Abstract
Simultaneous multi-element imaging using NanoSIMS (nanoscale secondary ion mass spectrometry), exploiting the novel combination of 195Pt and 15N in platinum-am(m)ine antitumour drugs, provides information on the internalisation and subcellular localisation of both metal and ligands, and allows identification of ligand exchange.
Understanding the subcellular distribution of metal-based anticancer drugs provides key insights in identifying their organelle and molecular targets. Platinum-based chemotherapeutics have been the subject of intense investigation for decades due to the clinical success of cisplatin (Chart 1).1 There have been numerous studies reporting the tagging of Pt complexes with fluorescent moieties to allow their intracellular distribution to be mapped using fluorescence microscopy,2, 3 as well as various reports on the direct mapping of Pt inside tumour cells by using either synchrotron techniques4 or electron microscopic methods.5 Herein we report on the novel use of simultaneous direct multi-element imaging by nanoscale secondary ion mass spectrometry (NanoSIMS) to monitor tracking and distribution of 15N-labelled Pt antitumour agents within cells. The results confirm the nucleolus as target of highly-charged polynuclear platinum drugs (PPCs), consistent with previous suggestions,3 and show distinct differences in processing compared to the mononuclear agents.6
Chart 1.

Structures of platinum antitumour compounds. Fully 15N-labelled cisplatin and TriplatinNC were used in this study.
Significant advances have been made in visualising cellular distribution of metal-based therapeutics through the application of highly sensitive surface analysis techniques such as secondary ion mass spectrometry (SIMS), to cellular imaging.7 SIMS has been used to study cisplatin-induced alterations in intracellular chemical composition in an established model (LLC-PK(1) cells) for studying renal injury.8 Nanoscale secondary ion mass spectrometry, a recent development in SIMS instrumentation, combines exquisite spatial resolution (50 nm), and the simultaneous detection of both heavy and light elements.9 In NanoSIMS, a high-energy ion beam (Cs+) is rastered across the sample surface, sputtering atoms from the topmost monolayers and generating negative secondary ions. The secondary ions are sorted according to their mass, producing a map of the sample surface showing the distribution of the selected ion species. Furthermore, the high mass resolution of NanoSIMS allows the simultaneous detection of multiple isotopes of the same element (e.g 15N/14N).9 We have previously reported the use of NanoSIMS to detect Au inside tumour cells following treatment with an antitumour Au(I) phosphine complex, resulting in the identification of molecular targets not previously considered.10 In this communication, we extend this technique to the dual imaging of both 15N and 195Pt inside cultured tumour cells following treatment with a 15N-labelled polynuclear Pt compound, TriplatinNC, a non-covalent analogue of the Phase II clinical agent BBR3464 (Chart 1). The results are compared with similar treatment with 15N-cisplatin.
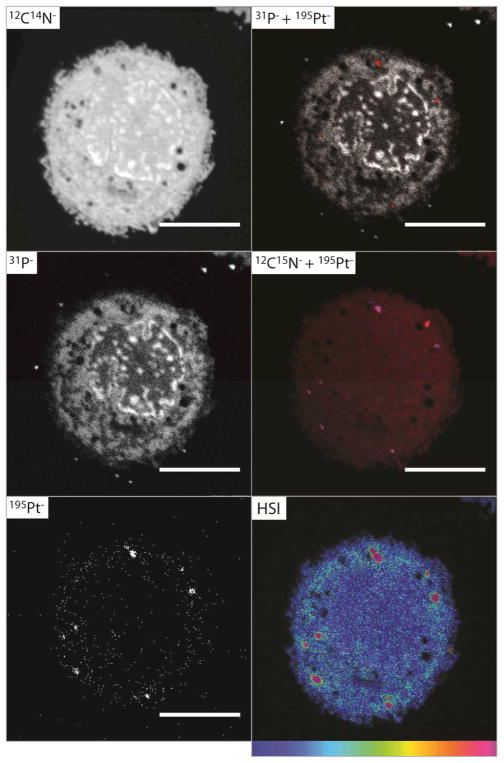
Fig. 1 (and Fig. S1, ESI†) shows NanoSIMS secondary ion images of a fixed section of a single human breast adenocarcinoma (MCF7) cell after 1 h exposure to TriplatinNC (20 μM). The subcellular morphology, nucleic acid and Pt distribution are visible in remarkable detail, and the morphology of the cell is unchanged in comparison to untreated control cells (Fig. S2 and Fig. S3, ESI†). At this early time-point, the 195Pt− ion map shows a clear accumulation of Pt and the formation of discrete ‘hotspots’, possibly endocytic vesicle-like structures, close to the perimeter of the cell. An overlay of the 31P− and 195Pt− secondary ion images reveals conclusively that the Pt is not associated with DNA, where the falsely coloured red spots (195Pt−) are independent of the high 31P− signal. As the Pt compound was fully 15N-labelled, both 14N and 15N counts were measured to determine regions where 15N was present in an amount exceeding the natural abundance. The hue-saturation-intensity (HSI) image allows the direct visualisation of 15N enrichment, where the value of the 15N/14N ratio is represented on a colour scale, and the intensity is an index of the statistical reliability.9 The HSI image in Fig. 1 clearly shows enrichment of 15N around the margin of the cell, and ‘hotspots’ in the cytoplasm (visible as pink).
Fig. 1.
Secondary ion maps acquired by NanoSIMS of fixed sections of an MCF7 cell treated with TriplatinNC (20 μM, 1 h). The 195Pt− (red) and + 31P− (greyscale) overlay shows no colocalisation of Pt and nucleic acids; the overlay of the 195Pt− (blue) 12C15N− (red) secondary ion maps shows Pt and 15N are mostly colocalised; the HSI representation of the 12C15N−/12C14N− ratio shows enrichment of 15N in the cytoplasm as well as ‘hotspots’; scale bars = 5 μm.
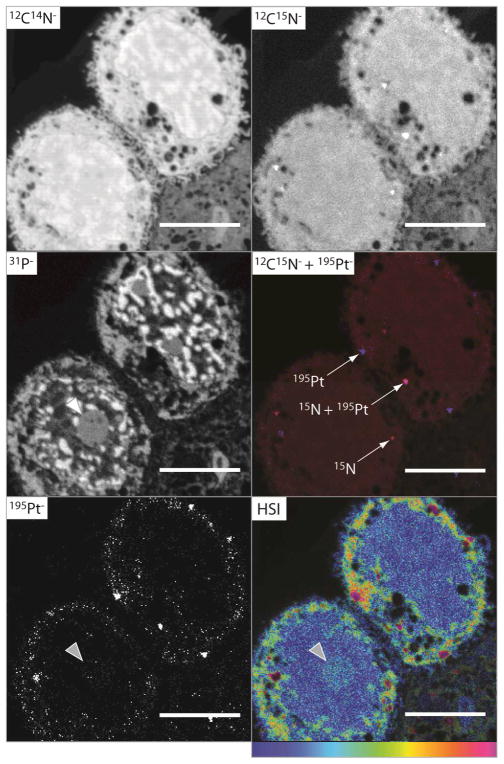
The cellular accumulation of TriplatinNC (20 μM) was also examined after 2 h treatment. Significant accumulation of both 195Pt− and 15N was observed; secondary ion images for two cells are shown in Fig. 2 (and Fig. S4, ESI†). In this case the Pt is located in the vesicle-like structures, and there is significant accumulation in the cytoplasm, reflecting greater accumulation with time. Notably, the HSI image shows 15N enrichment in the nucleolus (grey arrow), at the exclusion of the nucleus. Similar to the cells treated with TriplatinNC for 1 h (Fig. 1), the overlay of the 12C15N− (red) and the 195Pt− (blue) ion images in Fig. 2 shows that there is some colocalisation of 15N and Pt after 2 h, however, interestingly, some of the regions of high Pt counts do not correspond to high enrichment of 15N. This is more pronounced than at 1 h treatment, indicating some metabolism of TriplatinNC has occurred.
Fig. 2.
Secondary ion maps acquired by NanoSIMS of fixed sections of an MCF7 cell treated with TriplatinNC (20 μM, 2 h). The 195Pt− secondary ion map and the hue-saturation-intensity (HSI) representation of the 12C15N−/12C14N− ratio map, clearly show localisation of both 195Pt and 15N within the nucleolus (grey arrow); the overlay of the 195Pt− (blue) 12C15N− (red) secondary ion maps shows Pt and 15N are colocalised in some but not all instances; scale bars = 5 μm.
Secondary ion maps of fixed sections of single MCF7 cells treated with cisplatin under similar conditions (20 μM 1 h) were acquired and show that there are only very few Pt counts, the distribution of which is very diffuse (Fig. S5, ESI†), and that there is no detectable enrichment of 15N (Fig. S6, ESI†). These results show that the uptake of TriplatinNC is significantly different to that of cisplatin and mononuclear cisplatin analogues, where uptake is slower and localisation indiscriminate.6
The 15N enrichment was determined for whole cells and the individual subcellular compartments by extracting the data from the images, and is shown in Fig. S7 (ESI†), along with comparison to equivalent regions from the untreated control samples. These quantitative data support the observation of 15N enrichment in the nucleolus of the cells, but also indicate a small enrichment within the nucleus. The nucleoli of cells treated with TriplatinNC for 2h were found to be enriched up to 0.423 ±0.002 at%, compared to the mean control value of 0.381 ±0.002 at%, while the surrounding nucleus was enriched up to 0.395 ±0.001 at%. The 15N ‘hotspots’ are enriched by as much as 3.16 ±0.03 at% after 2 h. No 15N enrichment was detectable for whole cells or subcellular compartments for cells treated with cisplatin under similar conditions (20 μM for up to 2 h), again reiterating the different uptake mechanism for the polynuclear Pt compound. The enrichment of 15N in the nucleoli of the cells treated with TriplatinNC, in comparison to the surrounding nuclear regions, is demonstrated clearly by the plot in Fig. S7 (ESI†), and the localisation of 195Pt in the nucleolus suggests that a targeting mechanism is responsible. These results confirm what has been seen previously using a fluorophore-TriplatinNC conjugate inside HCT116 cells after 4 h, where the fluorescence originating from the conjugate was localised to the nucleolus and cytoplasm of the cells, with the exclusion of nuclear accumulation.11
The previous application of [1H, 15N] HSQC NMR methods has afforded significant insights into the hydrolysis and kinetics of DNA adduct formation of Pt drugs.12 In a novel extension of this isotopic labelling technique, we have shown that multi-element mapping of Pt-am(m)ine antitumour drugs by NanoSIMS provides a significant contribution to metal-based therapeutics imaging, given that the drugs can be studied in absence of fluorescent labels, which can potentially influence pharmacokinetics of the “parent” structure. Moreover, localisation to specific organelles can be studied without the need for colocalisation studies employing specific markers, such as Fibrillarin as a nucleolar marker. Clear differences in localisation and time-course between cisplatin and the polynuclear Pt drug TriplatinNC are readily apparent. It is notable that the cellular accumulation of polynuclear, but not mononuclear, Pt is mediated through Heparan Sulfate Proteo Glycan (HSPG) interactions,13 and the future use of NanoSIMS may contribute to understanding the internalisation of TriplatinNC upon HSPG binding. The identification of the nucleolus as a target for TriplatinNC confirms previous suggestions that this could be a novel target, with consequences for transcription inhibition.3 Metabolism of the drug is also evident, as presumably 15N in the absence of 195Pt and vice versa (e.g. Fig. 2) indicates dissociation of the drug. Given that TriplatinNC is a “non-covalent” analogue of the Phase II drug BBR3464, further comparisons (including use of fully and partially (end or central NH3 only) 15N-labelled BBR3464), may delineate “noncovalent” contributions to the metabolic fate of the promising anticancer drug in exquisite detail, as well as expanding the intrinsically interesting properties of TriplatinNC itself.1 Finally, simultaneous direct multi-element imaging by NanoSIMS is widely applicable to study of a wide range of other types of metal-based drugs, offering the exciting potential, demonstrated here, to probe ligand exchange reactions within intracellular compartments.
Supplementary Material
Acknowledgments
This work was supported by the Australian Research Council (Discovery grant to S.J.B-P and NF), and the National Institutes of Health (RO1-CA78754). We thank Erica Peterson and Luis Filgueira for helpful discussions and Ralph Kipping for synthesis and supply of 15N-TriplatinNC. The authors acknowledge the scientific and technical assistance of the Australian Microscopy & Microanalysis Research Facility at the Centre for Microscopy, Characterisation & Analysis, The University of Western Australia, a facility funded by the University, State and Commonwealth Governments.
Footnotes
Electronic Supplementary Information (ESI) available: Selected NanoSIMS data, cell culture and sample preparation methods, high mass resolution scans and image acquisition and processing details. See DOI: 10.1039/b000000x/
Contributor Information
Susan J. Berners-Price, Email: s.berners-price@griffith.edu.au.
Nicholas P. Farrell, Email: npfarrell@vcu.edu.
References
- 1.Farrell NP. Drugs of the Future. 2012;37:795–806. [Google Scholar]
- 2.Wilson JJ, Lippard SJ. Inorg Chim Acta. 2012;389:77–84. doi: 10.1016/j.ica.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]; Jansen BAJ, Wielaard P, Kalayda GV, Ferrari M, Molenaar C, Tanke HJ, Brouwer J, Reedijk J. J Biol Inorg Chem. 2004;9:403–413. doi: 10.1007/s00775-004-0539-y. [DOI] [PubMed] [Google Scholar]; Kalayda GV, Jansen BAJ, Molenaar C, Wielaard P, Tanke HJ, Reedijk J. J Biol Inorg Chem. 2004;9:414–422. doi: 10.1007/s00775-004-0540-5. [DOI] [PubMed] [Google Scholar]; Kalayda GV, Jansen BAJ, Wielaard P, Tanke HJ, Reedijk J. J Biol Inorg Chem. 2005;10:305–315. doi: 10.1007/s00775-005-0643-7. [DOI] [PubMed] [Google Scholar]; Kalayda GV, Zhang G, Abraham T, Tanke HJ, Reedijk J. J Med Chem. 2005;48:5191–5202. doi: 10.1021/jm050216h. [DOI] [PubMed] [Google Scholar]; Safaei R, Katano K, Larson BJ, Samimi G, Holzer AK, Naerdemann W, Tomioka M, Goodman M, Howell SB. Clin Cancer Res. 2005;11:756–767. [PubMed] [Google Scholar]; Liang XJ, Shen DW, Chen KG, Wincovitch SM, Garfield SH, Gottesman MM. J Cell Physiol. 2005;202:635–641. doi: 10.1002/jcp.20253. [DOI] [PubMed] [Google Scholar]; Gao J, Liu YG, Zingaro RA. Chem Res Toxicol. 2009;22:1705–1712. doi: 10.1021/tx900180v. [DOI] [PubMed] [Google Scholar]; New EJ, Duan R, Zhang JZ, Hambley TW. Dalton Trans. 2009:3092–3101. doi: 10.1039/b821603g. [DOI] [PubMed] [Google Scholar]; New EJ, Roche C, Madawala R, Zhang JZ, Hambley TW. J Inorg Biochem. 2009;103:1120–1125. doi: 10.1016/j.jinorgbio.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Bendetti BT, Peterson EJ, Kabolizadeh P, Martinez AR, Kipping R, Farrell NP. Mol Pharm. 2011;8:940–948. doi: 10.1021/mp2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mauthe RJ, Sideras-Haddad E, Turteltaub KW, Bench G. J Pharm Biomed Anal. 1998;17:651–663. doi: 10.1016/s0731-7085(97)00225-2. [DOI] [PubMed] [Google Scholar]; Hall MD, Alderden RA, Zhang M, Beale PJ, Cai Z, Lai B, Stampfl APJ, Hambley TW. J Struct Biol. 2006;155:38–44. doi: 10.1016/j.jsb.2006.01.011. [DOI] [PubMed] [Google Scholar]; Hall MD, Dillon CT, Zhang M, Beale P, Cai Z, Lai B, Stampfl APJ, Hambley TW. J Biol Inorg Chem. 2003;8:726–732. doi: 10.1007/s00775-003-0471-6. [DOI] [PubMed] [Google Scholar]; Harada S, Ehara S, Ishii K, Yamazaki H, Matsuyama S, Sato T, Oikawa S, Kamiya T, Arakawa K, Yokota W, Sera K, Ito J. Int J Radiat Oncol. 2009;75:455–462. doi: 10.1016/j.ijrobp.2009.02.082. [DOI] [PubMed] [Google Scholar]
- 5.Beretta GL, Righetti SC, Lombardi L, Zunino F, Perego P. Ultrastruct Pathol. 2002;26:331–334. doi: 10.1080/01913120290104610. [DOI] [PubMed] [Google Scholar]; Meijera C, van Luyn MJA, Nienhuis EF, Blom N, Mulder NH, de Vries EGE. Biochem Pharmacol. 2001;61:573–578. doi: 10.1016/s0006-2952(00)00584-0. [DOI] [PubMed] [Google Scholar]; Yang Z, Schumaker LM, Egorin MJ, Zuhowski EG, Guo Z, Cullen KJ. Clin Cancer Res. 2006;12:5817–5825. doi: 10.1158/1078-0432.CCR-06-1037. [DOI] [PubMed] [Google Scholar]
- 6.Hall MD, Okabe M, Shen DW, Liang XJ, Gottesman MM. Annu Rev Pharmacol. 2008;48:495–535. doi: 10.1146/annurev.pharmtox.48.080907.180426. [DOI] [PubMed] [Google Scholar]; Howell SB, Safaei R, Larson CA, Sailor MJ. Mol Pharmacol. 2010;77:887–894. doi: 10.1124/mol.109.063172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fletcher JS. Analyst. 2009;134:2204–2215. doi: 10.1039/b913575h. [DOI] [PubMed] [Google Scholar]
- 8.Chandra S. Methods Mol Biol. 2010;656:113–130. doi: 10.1007/978-1-60761-746-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lechene CP, Hillion F, McMahon G, Benson D, Kleinfeld AM, Kampf JP, Distel DL, Luyten Y, Bonventre J, Hentschel D, Park KM, Ito S, Schwartz M, Benichou G, Slodzian G. J Biol. 2006;5:20. doi: 10.1186/jbiol42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wedlock LE, Kilburn MR, Cliff JB, Filgueira L, Saunders M, Berners-Price SJ. Metallomics. 2011;3:917–925. doi: 10.1039/c1mt00053e. [DOI] [PubMed] [Google Scholar]
- 11.Peterson EJ, Menon VR, Kabolizadeh P, Benedetti BT, Kipping R, Ryan JJ, Povirk LF, Farrell NP. Unpublished work [Google Scholar]
- 12.Berners-Price SJ, Ronconi L, Sadler PJ. Prog Nucl Magn Reson Spectrosc. 2006;49:65–98. [Google Scholar]
- 13.Silva H, Frezard F, Peterson EJ, Kabolizadeh P, Ryan JJ, Farrell NP. Mol Pharm. 2012;9:1795–1802. doi: 10.1021/mp300098t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.