Abstract
Background
The red swamp crawfish, Procambarus clarkii, has become one of the most economically important cultured species in China. Currently, little is known about the gonadal development of this species. Isolation and characterization of genes are an initial step towards understanding gonadal development of P. clarkii.
Results
Using the 454 pyrosequencing technology, we obtained a total of 1,134,993 high quality sequence reads from the crawfish testis and ovary libraries. We aimed to identify different genes with a potential role in gonad development. The assembly formed into 22,652 isotigs, distributed by GO analysis across 55 categories in the three ontologies, ‘molecular function’, ‘cellular component’, and ‘biological processes’. Comparative transcript analysis showed that 1,720 isotigs in the ovary were up-regulated and 2138 isotigs were down-regulated. Several gonad development related genes, such as vitellogenin, cyclin B, cyclin-dependent kinases 2, Dmc1 and ubiquitin were identified. Quantitative real-time PCR verified the expression profiles of 14 differentially expressed genes, and confirmed the reliability of the 454 pyrosequencing.
Conclusions
Our findings provide an archive for future research on gonadal development at a molecular level in P. clarkii and other crustacean. This data will be helpful to develop new ideas for artificial regulation of the reproductive process in crawfish aquaculture.
Introduction
The red swamp crawfish, Procambarus clarkii, is a species of freshwater crawfish native to the Southern central United States and Northeastern Mexico [1], and has become one of the most widely introduced crawfish species in the world. This species was introduced from Japan to Nanjing, China, in 1929 [2]. Since the 1990s, P. clarkii have been farmed extensively as food source and have become one of the most economically important farmed species in China [3]. Due to its high market demand and high economic value, crawfish aquaculture has developed rapidly in the past decades, and its annual output has reached approximately 479,374 tons, accounting for 91.12% of the global production according to the 2009 United Nations Food and Agriculture Organization report [4]. Owing to the growing culture industry, a shortage of juvenile crawfish has become prevalent [5], leading to severe restriction in large-scale crawfish aquaculture.
With the aim of developing a method for controlling the quality and quantity of crawfish and their eggs in aquaculture, many studies have been investigated on crawfish gonadal development [6]–[8]. Knowledge of mechanisms governing gonadal development processes at the molecular level is crucial and could be directly applied to the crawfish industry. Recently, some genes have been shown as critical factors for gonadal development in crawfish and other crustaceans, including vasa [9], cyclin B [10], cell division cycle 2 [11], elongation factor 2 [12], heat shock protein 90 [13], cathepsin C [14], and Dmrt [15]. Generating gonad specific libraries facilitates an understanding of the molecular mechanisms of gonadal development.
DNA sequencing provides important data to study the regulation of gene expression. Although a large number of whole-genome sequencing studies have been performed for microbial [16] and vertebrate [17] in the past decades, crustacean genomes have only been sequenced in Daphnia pulex [18]. Transcriptome sequencing provides general representation of almost all transcripts expressed in specific cells or organs at particular conditions and stages. Because of its advantages of high throughput rates and low costs, the 454 pyrosequencing technology is regarded as the first choice for the identification of novel genes in organisms lacking a reference genome [19]. It has thus been employed for other crustacean species, including Litopenaeus vannamei [20], Macrobrachium rosenbergii [21], M. nipponense [22] and Euphausia superba [23].
Screening and identifying gonadal differentially expressed genes are an initial step towards understanding gonadal development in P. clarkii. In the present study, we performed a 454 pyrosequencing of P. clarkii using prepared cDNA from mRNA isolated from testis and ovary tissues. This was used to generate expression profiles and to discover differentially expressed genes in these two tissue types. Based on previous research, and employing bioinformatics tools, we aimed to generate a list of candidate genes that may be involved in the gonadal development of P. clarkii. This could provide a major resource for future studies of crawfish gonadal development, and in particular should help to establish new ideas for the artificial regulation of reproductive processes in crawfish aquaculture.
Materials and Methods
Ethics statement
The handling of crawfish was conducted in accordance with the guidelines on the care and use of animals for scientific purposes set by the Institutional Animal Care and Use Committee (IACUC) of Shanghai Ocean University, Shanghai, China.
Crawfish tissue samples collection
The crawfish used in this project were obtained from Jiangsu Xuyi Riverred Crawfish Eco-Park, Jiangsu Province, China. We collected healthy, sexually matured male and female crawfish in September, October and November 2012, respectively. Before tissue collection, the male and female crawfish were cultured in a tank, and maintained in continuously aerated freshwater at ambient temperature (28±1°C) for 72 h. For the transcriptome experiments, the crawfish were placed in an ice bath for 1–2 min until anesthetized. Then the gonad tissues were removed through surgery, immediately frozen in liquid nitrogen, and separately stored at −80°C until further use. Tissues from three different individuals were taken every month, and the nine tissue samples were equally pooled as a single sample for RNA extraction. In total, testis and ovary samples were separately taken from nine male and nine female individuals. All gonad samples were taken from fully mature individuals and were expected to cover all stages of germ cell development.
RNA extraction, cDNA synthesis, and sequencing
The total RNA of each testis or ovary sample were isolated using TRIzol Reagent (Invitrogen, USA) following the manufacturer’s instructions. The total RNA quality was determined using a NanoDrop (Thermo Scientific, USA), and the RNA integrity value (RIN) was checked using the RNA 6000 Pico LabChip of an Agilent 2100 Bioanalyzer (Agilent, USA). Total RNA of all samples was incubated with 10 U DNase I (Ambion, USA) at 37°C for 1 h, and purified with a MicroPloy (A) Purist Kit (Ambion, USA) following the manufacturer’s instructions. The purified mRNA was dissolved in RNA storage solution, and a NanoDrop (Thermo Scientific, USA) was used to determine the RNA final concentration.
Library construction and pyrosequencing was conducted by Beijing Genomics Institute (BGI; Shenzhen, China) on a 454 GS FLX system (Roche). The cDNA template for sequencing was generated using a cDNA library construction kit (Clontech, USA) according to the manufacturer’s protocol. The cDNA was fractionated into a range of 300–800 bp fragments. Specific adaptors were bound to the fragmented templates, and then used for purification, amplification, and sequencing steps. A half plate sequencing run was performed for each library on the 454 Genome Sequencer FLX instrument.
Data assembly, clustering, and bioinformatics analysis
Through basic requirement, the original image data generated from the sequencing were transformed into sequence data, called raw data or raw reads. Subsequent analyses were based on clean reads obtained after a series of data processing steps. Assembly analysis of the transcriptome was carried out using Newbler v2.5 (Roche). Overlapping reads were first assembled into contigs. Contigs with connecting reads were compiled into isotigs. For further analysis, we first used BLASTx (E-value<10−5) to search the isotigs against various protein databases, including the National Center for Biotechnology Information (NCBI) non-redundant (nr) database, Swiss-Prot, Kyoto Encyclopedia of Genes and Genomes (KEGG), and Clusters of Orthologous Groups (COG). If the results of different databases were contradictory, a priority order of alignments from the nr, Swiss-Prot, KEGG, and COG databases was followed to decide the sequence direction.
Sequence alignment and Phylogenetic analysis
Amino acid sequences related to vasa were downloaded from NCBI, sequence alignments were conducted with the BioEdit software [24] before construction of phylogenetic trees, and the result was imported to software MEGA5.0 [25]. Phylogenetic trees were generated using the neighbour-joining method and bootstrapped with 1000 iterations to evaluate the branch strength of the tree.
In situ hybridazation
In situ hybridization studies were conducted to determine the exact location of cyclin B and titin mRNA expression in the gonad of P. clarkii. DIG-labeled RNA probes were synthesized using a DIG RNA labeling kit (Roche, Germany). Fragments of cyclin B and titin were amplified by PCR and cloned into PGEM-T easy vector, and the plasmid clones were used as templates to synthesize both sense and antisense RNA probes by in vitro transcription. The transcriptions were performed from 1 µg linearized plasmid using either T7 or SP6 RNA polymerases.
Testis and ovary tissue sections were dewaxed with xylene, hydrated in ethanol gradient. The technique used RNase-free reagents as described by Braissant [26]. Rehydrated tissues underwent the following procedures: wash in PBS with 0.1% active DEPC (2×15 min); equilibration in 5×SSC (15 min); prehybridization, 50% formamide, 5×SSC (30 min at 50°C); hybridization with 400 ng/mL of DIG-labeled probe, in 50% deionized formamide, 5×SSC, 1×Denhardt's solution, 10 mg/mL tRNA, 10% dextran sulfate (12–18 h at 55°C); washed in 2×SSC (30 min at room temperature); incubation in 2×SSC (1 h at 55°C); incubation in 0.1×SSC (1 h at 55°C); equilibration in buffer 1 (Tris 100 mM/NaCl 150 mM, pH 7.5) (5 min); incubation with anti-DIG antibody, AP-coupled, diluted 1∶1000 in buffer 2 [buffer 1 with 0.5% of Blocking Solution (Roche)] (2 h); washed in buffer 1 (2×15 min); equilibration in buffer 3 (Tris 100 mM/NaCl 100 mM, pH 9.5) (5 min); stained in buffer 3 containing 20 µl NBT/BCIP Stock Solution (Roche) (overnight); washed in running tap water (15 min); dehydrated in alcohol series; after which slides were mounted with cover slips.
Identification of differentially expressed genes and quantitative real-time PCR analysis
The amount of isotig expression was calculated using the reads per kb per million reads method (RPKM), using the following formula:
According to the method proposed by Audic [27], we developed a rigorous algorithm to screen genes that were differentially expressed between the two libraries. To judge the significance of differences in expressed genes, we used the FDR (false discovery rate) method [28] to determine the threshold of the P-value in multiple tests. In this study, we used three criteria: FDR≤0.001, the absolute value of the log2Ratio≥1, and a P value<0.01, as threshold to judge the significance of gene expression.
Within each of the categories for up-regulated and down-regulated genes in the ovary, we selected 14 genes at random for quantitative real-time PCR (RT-PCR) validation, and the specific primers (Table S1) were designed using the Primer Premier 5.0 program [29]. The total RNA of testis from 4 individuals and the total RNA of ovary from 4 individuals were reverse transcribed using 1 µg RNA of each tissue and the PrimeScript RT-PCR Kit (TaKaRa, Dalian, China) according to the manual. RT-PCR were performed using the SYBR premix Ex Taq Kit (TaKaRa, Dalian, China). The thermal profile was 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 58°C for 30 s, and 72°C for 30 s. The mRNA expression of each gene was quantified relative to β-actin (Forward, 5′-AAATCACGGCTCTGGCTCCCT-3′; Reverse, 5′-GAAGCATTTGCGGTGGACGAT-3′). The average cycle thresholds (CT) were used to determine fold-change. The relative quantification of gene expression was reported as a relative quantity (RQ) to the control value, and all experiments were performed with three technological replicates. The relative expression levels were calculated using the equation 2−ΔΔCT (ΔCT = CT of target gene minus CT of β-actin, ΔΔCT = ΔCT of one gender sample minus ΔCT of opposite gender sample) [30].
Data deposition
The sequences reported in this paper were deposited into the National Center for Biotechnology Information (NCBI) Sequence Read Archive with accession numbers SRP035515.
Results and Discussion
RNA sequencing and reads assembly
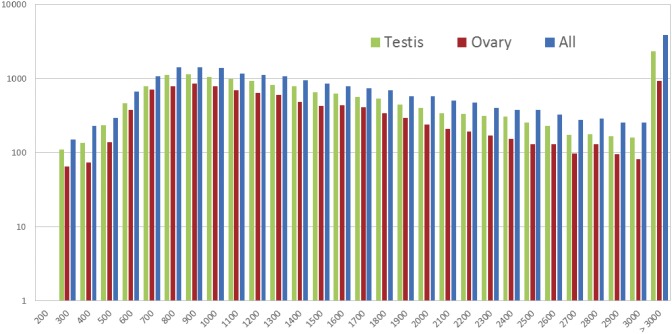
We conducted a 454 pyrosequencing for the testis and ovary cDNA libraries, resulting in 626,829 sequences with an average length of 775 bp and 508,144 sequences with an average length of 901 bp (Table 1), respectively. This produced a total of 943,840,902 nucleotides. Additionally, read lengths ranged from 55 to 1,780 bp in the testis library, and 53 to 1,780 bp in the ovary library. The assembly formed into 16,727 isotigs with an average length of 1827 bp in the testis library and 10,748 isotigs with an average length of 1544 bp in the ovary library. The length distribution of assembled isotigs is presented in Figure 1. The N50 isotig size of 2,255 bp in testis library was larger than that of 1,794 bp in ovary library. These assembly sequences represent the transcription and could be used for the further analysis.
Table 1. P. clarkii transcriptome assembly statistics.
| Category | Testis | Ovary | All |
| Number of reads | 626,829 | 508,144 | 1,134,993 |
| Total base pairs (bp) | 485,892,824 | 457,948,078 | 943,840,902 |
| Average read length (bp) | 775 | 901 | 832 |
| Maximum read length (bp) | 1,780 | 1,780 | 1,780 |
| Minimum read length (bp) | 55 | 53 | 53 |
| Number of assembled isotigs | 16,727 | 10,748 | 22,652 |
| Average isotig length (bp) | 1,827 | 1,544 | 1,921 |
| Maximum isotig length (bp) | 18,422 | 10,978 | 18,422 |
| Isotig N50 | 2,255 | 1,794 | 2,474 |
Figure 1. Assembly quality statistics of testis, ovary, and all isotigs from 454 sequencing.
Length distribution of de novo assemblies of isotigs (x-axis indicates the sequence size (nt), y-axis indicates the number of assembled isotigs).
Functional annotation
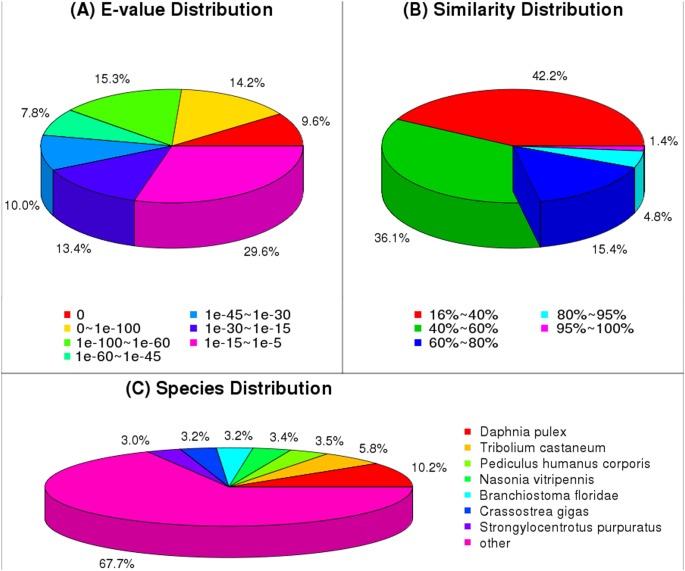
To understand the isotig functions, 22,652 isotigs were annotated using BLASTx alignment with an E-value smaller than 10E-5. A total of 15,667 (69.16%), 13,818 (61.00%), 12,419 (54.83%), 7,243 (31.98%), and 6,902 (30.47%) isotigs had significant matches with sequences in the NR, Swiss-Prot, KEGG, COG, and GO databases, respectively. Annotation results showed that many of the sequences have no homologous sequences in public databases due to the scarcity of species similar to P. clarkii in public databases. The E-value distribution of the top hits in the nr database showed that 39.1% of the mapped sequences had strong homology (less than 1.0E-60), whereas 60.9% of the homolog sequences ranged between 1.0E-5 and 1.0E-60 (Figure 2A). The similarity distribution showed a comparable pattern with 6.2% of the sequences having a similarity greater than 80%, while 93.8% of the sequences had a similarity ranging from 18% to 80% (Figure 2B). In terms of species distribution, similar isotigs were retrieved from Daphnia pulex (1,596, 10.19%), Tribolium castaneum (907, 5.79%), Pediculus humanus corporis (553, 3.53%), Nasonia vitripennis (530, 3.38%), Branchiostoma floridae (503, 3.21%), Crassostrea gigas (496, 3.17%), Strongylocentrotus purpuratus (473, 3.02%), and other species (10,608, 67.71%) (Figure 2C).
Figure 2. Characteristics of homology search of 454 sequences against nr database.
(A) E-value distribution of BLAST hits for each unique sequence with a cut-off E-value of 1.0E-5. (B) Similarity distribution of the top BLAST hits for each sequence. (C) Species distribution shown as a percentage of total homologous sequences with E-value of ≥1.0E-5.
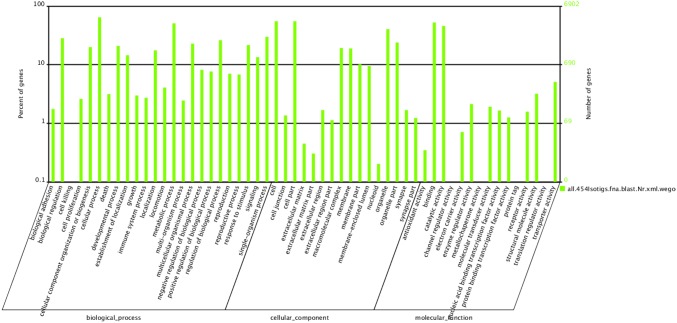
Gene ontology (GO) terms were used to classify the function of the predicted P. clarkii transcriptome. In total, 22,652 isotigs were analyzed with Blast2GO for GO classification [31] and distributed across 55 categories by WEGO [32] within the three ontologies ‘molecular function’, ‘cellular component’, and ‘biological processes’ (Figure 3). For the ontology ‘biological processes’, the most frequent categories were ‘cellular process’ (16.95%), ‘metabolic process’ (13.37%), ‘single-organism process’ (7.87%), and ‘biological regulation’ (7.46%), followed by ‘regulation of biological process’ (6.89%), ‘multicellular organismal process’ (6.02%), ‘response to stimulus’ (5.69%), ‘developmental process’ (5.52%), and ‘cellular component organization or biogenesis’ (5.25%). Other ‘biological process’ categories, such as ‘biological adhesion’ or ‘cell killing’ were present, but at lower percentages. In the ‘cellular component’ ontology, most of the terms were grouped into the ‘cell’ (22.94%) and ‘cell part’ (22.93%) categories, followed by ‘organelle’ (16.78%), ‘organelle part’ (9.94%), ‘macromolecular complex’ (8.01%), and ‘membrane’ (7.89%). Terms such as ‘cell junction’, ‘extracellular matrix’, ‘extracellular matrix part’, and ‘nucleoid’ were also present, but constituted a smaller proportion. The ‘molecular function’ ontology indicated that ‘binding’ and ‘catalytic activity’ contained over 84.52% of annotated sequences. Other categories, such as ‘translation regulator activity’, ‘channel regulator activity’, ‘metallochaperone activity’, and ‘protein tag’ were only present in small numbers.
Figure 3. Histogram of GO classifications.
Results summarized in three main categories: biological process, cellular component, and molecular function. Left y-axis indicates percentage of a specific category of genes in each main category. Right y-axis indicates number of genes in a category.
The Kyoto encyclopedia of genes and genomes (KEGG) is a knowledge base for systematic analysis of gene functions in terms of the networks and molecules [33]. According to KEGG annotation information, a total of 12,419 isotigs were assigned to 256 basic metabolic pathways. The number of isotigs in different pathways ranged from 1 to 1,501 (Table S2), and the major pathway of ‘metabolic pathways’ included 12.9% of the isotigs. All pathway predictions are likely to be useful for future research focusing on their specific processes during gonadal development in P. clarkii.
Sequence alignment and Phylogenetic analysis
In order to verify the accuracy of assembly, we observed sequences with a single complete open reading frame (ORF) as representative examples. Here we isolated the VASA sequences, for its critical functions in gonadal development. The sequence alignments and phylogenetic analysis results showed that P. clarkii vasa genes showed high identity with the homologues from other species, especially with that of crustaceans (Figure S1). It contained all nine conserved characteristic motifs, including AQTGSGKTAA, PTRELAVQ, GG, TPGR, DEAD, SAT, HGD, RGLD, and HRIGRTGR, similar to the other DEAD-box proteins. As shown in Figure S2, vasa of P. clarkii was remote from the vertebrate and mollusk vasa clade and clustered near the crustacean clade.
Localization of cyclin B and titin transcripts in testes and ovaries
In different developing stages of testis and ovary, the expression of cyclin B and titin transcripts are presented in different distribution patterns. In ovary, the positive signals of these two genes were detected in some follicle cells surrounding nearly mature oocytes and nucleus (Fig. S3, B and G). In testis, cyclin and titin mRNA are present in spermatocytes and spermatogonia (Fig. S3 D and I). No hybridization signal exists in the negative control group (Fig. S3 C, E, H and J).
Analysis of differentially expressed genes and verification using real-time PCR
In this study, statistical analysis of the differentially expressed genes between the testis and ovary libraries detected a list of 3,858 genes, of which 1,720 were up-regulated in the ovaries and 2,138 were down-regulated (Figure 4). These gonad specific up-regulated genes suggest their potential roles in gonadal development processes in P. clarkii. The most common GO terms associated with these features were ‘cell cycle regulation’ (36.8%), ‘muscle contraction and development’ (13.2%), and ‘transport’ (9.2%).
Figure 4. Scatter plots showing gene expression profiles in testis and ovary libraries of P. clarkii.
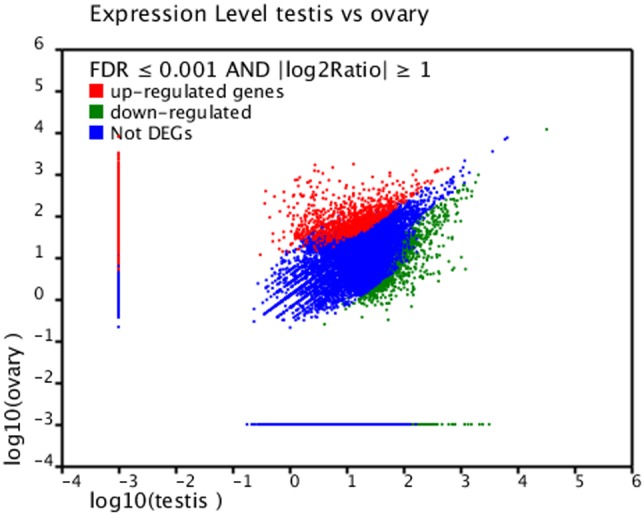
Limits defined by FDR ≤0.001 and |log2 ratio| ≥1. Blue dots represent genes with similar expression in testis and ovary; red and green dots indicate up-regulated and down-regulated genes in ovary, respectively (>1.5-fold change, black diagonals).
In order to ensure a reliable comparison between 454 sequencing and RT-PCR, we compared the mRNA levels of 14 candidate genes by RT-PCR. Of these 14 genes, eight were up-regulated and six were down-regulated in the ovary libraries. RT-PCR results verifying these differential expression profiles between testis and ovary libraries of P. clarkii are shown in Table S1 and Figure 5. Although the results of gene expression did not perfectly match to results detected by 454 sequencing, the up- and down-regulated trends were closely similar. Furthermore, it can be speculated that pooling of testis or ovary mRNAs from multiple individuals helped to level out potential variations. More genes will be validated in the future study.
Figure 5. Histogram, comparison of gene expression between RT-PCR and 454 sequencing analysis.
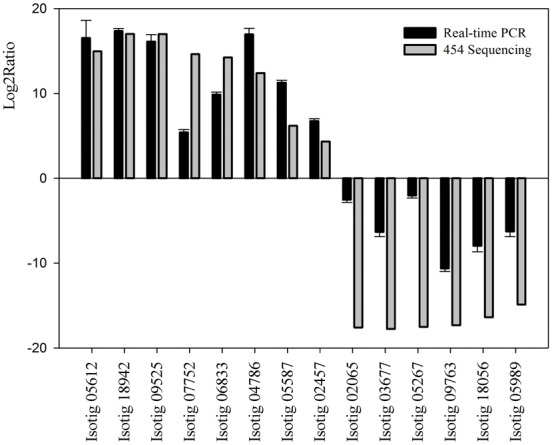
The RT-PCR are presented as the mean values of three repeats.
Putative regulatory genes involved in gonad development
It seems likely that the gonads contain more abundant transcription signals of gonadal development than its somatic tissue. We therefore constructed this database from testis and ovary libraries of P. clarkii. Based on the published literature and sequence annotation, several genes involved in gonad development were identified. These included ‘ovary development’, ‘ubiquitin proteasome pathway’, ‘cell cycle regulatory protein’ and ‘testis development’-related genes, such as vitellogenin(Vg), cyclin B, cyclin-dependent kinases 2, Dmc1 and ubiquitin. The genes in this list (Table 2) had been identified in previous studies to be essential for gonadal development.
Table 2. List of genes known to be involved in gonad development of P. clarkii transcriptome.
| Description | Matched species | Size | E value |
| Ovary development | |||
| Vg | Homarus americanus | 9,510 | 0 |
| VgR | Macrobrachium rosenbergii | 7,460 | 2.00E-07 |
| cathepsin C | Marsupenaeus japonicas | 2,614 | 0 |
| cathepsin L | Penaeus monodon | 1,195 | 6.00E-104 |
| PL10-like protein | Macrobrachium nipponense | 5,465 | 0 |
| vasa-like protein | Marsupenaeus japonicas | 3,099 | 1.00E-172 |
| EF2 | Eriocheir sinensis | 2,694 | 0 |
| EIF3 | Pediculus humanus corporis | 5,538 | 0 |
| PTGR1 | Penaeus monodon | 2,051 | 7.00E-107 |
| Cell cycle regulatory protein | |||
| cvclin A | Penaeus monodon | 2,173 | 0 |
| cyclin B | Marsupenaeus japonicas | 2,595 | 1.00E-152 |
| CDK2 | Macrobrachium rosenbergii | 1,461 | 3.00E-118 |
| CKS1 | Acromyrmex echinatior | 2,456 | 2.00E-32 |
| CDC2 | Penaeus monodon | 1,608 | 7.00E-168 |
| CKS1B | Scylla paramamosain | 2,307 | 1.00E-41 |
| Ubiquitin proteasome pathway | |||
| ubiquitin | Papilio xuthus | 807 | 3.00E-104 |
| ubiquitin-conjugating enzyme E2 G1 | Pediculus humanus corporis | 680 | 3.00E-88 |
| ubiquitin-conjugating enzyme E2 O | Camponotus floridanus | 1,872 | 4.00E-130 |
| UBR2 | Camponotus floridanus | 7,476 | 2.00E-83 |
| hect E3 ubiquitin ligase | Pediculus humanus corporis | 10,308 | 0 |
| UBE4B | Clonorchis sinensis | 1,526 | 1.00E-08 |
| SUMO-1 | Litopenaeus vannamei | 1,423 | 3.00E-46 |
| Testis development | |||
| dynactin subunit 5 | Penaeus monodon | 1,963 | 1.00E-99 |
| cyclophilin A | Eriocheir sinensis | 882 | 2.00E-83 |
| PCNA | Fenneropenaeus chinensis | 1,280 | 4.00E-135 |
| Nanos 2 | Podocoryna carnea | 3,107 | 1.00E-14 |
| Nuclear autoantigenic sperm protein | Penaeus monodon | 2,188 | 2.00E-147 |
| Dmc1 | Litopenaeus vannamei | 1,324 | 3.00E-175 |
| TEX14 | Taeniopygia guttata | 4,874 | 1.00E-16 |
| Sperm-associated antigen | Pediculus humanus corporis | 2,015 | 0 |
| PRDM9 | Bos taurus | 2,783 | 1.00E-26 |
Among these gonad-related genes, several ‘ovary development’ related genes were highly expressed in ovary tissues. As observed in many oviparous organisms, oocyte maturation depends on massive production of the egg yolk-precursor protein, Vg. Vitellogenin receptor (VgR) is involved in vg uptake by oocytes and plays a critical role in egg development [34]. The molecular characteristics of Vg and VgR have been described for many crustaceans [35]–[40]. In the current study, we found high occurrence of Vg and VgR in ovary of P. clarkii, and RT-PCR demonstrated that the expression level in ovary is significantly higher than in testis which provided some clues to further elucidate the function to ovary development of P. clarkii (Table S1). Cathepsins belong to a family of proteases that cleave other proteins and are ubiquitously present in almost all organisms [41], and involved in ovary maturation and embryo development [14], [42]. There are two kind of cathepsins: cathepsin C and L in our study, and were largely expressed in ovary libraries. Other ‘ovary development’ related genes including elongation factor 2 (EF2), prostaglandin reductase 1 (PTGR1), eukaryotic translation initiation factor 3 (EIF3) have been found in ovary library, suggested the possible contribution of ovarian maturation [12], [43]–[44]. In this study, we found partial cDNAs from two members of the DEAD-box family, one belonging to the vasa subfamily and the other to the PL10 subfamily. The vasa (and vasa-like) gene has been reported to play an essential role in germ cell development in higher metazoans, and localized in the oocyte cytoplasm throughout oogenesis [45]–[49]. Sequence and phylogenetic analysis results showed that P. clarkii vasa sequences present high identity with the homologues from other species, especially with that of crustaceans.
The cell cycle regulatory protein plays a key role in the oogenesis and oocyte development [10], [50]–[51]. The meiotic maturation of oocytes is regulated by the maturation promoting factor (MPF), a complex of cyclin dependent kinases (CDK), and cyclin B. In this project, a putative transcript of cyclin B and cyclin dependent kinases regulatory subunit 1 (CKS1) were characterized with a higher expression levels in the ovary than testis of P. clarkii, and RT-PCR results are consistent with it. Further demonstrated the important role in ovarian development of P. clarkii. In situ hybridization revealed that cyclin B mRNA were clearly localized in the testis and ovary, these results suggested that cyclin B may play essential roles in the oogenesis and spermatogenesis of the crawfish (Table S3). The high expression of the cyclin dependent kinases 2 (CDK2), cell division cycle 2 (CDC2) and the CDC28 protein kinase regulatory subunit 1B (CKS1B) plays an important role in the ovary development and gametogenesis in crustaceans [11], [52]–[53]. In our study, similar to other reported animals, we found high expression of CDK2, CDC2 and CKS1B in the P. clarkii testis and ovary libraries, show that it may be involved in the gametogenesis and gonad development of P. clarkii.
The ubiquitin proteasome pathway (UPP) is important for nearly every aspect of cellular life. It provides the cell with the ability to degrade proteins both specifically and temporally. The system generally includes three types of ubiquitin enzymes: ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin protein ligases (E3s). We found six kinds of ubiquitin enzymes in our current project, these ubiquitin-related homologous genes with different expression levels were found in the P. clarkii testis and ovary cDNA libraries, which suggests that ubiquitin proteins may play various roles in the reproductive process of P. clarkii. Recent studies have shown that UPP is involved in controlling the processes of gametogenesis, including meiosis control and reorganization of chromatin structure [54]. Ubiquitin-conjugating enzyme E2 G1 and ubiquitin-conjugating enzyme E2 O have a highly conserved catalytic domain that is common to all E2s with a special carboxy-terminal extension different from other E2s. E3 ubiquitin-protein ligase (UBR2) localizes to meiotic chromatin regions and functions together with the ubiquitin conjugating enzyme HR6B in histone H2A ubiquitination during male meiosis [55]. Hect E3 ubiquitin ligases is involved in the ubiquitin-mediated degradation pathway. Ubiquitin conjugation factor E4 B (UBR4B) play an important role in cellular regulation [56]. Small ubiquitin-related modifier1 (SUMO1) supports multiple roles in spermatogenesis [57]–[58].
Another interesting finding in the current study was the higher expression of genes in testis than ovary of P. clarkii, suggesting that involved in testicular development and spermatogenesis. Cyclophilin A and dynactin subunit 5 were discovered functionally involved in testicular development of P. monodon [59]. The proliferating cell nuclear antigen (PCNA) plays an important role in testis development, especially in the processes of mitosis and meiosis [55], and is involved in regulating spermatogenesis in the Japanese eel, Anguilla japonica [60]. Nanos2 is expressed in self-renewing spermatogonial stem cells and maintains the stem cell state during murine spermatogenesis [61]. Deleting Nanos2 may result in male sterility, owing to a loss of germ cells during fetal development [62]. The nuclear autoantigenic sperm protein provides the functional link between histones and cell cycle progression during meiosis [63]. The testis expressed 14 (TEX14) may play a key role in regulating gene expression or modulating nuclear events during mammalian spermatogenesis [64]. Dmc1 is a potentially useful indicator of the early stages of germ cell development [65], and plays a very important role during spermatogenesis. Research shows a possible association between PR domain 9 (PRDM9) and azoospermia by meiotic arrest [66]. Sperm associated antigen is commonly expressed in male germ cells and plays an important role during spermatogenesis [67].
Conclusions
The sequence data obtained in this study provide an archive for future research on gonadal development at a molecular level in P. clarkii. They could also serve as a theoretical foundation to solve problems in the breeding process. Here, we tried to elucidate their detailed role in the reproductive processes of P. clarkii. Future work will involve full-length amplification of candidate genes and validation of their functions.
Supporting Information
Alignment of the deduced amino acid sequences of vasa . The amino acids conserved across all the eighteens species are shown in asterisks at the bottom. The GenBank accession numbers of the sequences are as follows: M. japonicus AEB00819; L. vannamei AAY89069; P. monodon AEB00820; F. chinensis ABQ00071; C. hawaiensis ACH92926; S. paramamosain ADR51551; M. nipponense ADB28894; P. carinicauda AGF90963.
(TIF)
Neighbor-joining phylogenetic analysis of the vasa from P. clarkii .
(TIF)
Localization of cyclin B and titin transcripts in crawfish gonads. The results of In situ hybridization with DIG-labeled antisense RNA probe (B and D for cyclin B; G and I for titin) and sense probe as negative control (C and E for cyclin B; H and J for titin) were shown. Regular histological section was stained with hematoxylin and eosin (A and F). O: oogonium; Cy: cytoplasm; N: nucleus; Nu, nucleolus; Fc: follicle cells; Sg: spermatogonium; Sc: Spermatocyte; St: Spermatid; The scale bar indicates 100 µm.
(TIF)
Real-time PCR confirmation of differential expressed genes.
(DOCX)
KEGG biochemical pathways of P. clarkii isotig sequences.
(XLSX)
List of the up-regulated and down-regulated genes between male and female libraries.
(XLSX)
Acknowledgments
We thank the staff of Jiangsu Xuyi Riverred Crawfish Eco-Park Co., Ltd, Jiangsu Province, China for their help in the culturing and collecting of red swamp crawfish. We also thank lab members, Xilei Li and Hongyan Yu (Key Laboratory of Freshwater Aquatic Genetic Resources Certificated by Ministry of Agriculture, College of Fisheries and Life Science, Shanghai Ocean University) for their help in tissues collection and RNA extraction.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. The sequences reported in this paper were deposited into the National Center for Biotechnology Information (NCBI) Sequence Read Archive with accession numbers SRP035515.
Funding Statement
This study was supported by the Project of Shanghai Engineering and Technology Center for Promoting Ability (13DZ2280500) and Shanghai Universities Knowledge Service Platform (ZF1206). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Huner JV, Holdich DM, Lowery RS (1988) Procambarus in North America and elsewhere. Freshwater crayfish: biology, management and exploitation: 239–261p.
- 2.Li JL, Dong ZG, Li YS, Wang CH (2007) Invasive aquatic species in China. Shanghai: Shanghai Science and Technology Publisher. 104 p. [Google Scholar]
- 3. Wang W, Gu W, Ding ZF, Ren Y, Chen JX, et al. (2005) A novel Spiroplasma pathogen causing systemic infection in the crayfish Procambarus clarkii (Crustacea: Decapod), in China. FEMS microbiology letters 249: 131–137. [DOI] [PubMed] [Google Scholar]
- 4.FAO (2009) Food and Agriculture Organization of the United Nations. FAO Statistical Databases.
- 5. Song L, Zhang JP, Han XL, Xu JR (2011) Analysis of Breeding Situation and Countermeasures of Procambarus clarkia . Journal of Changshu Institute Technology (Natural Sciences) 25: 85–87. [Google Scholar]
- 6. Xia AJ, Pang L, Yan WH, Tang JQ, Huang C (2008) Study on the Mathematic Model of Gonad Development and Annual Changes of Relative Coefficients of Female Procambarus clarkii . Freshwater Fisheries 38: 12–15. [Google Scholar]
- 7. Yue GH, Li JL, Wang CM, Xia JH, Wang GL, et al. (2010) High prevalence of multiple paternity in the invasive crayfish species, Procambarus clarkii . International journal of biological sciences 6: 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shui Y, Guan ZB, Xu ZH, Zhao CY, Liu DX, et al. (2012) Proteomic identification of proteins relevant to ovarian development in the red swamp crayfish Procambarus clarkii . Aquaculture (370) 14–18. [Google Scholar]
- 9. Wang YL, Chen YD, Han KH, Zou ZH, Zhang ZP (2012) A vasa gene from green mud crab Scylla paramamosain and its expression during gonadal development and gametogenesis. Molecular biology reports 39: 4327–4335. [DOI] [PubMed] [Google Scholar]
- 10. Visudtiphole V, Klinbunga S, Kirtikara K (2009) Molecular characterization and expression profiles of cyclin A and cyclin B during ovarian development of the giant tiger shrimp Penaeus monodon . Comparative Biochemistry and Physiology-Part A: Molecular & Integrative Physiology 152: 535–543. [DOI] [PubMed] [Google Scholar]
- 11. Phinyo M, Visudtiphole V, Roytrakul S, Phaonakrop N, Jarayabhand P, et al. (2013) Characterization and expression of cell division cycle 2 (Cdc2) mRNA and protein during ovarian development of the giant tiger shrimp Penaeus monodon . General and comparative endocrinology 193: 103–111. [DOI] [PubMed] [Google Scholar]
- 12. Qiu LH, Jiang SG, Zhou FL, Zhang DC, Huang JH, et al. (2008) Molecular cloning of the black tiger shrimp (Penaeus monodon) elongation factor 2 (EF-2): sequence analysis and its expression on the ovarian maturation stage. Molecular biology reports 35: 431–438. [DOI] [PubMed] [Google Scholar]
- 13. Zhao WH, Chen LQ, Qin JG, Wu P, Zhang FY, et al. (2011) MnHSP90 cDNA characterization and its expression during the ovary development in oriental river prawn, Macrobrachium nipponense . Molecular biology reports 38: 1399–1406. [DOI] [PubMed] [Google Scholar]
- 14. Qiu GF, Yamano K, Unuma T (2005) Cathepsin C transcripts are differentially expressed in the final stages of oocyte maturation in kuruma prawn Marsupenaeus japonicus . Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology 140: 171–181. [DOI] [PubMed] [Google Scholar]
- 15. Zhang EF, Qiu GF (2010) A novel Dmrt gene is specifically expressed in the testis of Chinese mitten crab, Eriocheir sinensis . Development genes and evolution 220: 151–159. [DOI] [PubMed] [Google Scholar]
- 16. Cole S, Brosch R, Parkhill J, Garnier T, Churcher C, et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393: 537–544. [DOI] [PubMed] [Google Scholar]
- 17. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- 18. Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, et al. (2011) The ecoresponsive genome of Daphnia pulex . Science 331: 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li CZ, Weng SP, Chen YG, Yu XQ, Lü L, et al. (2012) Analysis of Litopenaeus vannamei Transcriptome Using the Next-Generation DNA Sequencing Technique. PloS one 7: e47442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jung H, Lyons RE, Dinh H, Hurwood DA, McWilliam S, et al. (2011) Transcriptomics of a giant freshwater prawn (Macrobrachium rosenbergii): de novo assembly, annotation and marker discovery. PloS one 6: e27938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma KY, Qiu GF, Feng J, Li JL (2012) Transcriptome analysis of the oriental river prawn, Macrobrachium nipponense using 454 pyrosequencing for discovery of genes and markers. PloS one 7: e39727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Clark MS, Thorne MA, Toullec JY, Meng Y, Peck LS, et al. (2011) Antarctic krill 454 pyrosequencing reveals chaperone and stress transcriptome. PLoS One 6: e15919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall T (1999) BioEdit software, version 5.0. 9. North Carolina State University, Raleigh, NC.
- 25. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Braissant O, Wahli W (1998) A simplified in situ hybridization protocol using non-radioactively labeled probes to detect abundant and rare mRNAs on tissue sections. 1: 10–16. [Google Scholar]
- 27. Audic S, Claverie JM (1997) The significance of digital gene expression profiles. Genome research 7: 986–995. [DOI] [PubMed] [Google Scholar]
- 28.Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Annals of statistics: 1165–1188.
- 29. Lalitha S (2000) Primer premier 5. Biotech Software & Internet Report: The Computer Software Journal for Scient 1: 270–272. [Google Scholar]
- 30. Livak KJ, Schmittgen TD (2001) Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 31. Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 32. Ye J, Fang L, Zheng HK, Zhang Y, Chen J, et al. (2006) WEGO: a web tool for plotting GO annotations. Nucleic acids research 34: W293–W297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, et al. (1999) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic acids research 27: 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mizuta H, Luo W, Ito Y, Mushirobira Y, Todo T, et al. (2013) Ovarian expression and localization of a vitellogenin receptor with eight ligand binding repeats in the cutthroat trout (Oncorhynchus clarki). Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology 166: 81–90. [DOI] [PubMed] [Google Scholar]
- 35. Yang F, Xu HT, Dai ZM, Yang WJ (2005) Molecular characterization and expression analysis of vitellogenin in the marine crab Portunus trituberculatus . Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology 142: 456–464. [DOI] [PubMed] [Google Scholar]
- 36. Phiriyangkul P, Utarabhand P (2006) Molecular characterization of a cDNA encoding vitellogenin in the banana shrimp, Penaeus (Litopenaeus) merguiensis and sites of vitellogenin mRNA expression. Molecular reproduction and development 73: 410–423. [DOI] [PubMed] [Google Scholar]
- 37. Raviv S, Parnes S, Segall C, Davis C, Sagi A (2006) Complete sequence of Litopenaeus vannamei (Crustacea: Decapoda) vitellogenin cDNA and its expression in endocrinologically induced sub-adult females. General and comparative endocrinology 145: 39–50. [DOI] [PubMed] [Google Scholar]
- 38. Xie S, Sun LN, Liu FS, Dong B (2009) Molecular characterization and mRNA transcript profile of vitellogenin in Chinese shrimp, Fenneropenaeus chinensis . Molecular biology reports 36: 389–397. [DOI] [PubMed] [Google Scholar]
- 39. Tiu SHK, Benzie J, Chan SM (2008) From hepatopancreas to ovary: molecular characterization of a shrimp vitellogenin receptor involved in the processing of vitellogenin . Biology of reproduction 79: 66–74. [DOI] [PubMed] [Google Scholar]
- 40. Roth Z, Khalaila I (2012) Identification and characterization of the vitellogenin receptor in Macrobrachium rosenbergii and its expression during vitellogenesis. Molecular Reproduction and Development 79: 478–487. [DOI] [PubMed] [Google Scholar]
- 41. Turk B, Turk V, Turk D (1996) Structural and functional aspects of papain-like cysteine proteinases and their protein inhibitors. Biological chemistry 378: 141–150. [PubMed] [Google Scholar]
- 42. Zhao W, Chen L, Zhang F, Wu P, Li E, et al. (2012) Molecular characterization of cathepsin L cDNA and its expression during oogenesis and embryogenesis in the oriental river prawn Macrobrachium nipponense (Palaemonidae). Genetics and molecular research: GMR 12: 5215–5225. [DOI] [PubMed] [Google Scholar]
- 43. Prasertlux S, Sittikankaew K, Chumtong P, Khamnamtong B, Klinbunga S (2011) Molecular characterization and expression of the Prostaglandin reductase 1 gene and protein during ovarian development of the giant tiger shrimp Penaeus monodon . Aquaculture 322: 134–141. [Google Scholar]
- 44. Chen L, Zhang DC, Yang LS, Huang JH, Yang QB, et al. (2009) Sequence analysis of eukaryotic translation initiation factor 3 subunit G (eIF3g) of black tiger shrimp (Penaeus monodon) and its mRNA tissue expression. South China Fisheries Science 5: 1–9. [Google Scholar]
- 45. Aflalo ED, Bakhrat A, Raviv S, Harari D, Sagi A, et al. (2007) Characterization of a vasa-like gene from the pacific white shrimp Litopenaeus vannamei and its expression during oogenesis. Molecular reproduction and development 74: 172–177. [DOI] [PubMed] [Google Scholar]
- 46. Feng ZF, Zhang ZF, Shao MY, Zhu W (2011) Developmental expression pattern of the Fc-vasa-like gene, gonadogenesis and development of germ cell in Chinese shrimp, Fenneropenaeus chinensis . Aquaculture 314: 202–209. [Google Scholar]
- 47. Zhu XL (2010) Cloning of two vasa/PL10 genes and their expression during oogenesis in prawn Macrobrachium nipponense . Marine Fisheries 32: 132–140. [Google Scholar]
- 48. Sellars MJ, Lyons RE, Grewe PM, Vuocolo T, Leeton L, et al. (2007) A PL10 vasa-like gene in the kuruma shrimp, Marsupenaeus japonicus, expressed during development and in adult gonad. Marine Biotechnology 9: 377–387. [DOI] [PubMed] [Google Scholar]
- 49. Olsen LC, Aasland R, Fjose A (1997) A vasa-like gene in zebrafish identifies putative primordial germ cells. Mechanisms of development 66: 95–105. [DOI] [PubMed] [Google Scholar]
- 50. Yu QS, Wu J (2008) Involvement of cyclins in mammalian spermatogenesis. Molecular and cellular biochemistry 315: 17–24. [DOI] [PubMed] [Google Scholar]
- 51. Ganoth D, Bornstein G, Ko TK, Larsen B, Tyers M, et al. (2001) The cell cycle regulatory protein CkS1 is required for SCFSkp2-mediated ubiquitinylation of p27. Nature cell biology 3: 321–324. [DOI] [PubMed] [Google Scholar]
- 52.Chen J, Liu P, Li Z, Chen Y, Qiu GF (2013) The cloning of the cdk2 transcript and the localization of its expression during gametogenesis in the freshwater giant prawn, Macrobrachium rosenbergii. Molecular biology reports: 1–10. [DOI] [PubMed]
- 53. Hadwiger J, Wittenberg C, Mendenhall M, Reed S (1989) The Saccharomyces cerevisiae CKS1 gene, a homolog of the Schizosaccharomyces pombe suc1 gene, encodes a subunit of the CDC28 protein kinase complex. Molecular and cellular biology 9: 2034–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Baarends WM, Roest HP, Grootegoed JA (1999) The ubiquitin system in gametogenesis. Molecular and cellular endocrinology 151: 5–16. [DOI] [PubMed] [Google Scholar]
- 55. Zhang ZP, Shen BL, Wang YL, Chen Y, Wang GD, et al. (2010) Molecular cloning of proliferating cell nuclear antigen and its differential expression analysis in the developing ovary and testis of penaeid shrimp Marsupenaeus japonicus . DNA and Cell Biology 29: 163–170. [DOI] [PubMed] [Google Scholar]
- 56. Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, et al. (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96: 635–644. [DOI] [PubMed] [Google Scholar]
- 57. Vigodner M, Morris PL (2005) Testicular expression of small ubiquitin-related modifier-1 (SUMO-1) supports multiple roles in spermatogenesis: silencing of sex chromosomes in spermatocytes, spermatid microtubule nucleation, and nuclear reshaping. Developmental biology 282: 480–492. [DOI] [PubMed] [Google Scholar]
- 58. Vigodner M, Ishikawa T, Schlegel PN, Morris PL (2006) SUMO-1, human male germ cell development, and the androgen receptor in the testis of men with normal and abnormal spermatogenesis. American Journal of Physiology-Endocrinology and Metabolism 290: E1022–E1033. [DOI] [PubMed] [Google Scholar]
- 59. Leelatanawit R, Sittikankeaw K, Yocawibun P, Klinbunga S, Roytrakul S, et al. (2009) Identification, characterization and expression of sex-related genes in testes of the giant tiger shrimp Penaeus monodon . Comparative Biochemistry and Physiology-Part A: Molecular & Integrative Physiology 152: 66–76. [DOI] [PubMed] [Google Scholar]
- 60. Miura C, Miura T, Yamashita M (2002) PCNA protein expression during spermatogenesis of the Japanese eel (Anguilla japonica). Zoological science 19: 87–91. [DOI] [PubMed] [Google Scholar]
- 61. Sada A, Suzuki A, Suzuki H, Saga Y (2009) The RNA-binding protein Nanos2 is required to maintain murine spermatogonial stem cells. Science 325: 1394–1398. [DOI] [PubMed] [Google Scholar]
- 62. Barrios F, Filipponi D, Pellegrini M, Paronetto MP, Siena SD, et al. (2010) Opposing effects of retinoic acid and FGF9 on Nanos2 expression and meiotic entry of mouse germ cells. Journal of cell science 123: 871–880. [DOI] [PubMed] [Google Scholar]
- 63. Alekseev OM, Richardson RT, Michael G (2009) Linker histones stimulate HSPA2 ATPase activity through NASP binding and inhibit CDC2/Cyclin B complex formation during meiosis in the mouse. Biology of reproduction 81: 739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu MH, Rajkovic A, Burns KH, Yan W, Lin YN, et al. (2003) Sequence and expression of testis-expressed gene 14 Tex14: a gene encoding a protein kinase preferentially expressed during spermatogenesis. Gene expression patterns 3: 231–236. [DOI] [PubMed] [Google Scholar]
- 65. Okutsu T, Kang BJ, Miwa M, Yoshizaki G, Maeno Y, et al. (2010) Molecular cloning and characterization of Dmc1, a gene involved in gametogenesis, from the whiteleg shrimp Litopenaeus vannamei . Fisheries Science 76: 961–969. [Google Scholar]
- 66. Miyamoto T, Koh E, Sakugawa N, Sato H, Hayashi H, et al. (2008) Two single nucleotide polymorphisms in PRDM9 (MEISETZ) gene may be a genetic risk factor for Japanese patients with azoospermia by meiotic arrest. Journal of assisted reproduction and genetics 25: 553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wu HY, Chen YC, Miao SY, Zhang CY, Zong SD, et al. (2010) Sperm associated antigen 8 (SPAG8), a novel regulator of activator of CREM in testis during spermatogenesis. FEBS letters 584: 2807–2815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of the deduced amino acid sequences of vasa . The amino acids conserved across all the eighteens species are shown in asterisks at the bottom. The GenBank accession numbers of the sequences are as follows: M. japonicus AEB00819; L. vannamei AAY89069; P. monodon AEB00820; F. chinensis ABQ00071; C. hawaiensis ACH92926; S. paramamosain ADR51551; M. nipponense ADB28894; P. carinicauda AGF90963.
(TIF)
Neighbor-joining phylogenetic analysis of the vasa from P. clarkii .
(TIF)
Localization of cyclin B and titin transcripts in crawfish gonads. The results of In situ hybridization with DIG-labeled antisense RNA probe (B and D for cyclin B; G and I for titin) and sense probe as negative control (C and E for cyclin B; H and J for titin) were shown. Regular histological section was stained with hematoxylin and eosin (A and F). O: oogonium; Cy: cytoplasm; N: nucleus; Nu, nucleolus; Fc: follicle cells; Sg: spermatogonium; Sc: Spermatocyte; St: Spermatid; The scale bar indicates 100 µm.
(TIF)
Real-time PCR confirmation of differential expressed genes.
(DOCX)
KEGG biochemical pathways of P. clarkii isotig sequences.
(XLSX)
List of the up-regulated and down-regulated genes between male and female libraries.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. The sequences reported in this paper were deposited into the National Center for Biotechnology Information (NCBI) Sequence Read Archive with accession numbers SRP035515.