Abstract
Vibrio fischeri is a typical quorum-sensing bacterium for which lux box, luxR, and luxI have been identified as the key elements involved in quorum sensing. To decode the quorum-sensing mechanism, an artificially constructed cell–cell communication system has been built. In brief, the system expresses several programmed cell-death BioBricks and quorum-sensing genes driven by the promoters lux pR and PlacO-1 in Escherichia coli cells. Their transformation and expression was confirmed by gel electrophoresis and sequencing. To evaluate its performance, viable cell numbers at various time periods were investigated. Our results showed that bacteria expressing killer proteins corresponding to ribosome binding site efficiency of 0.07, 0.3, 0.6, or 1.0 successfully sensed each other in a population-dependent manner and communicated with each other to subtly control their population density. This was also validated using a proposed simple mathematical model.
Introduction
The marine bacterium Vibrio fischeri lives symbiotically in light organs of some fishes and squids, and exhibits bioluminescence at high population densities [1]–[3]. Nealson et al. [4] reported that this bioluminescence was due to the accumulation of an autoinducer, and that the intensity of bioluminescence positively correlated with bacterial population density. This phenomenon is called “quorum sensing” [5], and considerable attention has been paid to determine the signaling molecules involved in it. N-3-oxohexanoyl-L-homoserine lactone, a type of acyl-homoserine lactone (AHL), was the first identified molecule in the process [6]. Interestingly, AHL was found to freely diffuse through cell membranes and to play a role in cell–cell communication [7].
As reported, at low bacterial population density, the concentration of AHLs remains low; however, their concentration increases with an increase in bacterial population density. At or above a particular threshold, they effectively bind to LuxR, which has been identified as an intracellular regulatory protein that mediates the formation of the LuxR:AHL complex. This dimer complex subsequently binds to the promoter of lux box and induces the expression of downstream genes such as luxC, luxD, luxA, luxB and luxE, which generates bioluminescence. V. fischeri lux box contains the promoters lux pL and lux pR that regulate the transcription of luxR and luxI, respectively.
luxI encodes a synthase, LuxI, which produces signaling molecule AHLs. luxI expression is upregulated by the interaction between the LuxR:AHL complex and lux box. Therefore, a positive feedback is involved in sensing signals [8]. In brief, quorum sensing in bacterial bioluminescence is tightly controlled by several key elements such as lux box, luxR, and luxI. This provides the basis for building an artificial quorum-sensing circuit.
You et al. [9] reported a programmed cell-death circuit by cloning luxR and luxI under the control of a synthetic promoter Plac/ara-1, and cotransforming the bacterial lethal gene ccdB into Escherichia coli cells. These engineered bacteria could successfully produce and release autoinducer AHLs, which mediated luxR and ccdB expression in a feedback manner. As these engineered bacteria grew, they continuously released autoinducer AHLs into their environment. When AHL concentration reached a particular threshold, AHLs combined with LuxR and activated ccdB expression to produce CcdB. Consequently, CcdB poisoned bacterial gyrase and resulted in the death of numerous bacterial cells [10]–[12]. However, when bacterial population density decreased, the concentration of LuxR:AHL dimer complexes required to activate lux pR was insufficient, and resulted in termination of cell death. Thereafter, the cells entered into a new round of growth. Once population density reached the abovementioned threshold, another round of cell death was triggered. This system successfully mimicked the situation of quorum sensing, in which bacterial cell growth and death could be regulated through bacterial population density.
However, this system was cotransformed using two separate plasmids under the control the promoters Plac/ara-1 and PluxI. This could be problematic because of an imbalance in the copy numbers of the two plasmids, making it difficult to accurately investigate cellular communication on a genetic basis. In addition, it is difficult to quantitatively manipulate population density at different levels.
Synthetic biology combines biology with engineering to design and build novel biomolecular components, networks, and pathways. It is well known that the features important for synthetic biology include modularity, standardization, and rigorously predictive models [13]–[18]. In recent years, an increasing number of studies in synthetic biology have focused on constructing simple synthetic gene circuits that exhibit desired properties. Meanwhile, a large variety of functional genetic parts are characterized and assembled to construct biological circuits to program new biological behaviors, dynamics, and logical control [19]–[21]. Standard biological parts, such as BioBricks from the “Registry of Standard Biological Parts” (RSBP) at MIT, USA (http://partsregistry.org), provide the foundation for designing and constructing synthetic biological systems [22]. Therefore, a synthetic biological system can be achieved using synthetic biological methods and BioBricks for which gene expression can be precisely regulated and quantitatively and accurately measured [23].
Ribosome binding site (RBS) is a sequence where translation of mRNA into protein starts with. RBS efficiency can be different with binding strengths of various sequences with a ribosome, as a result, yields of protein will be different [24].
In this study, we investigated a few RBSs with different binding strengths (RBS0.07, RBS0.3, RBS0.6, and RBS1.0). Our aim was to combine the assembly of BioBricks and a bacterial quorum-sensing mechanism to build synthetic circuits to subtly control bacterial population density at different levels.
Materials and Methods
Ethics statement
Our research did not involve human subjects; thus, no ethics statement is required.
Strains and plasmids
The E. coli DH5α and BL21(DE3) strains were preserved in our laboratory. The plasmids used in this study included pSB1A2-PlacO-1, pSB1A2-lux pL-RBS1.0-luxR-TT-lux pR, pSB1A2-RBS0.07, pSB1A2-RBS0.3, pSB1A2-RBS0.6, pSB1A2-RBS1.0, pSB1AK3-RBS1.0- luxI-TT, pSB1A3-lacZα-ccdB, and pSB1AK3-TT, and were constructed in our laboratory based on the BioBrick plasmids from RSBP with the BioBrick standard assembly method [25], [26].
Polymerase Chain Reaction (PCR) primers
Primers were designed based on the sequences of target genes. The sequences for five restrictive enzymes (EcoRI, XbaI, PstI, NotI and SpeI) and RBS1.0 were introduced at the end of the individual primers for subcloning. The primers were purchased from Shanghai Boya Biotechnology Co., Ltd. and had the following sequences:
2621-F: 5′-GCTCTAGAGAAAGAGGAGAAATACT-3′
2621-R: 5′-AAAACTGCAGCGGCCGCTACTA-3′
Construction of BioBrick plasmids
To construct BioBrick plasmids, inserts were amplified with 2621-F and 2621-R primers at 94°C for 5 min, followed by 30 cycles at 94°C for 45 s, 59.3°C for 45 s, and 72°C for 1 min, with a final extension at 72°C for 10 min. PCR products were purified using PCR purification kits, followed by digestion with the restriction enzymes EcoRI and PstI, ligation with a plasmid vector, transformation into competent cells, and culture on agar plates. Positive clones were confirmed by restriction enzyme digestion and verified by Sanger sequencing. Taq polymerase, restriction enzymes, and other related reagents were purchased from TaKaRa Biotechnology (Dalian) Co., Ltd.
Growth kinetic analysis
Bacteria transformed with plasmids to express RBS0.07, RBS0.3, RBS0.6, or RBS1.0 artificial quorum-sensing circuits were grown in standard Luria-Bertani medium. To assess their growth kinetics, cultures were incubated at 37°C until their optical density at 600 nm reached 0.6–0.8. To induce the expression of inserts, isopropyl β-D-1-thiogalactopyranoside (IPTG) was added at a final concentration of 1 mM. Sample cultures were taken at sequential time intervals, diluted for plating onto agar plates, and incubated overnight at 37°C. Next, the number of clones was counted. Each experiment was repeated at least three times.
Mathematical modeling
Mathematical modeling has been shown to play an important role in predicting how well a network works, and can be used to optimize the design of synthetic biological models, thereby eliminating the need for a large amount of experimental work [27]–[32]. For this study, we used a mathematical model developed by You et al. [9] that has been validated by experiments and successfully applied to monitor the associations among population density, killer protein concentration and AHL signals. Based on this model, we sought to develop a model to predict the transient dynamics of our synthetic organisms that incorporated artificial population- control circuits.
Results
Recombination of BioBrick components
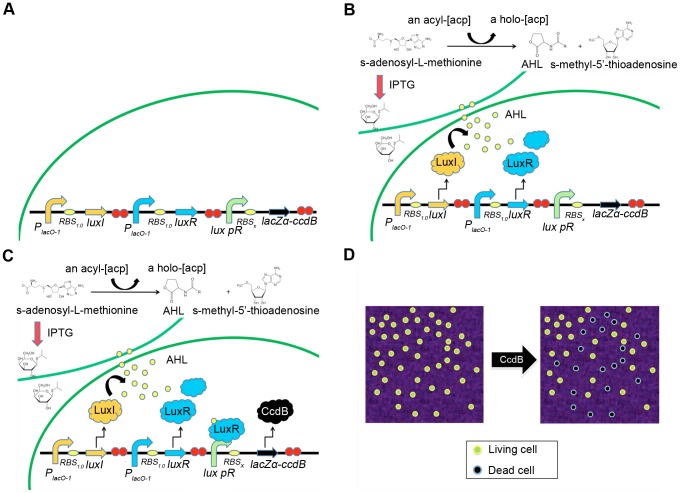
To build a functional system, a construct was made using multiple BioBrick gene components, which included luxI, luxR, and RBS sequences having different binding strengths (RBS0.07, RBS0.3, RBS0.6, or RBS1.0). To investigate the control of bacterial population density using a bacterial quorum-sensing system, a lethal fusion gene, lacZa-ccdB, driven by the promoter lux pR was cloned into the same construct. Furthermore, four types of RBSs with different binding strengths were assembled separately, upstream of ccdB to subtly control its expression strength. In addition, the promoter for the original vector, pSB1A2, was replaced with an IPTG-inducible promoter, Placo-1, so that the expression of the construct genes could be switched on or off as required. The essential components and mechanisms of this artificial quorum-sensing circuit are shown in Figure 1. The essential components are PlacO-1-RBS1.0-luxI-TT-PlacO-1-RBS1.0-luxR-TT-lux pR-RBSx-lacZα-ccdB-TT. The four panels of Figure 1 illustrate the regulation mechanism of each stage of the quorum-sensing process. Each panel indicates a regulation stage. For example, Figure 1A represents the cell state before the inducer IPTG is added to a culture, during which the circuit is switched off. Figure 1B–1D represent the cell states after the inducer IPTG is added to a culture, when the circuit is switched on. Figure 1B indicates that the promoter Placo-1 triggers luxI and luxR expression, producing LuxI and LuxR. LuxI produces the signaling molecule AHL. Figure 1C indicates that when the concentration of AHLs reaches a certain threshold, AHLs will effectively bind to LuxR. The LuxR:AHL complex will subsequently bind to the promoter lux pR and induce ccdB expression to produce killer protein CcdB. Figure 1D indicates that because more and more CcdB accumulates in the environment to kill large numbers of cells, cell population density will decrease. Plasmids used for expressing luxI, luxR, and ccdB corresponding to different RBS binding strengths were successfully generated. Positive clones were confirmed by digestion with restriction enzymes and verified by Sanger sequencing (data not shown).
Figure 1. Essential components and mechanism of an artificial quorum-sensing circuit.
Note: the subscript “x” in RBSx represents 0.07, 0.3, 0.6, or 1.0. (A) The essential components of an artificial quorum-sensing circuit are PlacO-1-RBS1.0-luxI- TT-PlacO-1-RBS1.0-luxR-TT-lux pR-RBSx-lacZα-ccdB-TT. (B–D) The mechanism of an artificial quorum-sensing circuit is the following: (B) When IPTG is added to a culture, the promoter Placo-1 will trigger luxI and luxR expression, producing synthase LuxI (which produces the signaling molecule AHL) and an intracellular regulatory protein, LuxR, respectively. (C) AHLs will accumulate within and outside cells, and at or above a certain threshold, AHLs will effectively bind to LuxR. The LuxR:AHL complex will subsequently bind to the promoter lux pR and induce downstream expression of the lethal gene, ccdB, to produce killer protein CcdB. (D) Because more and more killer protein CcdB accumulates in the environment to kill large numbers of cells, the cell population density will decrease.
Bacterial population density control by different RBS artificial quorum-sensing circuits
Different RBS sequences upstream of the lethal gene will influence its expression which has a direct effect on the performances of standardized programmed bacterial death quorum-sensing circuits. Therefore, we constructed four types of RBS artificial quorum-sensing circuits and assessed cell growth kinetics to investigate the effect of different types of circuits on controlling bacterial cell growth and manipulating bacterial population density. These results indicated that an RBS0.07-expressing circuit could maintain a lower population density than the control bacteria (Figure 2A). This could be achieved by slowing bacterial growth or by coordinating the ratio between cell growth and death so that population density could be controlled. In addition, we chose to determine population density of bacterial growth to 22, 26, and 30 h, when bacteria had been at steady state, to indicate the control effect of engineered bacteria containing different RBS (RBS0.07, RBS0.3, RBS0.6, or RBS1.0) quorum-sensing circuits on population density (Figure 2B). In order to more intuitively show the relationship between RBS efficiency and bacterial population density, we took the average values of viable cell numbers of the four strains of engineered bacteria from Figure 2B to prepare the histogram (Figure 2C). Both Figure 2B and Figure 2C indicated that with an increase in RBS efficiency, viable cell numbers gradually decreased. Thus, the constructed RBS0.07, RBS0.3, RBS0.6, and RBS1.0 artificial quorum-sensing circuits could successfully control bacterial population density at different levels. In brief, the results verified the effectiveness of the proposed mechanism of the artificial quorum-sensing circuits and the circuits were functional.
Figure 2. Bacterial population density controlled by different RBS artificial quorum-sensing circuits.
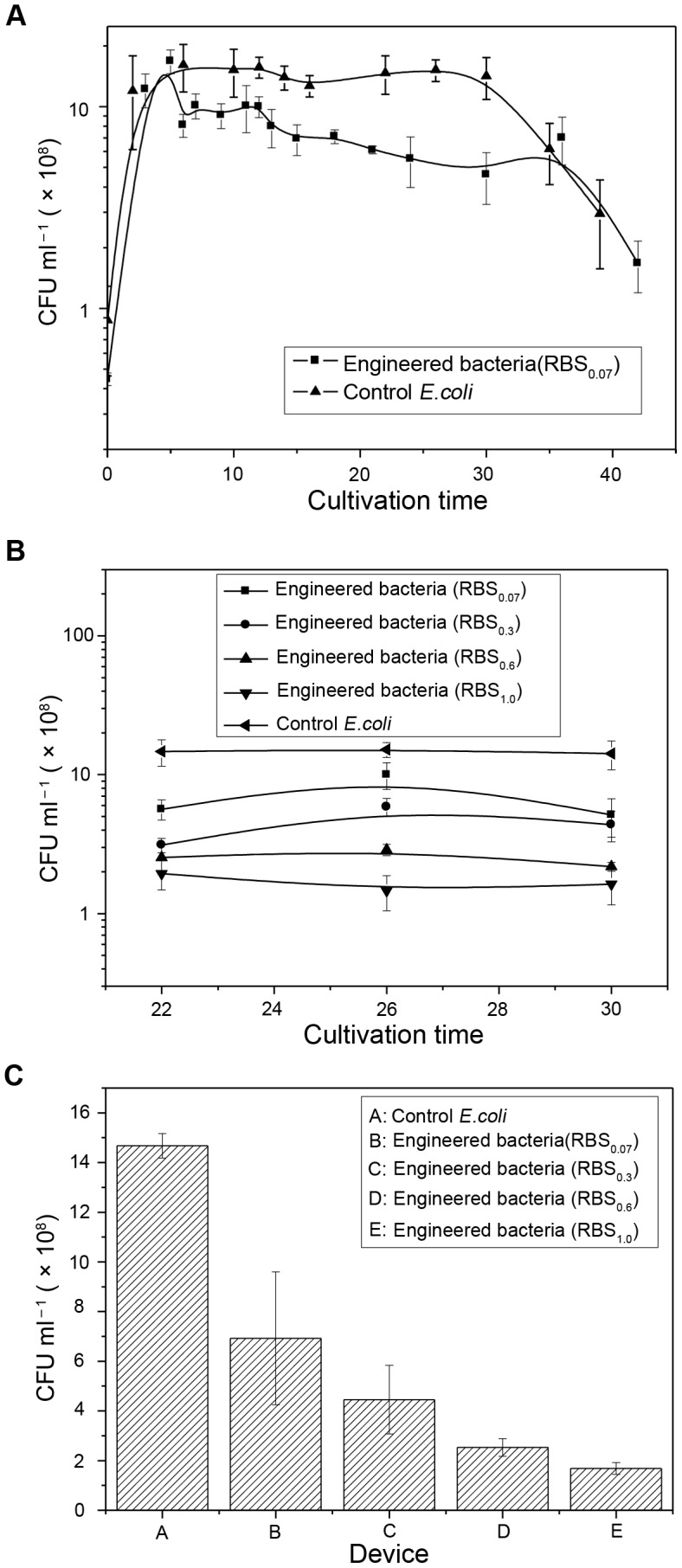
Note: The error bars represent the standard deviation of at least three replicates. (A) Curves for viable cell numbers of the engineered bacteria E. coli BL21(DE3) containing an RBS0.07 programmed bacterial death quorum-sensing circuit and control E. coli. The curve for engineered bacteria was different from that for control E. coli. And these differences were significant (t = 1.98 > t0.90, 17 = 1.74) during 6–30 h with 90% confidence interval using an independent samples t-test. At approximately 5 h, there was a dramatic decline in the number of viable engineered cells compared with control cells, after which the growth curve tended to level off after two small amplitude oscillations. The lower population density of the RBS0.07 engineered bacteria remained until its growth rate decreased after >30 h of culture. (B) Viable cell numbers at different times of steady state for four strains of engineered bacteria containing different RBS (RBS0.07, RBS0.3, RBS0.6, or RBS1.0) quorum-sensing circuits. (C) Final histogram for viable cell numbers of the four strains of engineered bacteria. This indicated that with an increase in RBS efficiency, viable cell numbers gradually decreased.
Mathematical model and modeling analysis
The major purpose of the present study was to determine the roles and functions of the different elements in our system. In brief, the study focused on how it maintains its population density at a reasonable level by producing substances, such as killer protein, LuxI, and LuxR, and how these different proteins function together in the complex process. To further optimize this system, we applied a mathematical model to monitor the performance of our system and to analyze and predict the growth rules for bacteria with a programmed cell-death circuit. This was derived as follows.
As developed by You et al. [9], a model is
| (1) |
The variables and parameters in equation (1) are
NS: viable cell density at steady state (ml−1);
Nm: cell carrying capacity in limited medium without a cell-death circuit (ml−1);
dA: degradation rate constant for AHL (h−1);
dE: degradation rate constant for killer protein (h−1);
k: cell growth rate (h−1);
vA: production rate constant for AHL (nM ml h−1);
kE: production rate constant for killer protein (h−1);
d: death rate constant for circuit-regulated cells (nM−1 h−1).
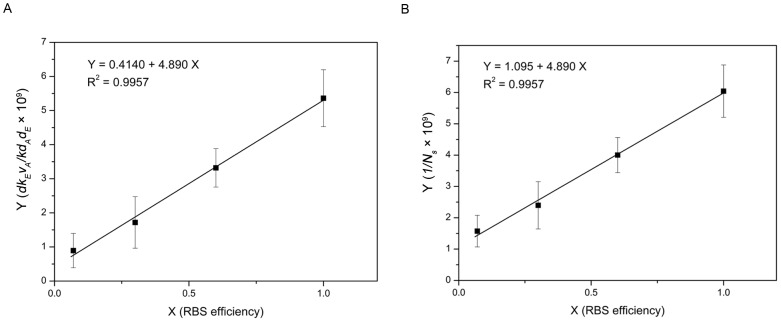
To characterize and compare the performances of different components of our constructed circuits, we determined cell density at steady state along with the expression of different components, which were considered to be involved in controlling bacterial population density. Based on equation (1), we observed that the value in 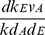 for
RBS0.07, RBS0.3,
and
RBS0.6 circuits (Figure 2B, 2C) was 0.14, 0.30, 0.62, while the value for the RBS1.0 circuit was defined as 1.00 (Table 1). Through computational analysis using Origin 7.0 software, a linear relationship was observed between RBS efficiency (X) and
for
RBS0.07, RBS0.3,
and
RBS0.6 circuits (Figure 2B, 2C) was 0.14, 0.30, 0.62, while the value for the RBS1.0 circuit was defined as 1.00 (Table 1). Through computational analysis using Origin 7.0 software, a linear relationship was observed between RBS efficiency (X) and 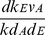 (Y), as shown in Figure 3A (Y = 0.4140+4.890X; R2 = 0.9957).
(Y), as shown in Figure 3A (Y = 0.4140+4.890X; R2 = 0.9957).
Table 1. Quantitative relationship between RBS efficiency of a bacterial population control circuit and steady-state cell density.
| NS | RBS efficiency |
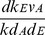
|
| 6.93×108 | 0.07 | 0.14 |
| 4.45×108 | 0.3 | 0.30 |
| 2.53×108 | 0.6 | 0.62 |
| 1.68×108 | 1.0 | 1.00 |
Figure 3. Linear relationship between RBS efficiency and model parameters.
Note: Ns is the viable cell density of engineered bacteria at steady state. The error bars represent the standard deviation of at least three replicates. (A) RBS efficiency (X) showed a linear relationship with the combined parameter 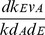 (Y): Y = 0.4140+4.890X; R2 = 0.9957. (B) RBS efficiency (X) also had a linear relationship with the parameter
(Y): Y = 0.4140+4.890X; R2 = 0.9957. (B) RBS efficiency (X) also had a linear relationship with the parameter 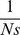 (Y): Y = 1.095+4.890X; R2 = 0.9957.
(Y): Y = 1.095+4.890X; R2 = 0.9957.
Based on these results, we derived the mathematical model shown in equation (2) to describe the relationship between viable cell density at steady state (NS) and RBS efficiency for ccdB (D).
| (2) |
KS is a constant for how much RBS efficiency affects viable cell density. The reciprocal of equation (2) is
| (3) |
Based on the data (Table 1) for viable cell density of engineered bacteria at steady state and RBS efficiency, the linear relationship between RBS efficiency (D, i.e., X) and 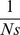 (Y) is shown in Figure 3B (Y = 1.095+4.890X; R2 = 0.9957).
(Y) is shown in Figure 3B (Y = 1.095+4.890X; R2 = 0.9957).
According to our mathematical model, we predicted that when bacteria were growing at steady state, a Ks of 4.47 and 1/Nm of 1.095 would give Nm of 0.91×109, which was close to the experimentally measured Nm of 1.47×109. Therefore, the mathematical model given here reflected the relationship between RBS efficiency and viable cell density of engineered bacteria at steady state. Thus, this model could be used to predict viable cell density of engineered bacteria at steady state through RBS efficiency values of a synthetic circuit.
Discussion
Our work highlighted constructing several standardized programmed bacterial cell-death quorum-sensing circuits by cloning a series of BioBrick genes and splicing them with different RBS forms along with a lethal fusion gene, lacZa-ccdB using standard assembly methods. Compared with the reported research [9], the outstanding of our work is that the constructed RBS0.07, RBS0.3, RBS0.6, and RBS1.0 artificial quorum-sensing circuits functioned successfully to subtly control bacterial population density at different levels by regulating ccdB expression.
To monitor and validate the performance of our system, we developed a mathematical model on the basis of the model of You et al. [9]. According to our results, there was a strong linear relationship between the reciprocal of viable cell density at steady state and RBS efficiency for ccdB. Thus, viable cell density at steady state could be quantitatively controlled by interchanging RBS sequences of different strengths in a population density control circuit. In addition, this model could predict the roles of the artificially constructed components in our system for regulating population density. For example, on account of this model, by determining the dynamics of population density, expression levels, and coordination of luxR, luxI, and ccdB in this system, we will know if the artificially constructed components are functioning normally and which parts need to be improved. Our system would be better optimized if the regulation levels of RBSs could be standardized so that their functions and roles could be more precisely predicted and validated.
Moreover, our current investigation systematically studied the functions of different components in regulating bacterial population density through quorum-sensing circuits. Therefore, our system provides an example and model for modulation of cell density in other field, such as fermentation industry, which requires to maintain bacterial growth at a desired density for a long time or to prolong the stationary phase of bacterial growth, so that the production of bacterial metabolites can be achieved.
Synthetic biology holds promise for developing artificial systems to address challenges posed by major global problems [33], [34] such as health [35], [36], environment [37]–[40], materials [41], and energy [42], [43]. It is advancing from the development of proof-of-concept designs to a focus on core platforms, including DNA construction, parts libraries, computational design tools, and interfaces for manipulating and probing synthetic circuits [18]. We generated a series of engineered E. coli cells by cloning all the essential components of a quorum-sensing circuit into the same plasmid of each engineered E. coli cell using the BioBrick standard assembly method. As a result, luxR and luxI expression levels in this system were regulated and coordinated by the promoter PlacO-1. Since it is an artificial simplified system, it can be an ideal platform to investigate how the quorum-sensing process works, such as by exploring its molecular signal pathway.
Furthermore, the quorum-sensing process involves in well controlled regulations and responses, so that the sensor circuit can monitor subtle change of environmental stimuli to control its own population density. Therefore, it can be potentially used as a testing or alarming system to sense environmental change, such as pollution, or microbes outbreak.
Acknowledgments
We thank Dr. Yuanxin Hu (Washington University in St. Louis) and Professor Jianjiang Zhong (Shanghai Jiao Tong University) for critically reading and revising our manuscript.
Funding Statement
Financial support for this work was provided by the National Natural Science Foundation of China (No. 30770059, No. 21336009) and the Fundamental Research Funds for the Central Universities in China (No. 2010121050). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kempner E, Hanson F (1968) Aspects of light production by Photobacterium fischeri . J Bacteriol 95: 975–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ruby EG, Nealson KH (1976) Symbiotic association of Photobacterium fischeri with the marine luminous fish Monocentris japonica: a model of symbiosis based on bacterial studies. Biol Bull 151: 574–586. [DOI] [PubMed] [Google Scholar]
- 3. Ruby EG, McFall-Ngai MJ (1999) Oxygen-utilizing reactions and symbiotic colonization of the squid light organ by Vibrio fischeri . Trends Microbiol 7: 414–420. [DOI] [PubMed] [Google Scholar]
- 4. Nealson KH, Platt T, Hastings J (1970) Cellular control of the synthesis and activity of the bacterial luminescent system. J Bacteriol 104: 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fuqua WC, Winan SC, Greenberg E (1994) Quorum sensing in bacteria: the luxR-luxI family of cell density-responsive transcriptional regulators. J Bacteriol 176: 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, et al. (1981) Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochem 20: 2444–2449. [DOI] [PubMed] [Google Scholar]
- 7. Kaplan HB, Greenberg E (1985) Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol 163: 1210–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collin M, Schuch R (2009) Bacterial Sensing and Signaling. Basel: Karger Publishers. 1–32 pp.
- 9. You L, Cox RS, Weiss R, Arnold FH (2004) Programmed population control by cell-cell communication and regulated killing. Nature 428: 868–871. [DOI] [PubMed] [Google Scholar]
- 10. Kampranis SC, Howells AJ, Maxwell A (1999) The interaction of DNA gyrase with the bacterialtoxin CcdB: evidence for the existence of two gyrase-CcdB complexes. J Mol Biol 293: 733–744. [DOI] [PubMed] [Google Scholar]
- 11. Dwyer DJ, Kohanski MA, Hayete B, Collins JJ (2007) Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli . Mol Syst Biol 3: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Galhardo RS, Do R, Yamada M, Friedberg EC, Hastings PJ, et al. (2009) DinB upregulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli . Genetics 182: 55–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee HC, Chen BS (2010) Synthetic biology: Construction of genetic circuits for engineering applications. Int J Syst Synth Biol 1: 71–86. [Google Scholar]
- 14. Serrano L (2007) Synthetic biology promises and challenges. Mol Syst Biol 3: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haseloff J, Ajioka J (2009) Synthetic biology: History, challenges and prospects. J R Soc Interface 6: S389–S391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Agapakis CM, Silver PA (2009) Synthetic biology: Exploring and exploiting genetic modularity through the design of novel biological networks. Mol Biosyst 5: 704–713. [DOI] [PubMed] [Google Scholar]
- 17. Mukherji S, Oudenaarden A (2009) Synthetic biology: Understanding biological design from synthetic circuits. Nat Rev Genet 10: 859–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng A, Lu TK (2012) Synthetic biology: An emerging engineering discipline. Annu Rev Biomed Eng 14: 155–78. [DOI] [PubMed] [Google Scholar]
- 19. Chandran D, Copeland WB, Sleight SC, Sauro HM (2008) Mathematical modeling and synthetic biology. Drug Discov Today Dis Model 5: 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leonard E, Nielsen D, Solomon K, Prather KJ (2008) Engineering microbes with synthetic biology frameworks. Trends Biotechnol 26: 674–681. [DOI] [PubMed] [Google Scholar]
- 21. Slusarczyk AL, Lin A, Weiss R (2012) Foundations for the design and implementation of synthetic genetic circuits. Nat Rev Genet 13: 406–420. [DOI] [PubMed] [Google Scholar]
- 22. Anderson JC, DueberJE, Leguia M, Wu GC, Goler JA, et al. (2010) BglBricks: A flexible standard for biological part assembly. J Biol Eng 4: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lucks JB, Qi L, Whitaker WR, Arkin AP (2008) Toward scalable parts families for predictable design of biological circuits. Curr Opin Microbiol 11: 567–573. [DOI] [PubMed] [Google Scholar]
- 24. Norville JE, Derda R, Gupta S, Drinkwater KA, Belcher AM, et al. (2010) Introduction of customized inserts for streamlined assembly and optimization of BioBrick synthetic genetic circuits. J Biol Eng 4: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knight TF (2003) Idempotent vector design for standard assembly of BioBricks. MIT Synthetic Biology Working Group Technical Reports.
- 26. Speer MA, Richard TL (2011) Amplified insert assembly: An optimized approach to standard assembly of BioBrick genetic circuits. J Biol Eng 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kambam PKR, Henson MA, Sun LH (2008) Design and mathematical modelling of a synthetic symbiotic ecosystem. IET Syst Biol 2: 33–38. [DOI] [PubMed] [Google Scholar]
- 28. Chandran D, Copeland WB, Sleight SC, Sauro HM (2008) Mathematical modeling and synthetic biology. Drug Discov Today Dis Model 5: 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Y, Sriram G (2010) Mathematical Modeling: Bridging the gap between concept and realization in synthetic biology. J Biomed Biotechnol 2010: doi:10.1155/2010/541609. [DOI] [PMC free article] [PubMed]
- 30. Ceroni F (2010) A computational model of gene expression in an inducible synthetic circuit. Pac Symp Biocomput 15: 409–420. [DOI] [PubMed] [Google Scholar]
- 31. Endler L, Rodriguez N, Juty N, Chelliah V, Laibe C, et al. (2009) Designing and encoding models for synthetic biology. J R Soc Interface 6: S405–S417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gendrault Y, Madec M, Lallement C, Pecheux F, Haiech J (2011) Synthetic biology methodology and model refinement based on microelectronic modeling tools and languages. Biotechnol J 6: 796–806. [DOI] [PubMed] [Google Scholar]
- 33. Redford KH, Adams W, Mace GM (2013) Synthetic biology and conservation of nature: wicked problems and wicked solutions. PLoS Biol 11: e1001530 doi:10.1371/journal. pbio.1001530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang YH, Wei KY, Smolke CD (2013) Synthetic biology: Advancing the design of diverse genetic systems. Annu Rev Chem Biomol Eng 4: 69–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anderson JC, Clarke EJ, Arkin AP, Voigt CA (2012) Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol 355: 619–627. [DOI] [PubMed] [Google Scholar]
- 36. Neumann H, Neumann-Staubitz P (2012) Synthetic biology approaches in drug discovery and pharmaceutical biotechnology. Appl Microbiol Biotechnol 87: 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kirby JR (2012) Synthetic biology: Designer bacteria degrades toxin. Nat Chem Biol 6: 398–399. [DOI] [PubMed] [Google Scholar]
- 38. Lorenzo VD (2008) Systems biology approaches to bioremediation. Curr Opin Biotechnol 19: 579–589. [DOI] [PubMed] [Google Scholar]
- 39. Nicolaou SA, Gaida SM, Papoutsakis ET (2010) A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: From biofuels and chemicals, to biocatalysis and bioremediation. Metab Eng 12: 307–331. [DOI] [PubMed] [Google Scholar]
- 40. Checa SK, Zurbriggen MD, Soncini FC (2012) Bacterial signaling systems as platforms for rational design of new generations of biosensors. Curr Opin Biotechnol 23: 766–772. [DOI] [PubMed] [Google Scholar]
- 41. Becker J, Wittmann C (2011) Bio-based production of chemicals, materials and fuels–Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol 23: 1–10. [DOI] [PubMed] [Google Scholar]
- 42. Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, et al. (2010) Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463: 559–562. [DOI] [PubMed] [Google Scholar]
- 43. Connor RM, Atsumi S (2010) Synthetic biology guides biofuel production. J Biomed Biotechnol 2010: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]