAGEN KK, Tokyo, Japan).
Clinical Outcomes
We retrospectively analyzed the clinical characteristics, response rate (RR), disease control rate (DCR), and toxicity of EGFR-TKI in patients with and without BIM deletion polymorphism. Toxicity was assessed according to National Cancer Institute Common Terminology Criteria for Adverse Events, version 3.
We estimated PFS and overall survival (OS) in patients with and without BIM deletion polymorphism. The PFS of patients treated with EGFR-TKI was assessed from the date EGFR-TKI therapy was started to the earliest sign of disease progression as determined by findings from computed tomography or magnetic resonance imaging, according to the Response Evaluation Criteria in Solid Tumors. OS was defined as the period from the date of diagnosis until death from any cause.
Statistical Analysis
Statistical analyses were conducted using SPSS software for Windows, version 12.0 (SPSS, Tokyo, Japan). Differences in clinical characteristics, RR, DCR, and adverse events between patients with and without BIM deletion polymorphism were compared using Fisher’s exact test. Survival curves were drawn by the Kaplan-Meier method, and statistical analysis was performed using the log-rank test.
We used univariate analysis and multivariate Cox regression analysis to identify factors associated with shorter PFS. The investigated prognostic factors were age, sex (male versus female), performance status (2 versus 1 versus 0), brain metastasis (yes versus no), bone metastasis (yes versus no), pulmonary metastasis (yes versus no), liver metastasis (yes versus no), lymph node metastasis (yes versus no), EGFR mutation (major mutations [L858R and exon 19 deletion] versus minor mutations [other mutations]), EGFR-TKI response (partial response versus stable disease), smoking history (pack-years), and BIM deletion (yes versus no).
This single-center study was conducted at Toho University Omori Medical Center (Tokyo, Japan) and was approved by its Human Genome/Gene Analysis Research Ethical Committee (Authorization number; 24-1).
RESULTS
Detection of BIM Deletion in Cell Lines and Healthy Volunteers
Using the two types of PCR analysis, we analyzed 20 cell lines and 30 DNA samples from healthy Japanese volunteers. Among the 20 cell lines, only KCL-22 showed BIM deletion. As for DNA samples, BIM deletion polymorphism was present in six of the 30 (20%) healthy volunteers. There was no discordance between the two types of PCR analysis.
Validation between Blood Samples and FFPE Slides
We confirmed the validity between blood samples (leukocyte DNA in 12 and cell-free DNA in four) and FFPE slides of surgical specimens (lung tissue in 12): BIM deletion was detected in three of 12 patients (25%). There was no discordance between the two sample types.
Detection of BIM Deletion on EGFR-Positive NSCLC
We analyzed BIM deletion polymorphism in 70 patients with EGFR mutation-positive NSCLC who were treated with EGFR-TKI. BIM deletion polymorphism was present in 13 of the 70 patients (18.6%); homozygous deletion was noted in one and heterozygous deletion in 12. For the one case of homozygous deletion, PCR analysis using the primer set for the wild-type allele showed no amplification (Table 1).
TABLE 1.
Presence of BIM Deletion in Patients with EGFR Mutation-Positive NSCLC (n = 70)

Comparison between Patients with and without BIM Deletion Polymorphism
There were no significant differences in the clinical characteristics, RR, DCR, or incidence of adverse events between patients with (n = 13) and without (n = 57) BIM deletion polymorphism (Tables 2 and 3).
TABLE 2.
Patient Characteristics (n = 70)

TABLE 3.
Comparison of Clinical Response and Adverse Events after EGFR-TKI Therapy

Survival and Indicators of Shorter PFS
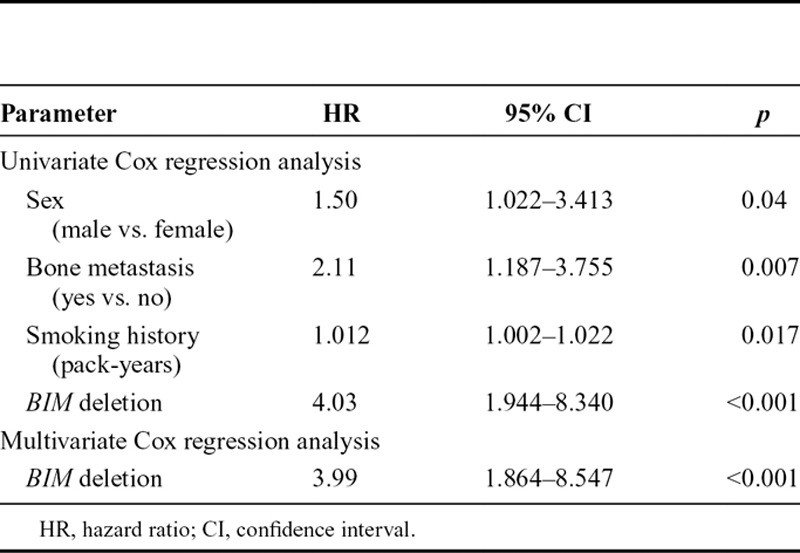
We estimated PFS and OS in patients with and without BIM deletion polymorphism. The patients with BIM deletion polymorphism had significantly shorter PFS than did those without BIM deletion polymorphism (median, 227 versus 533 days; p < 0.001; Fig. 1). There was no significant difference in OS (median, 1176 versus 1363 days; p = 0.27; Fig. 2). Multivariate Cox regression analysis showed that BIM deletion polymorphism was the strongest independent indicator of shorter PFS (hazard ratio [HR], 3.99; 95% confidence interval [CI], 1.864–8.547; p < 0.001; Table 4).
FIGURE 1.

Patients with BIM deletion polymorphism had significantly shorter progression-free survival than did those without BIM deletion polymorphism (median, 227 versus 533 days; p < 0.001).
FIGURE 2.

There was no significant difference in overall survival between patients with and without BIM deletion polymorphism (median, 1176 versus 1363 days; p = 0.27).
TABLE 4.
Indicators of Shorter PFS after EGFR-TKI Treatment
DISCUSSION
BIM deletion polymorphism is a germline alteration that affects EGFR-TKI–related apoptosis.17,18 In a study that screened 2597 healthy individuals, BIM deletion polymorphism was present in 12.3% of East Asians but absent in Africans and Europeans.22 In the present study, BIM deletion polymorphism was present in 13 of 70 Japanese patients (18.6%) with EGFR mutation-positive NSCLC and in six of 30 healthy Japanese volunteers (20%), a statistically insignificant difference. The overall frequency of BIM deletion polymorphism in our study (19%, n = 100) was higher than that noted in a previous report.22
Tagawa et al.23 reported homozygous BIM deletions in patients with mantle-cell lymphoma, and homozygous BIM deletion was found in 0.5% of East Asians.24 Among the present 70 Japanese patients with NSCLC, one (1.4%) had homozygous deletion and 12 had heterozygous deletion. Future studies should investigate the characteristics of patients with homozygous BIM deletion polymorphism to determine if this genotype results in worse clinical outcomes when compared with heterozygous BIM deletion.
There were no significant differences between clinical characteristics, response to EGFR-TKI, or incidences of adverse events due to EGFR-TKI among patients with and without BIM deletion polymorphism. Thus, it is difficult to distinguish between patients with and without BIM deletion polymorphism on the basis of clinical characteristics alone. No patient with BIM deletion developed EGFR-TKI–related pneumonitis. BIM knockdown was reported to prevent FOXO3 (i.e., FKHRL1, a member of the forkhead transcription factor subfamily)-mediated overproduction of reactive oxygen species and apoptosis.25 BIM deletion polymorphism might affect EGFR-TKI–related lung injury by preventing overproduction of reactive oxygen species. Further studies are needed to clarify the relationship between EGFR-TKI–related pneumonitis and BIM.
BIM deletion polymorphism, a germline alteration, is thought to be associated with intrinsic resistance to EGFR-TKI and would likely result in primary resistance and no response to treatment. However, the present clinical outcomes were probably due to acquired resistance: when compared with patients without BIM polymorphism, those with BIM deletion polymorphism had similar RRs and DCRs but shorter PFS. The reasons for these findings remain to be investigated. It has been hypothesized that EGFR-TKI–induced apoptosis does not completely depend on the BIM pathway and that tumor response to EGFR-TKI in patients with BIM deletion might depend on other proapoptotic regulators, which could have less-prolonged clinical activity than those of the BIM pathway. A second hypothesis is that BIM deletion polymorphism itself results in relative resistance to EGFR-TKI. Kuroda et al.26 showed that cancer cells were sensitive to small changes in BIM protein concentrations and that BIM protein concentration exerted a dose-dependent effect on apoptosis and the degree of TKI resistance.26 In a report by Faber et al.,21 PFS was shorter (4.7 versus 13.7 mo, p = 0.007) among patients with low BIM RNA expression, which appeared to correlate with high BIM protein expression on immunohistochemistry. The RR after EGFR-TKI was worse among patients with low BIM RNA expression (44%) than among those with high BIM RNA expression (77%), although the difference was not statistically significant. Patients with BIM deletion polymorphism could be regarded as “carriers” that have varied BIM expression and clinical responses that are modulated by genetic or epigenetic interactions, a possibility that warrants further study. Although cells with BIM deletion polymorphism show decreased induction of exon-4–containing transcripts after TKI exposure,22 the response after prolonged TKI exposure should be investigated.
Ng et al.22 reported that patients with BIM deletion polymorphism had significantly shorter PFS than did patients without BIM deletion polymorphism after EGFR-TKI treatment (6.6 versus 11.9 mo, p = 0.0027), but they did not report RR or OS. Our present study in a Japanese population yielded similar results: BIM deletion polymorphism was an independent indicator of shorter PFS. However, there was no significant difference in OS among patients with and without BIM deletion polymorphism. Multivariate Cox regression analysis showed that indicators of shorter OS were EGFR-TKI–related pneumonitis (HR, 3.52; 95% CI, 1.190–3.860; p = 0.023), brain metastasis (HR, 2.14; 95% CI, 1.099–4.165; p = 0.025), and smoking history (HR, 1.001; 95% CI, 1.000–1.001; p = 0.026). EGFR-TKI–related pneumonitis developed only in patients without BIM deletion polymorphism (n = 8, 14%) but has a detrimental effect on chemotherapy given after pneumonitis. Thus, EGFR-TKI–related pneumonitis might have reduced OS among the present patients without BIM deletion, which possibly explains the lack of a significant difference in OS between patients with and without BIM deletion polymorphism in the present study.
BH3-mimetic drugs22 and histone deacetylase inhibitors24 may be able to surmount BIM-associated resistance to EGFR-TKI. Our findings suggest that although there was no significant difference in RR or OS among patients with and without BIM deletion polymorphism, the addition of these drugs might prolong PFS. However, this study was a retrospective study at a single center. A prospective multicenter study should be conducted to investigate the clinical significance of BIM deletion polymorphism on EGFR-TKI therapy. In addition, EGFR-TKI–related pneumonitis should be considered in any randomized prospective study of the clinical benefit of BH3-mimetic drugs or histone deacetylase inhibitors for patients with BIM deletion polymorphism.
In conclusion, BIM deletion polymorphism, a germline alteration, was successfully detected by PCR analysis of samples of peripheral blood and FFPE slides of surgical specimens, thus providing a minimally invasive and convenient detection method. BIM deletion polymorphism was the strongest indicator of shorter PFS among patients with EGFR mutation-positive NSCLC treated with EGFR-TKI. Our results indicate that new treatment strategies should be established for patients with BIM deletion polymorphism.
ACKNOWLEDGMENTS
We thank Atsushi Kakimoto, Hiroki Todoroki, and Satoshi Natsume of SRL (Tokyo, Japan) and Hiroyuki Mano and Manabu Soda of Jichi Medical University (Tochigi, Japan) for their help. We are also grateful to R. J. Turner of Toho University and David Kipler for their review of the language of this article.
Footnotes
Disclosure: The authors declare no conflict of interest.
REFERENCES
- 1.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 4.Bell DW, Lynch TJ, Haserlat SM, et al. Epidermal growth factor receptor mutations and gene amplification in non-small-cell lung cancer: molecular analysis of the IDEAL/INTACT gefitinib trials. J Clin Oncol. 2005;23:8081–8092. doi: 10.1200/JCO.2005.02.7078. [DOI] [PubMed] [Google Scholar]
- 5.Ettinger DS, Akerley W, Bepler G, et al. NCCN Non-Small Cell Lung Cancer Panel Members. Non-small cell lung cancer. J Natl Compr Canc Netw. 2010;8:740–801. doi: 10.6004/jnccn.2010.0056. [DOI] [PubMed] [Google Scholar]
- 6.Azzoli CG, Temin S, Aliff T, et al. American Society of Clinical Oncology. 2011 focused update of 2009 American Society of Clinical Oncology clinical practice guideline update on chemotherapy for stage IV non-small-cell lung cancer. J Clin Oncol. 2011;29:3825–3831. doi: 10.1200/JCO.2010.34.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 8.Maemondo M, Inoue A, Kobayashi K, et al. North-East Japan Study Group. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 9.Mitsudomi T, Morita S, Yatabe Y, et al. West Japan Oncology Group. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 10.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 11.Ohashi K, Maruvka YE, Michor F, Pao W. Epidermal growth factor receptor tyrosine kinase inhibitor-resistant disease. J Clin Oncol. 2013;31:1070–1080. doi: 10.1200/JCO.2012.43.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 13.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 15.Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci U S A. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 18.Faber AC, Ebi H, Costa C, Engelman JA. Apoptosis in targeted therapy responses: the role of BIM. Adv Pharmacol. 2012;65:519–542. doi: 10.1016/B978-0-12-397927-8.00016-6. [DOI] [PubMed] [Google Scholar]
- 19.Costa DB, Halmos B, Kumar A, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 2007;4:1669–1679; discussion 1680. doi: 10.1371/journal.pmed.0040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong Y, Somwar R, Politi K, et al. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med. 2007;4:e294. doi: 10.1371/journal.pmed.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faber AC, Corcoran RB, Ebi H, et al. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011;1:352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng KP, Hillmer AM, Chuah CT, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18:521–528. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- 23.Tagawa H, Karnan S, Suzuki R, et al. Genome-wide array-based CGH for mantle cell lymphoma: identification of homozygous deletions of the proapoptotic gene BIM. Oncogene. 2005;24:1348–1358. doi: 10.1038/sj.onc.1208300. [DOI] [PubMed] [Google Scholar]
- 24.Nakagawa T, Takeuchi S, Yamada T, et al. EGFR-TKI resistance due to BIM polymorphism can be circumvented in combination with HDAC inhibition. Cancer Res. 2013;73:2428–2434. doi: 10.1158/0008-5472.CAN-12-3479. [DOI] [PubMed] [Google Scholar]
- 25.Hagenbuchner J, Kuznetsov A, Hermann M, Hausott B, Obexer P, Ausserlechner MJ. FOXO3-induced reactive oxygen species are regulated by BCL2L11 (Bim) and SESN3. J Cell Sci. 2012;125(Part 5):1191–1203. doi: 10.1242/jcs.092098. [DOI] [PubMed] [Google Scholar]
- 26.Kuroda J, Puthalakath H, Cragg MS, et al. Bim and Bad mediate imatinib-induced killing of Bcr/Abl+ leukemic cells, and resistance due to their loss is overcome by a BH3 mimetic. Proc Natl Acad Sci U S A. 2006;103:14907–14912. doi: 10.1073/pnas.0606176103. [DOI] [PMC free article] [PubMed] [Google Scholar]


