Abstract
Objective: This study aimed to investigate the methylation status of promoter 1 region of glial cell line-derived neurotrophic factor (GDNF) in human glioma cells and to explore the effect of GDNF methylation on the expression of GDNF in glioma. Methods: GDNF gene mutation was detected by sequencing in 10 patients with glioma and 5 healthy controls. Bisulfite modification for analysis of DNA methylation was done to detect the methylation status of promoter 1 region of GDNF in 20 patients with glioma (10 with poorly differentiated and 10 with well differentiated) and 5 healthy controls. Results: There was no mutation at the promoter 1 region of GDNF gene in glioma. The incidence of methylation of GDNF gene at the promoter 1 region in healthy control, patients with poorly differentiated glioma and those with well differentiated glioma was 72.25%, 86.25% and 86.75%. The incidence of GDNF methylation in glioma was significantly higher than that in the normal brain (P<0.05); while there was no significant difference between well differentiated glioma and poorly differentiated glioma. Conclusions: Hypermethylation occurs in the promoter 1 region of GDNF and may influence the expression of GDNF in glioma.
Keywords: Glioma cell, glial cell line-derived neurotrophic factor, promoter, methylation
Introduction
Glioma is the most common primary tumor in the brain [1,2], and its occurrence and development are complex. Abnormal or silence expression of genes secondary to gene mutation and epigenetic modifications play important roles in the occurrence and development of tumors [3]. Of these modifications, methylation of the promoter of genes related tumors is often found to affect the expression of these genes. In normal cells of humans, the CpG island of the promoter is usually in a non-methylated status, which assures the normal expression of a specific gene [4]. However, sporadic CpG dinucleotides in the genome are often methylated, which is better to maintain the stability of chromosome structure. The methylation status is relatively stable in the genome, but it changes significantly in cancer cells [5]. To elucidate the methylation status of promoter 1 of glial cell line-derived neutrophic factor (GDNF), the initial sequence of promoter 1 of GDNF and that after bisulfite modification were compared between glioma and normal brain tissues, and the change in the methylation status of promoter 1 of GDNF was further analyzed. Our findings may provide evidence for further elucidation of change in the epigenetic modification of GDNF in the glioma.
Materials and methods
Sample collection and reagents
The normal brain tissues were collected from 5 patients with acute brain trauma who received intracranial decompression and glioma tissues was collected from 20 patients with glioma (WHO grades 1-4) in the Central Hoapital of Wenzhou City. Primary sequencing for SNPs and for BSP methylation was done in samples randomly collected from patients who did not receive pharmacotherapy. Glioma of pathologic grade 1-2 was classified as low grade one and that of pathologic grade 3-4 as high grade one. Following reagents were used in the present study: QIAamp DNA Mini andBlood Mini Handbook (QIAGEN; Germany), DNA Gel Extraction Kit (Shanghai Genebase Gene-Tech Co., Ltd), EZ DNA Methylation-GoldTM Kit (Zymo Research Biotech, USA).
Sequencing for SNP analysis
QIAamp DNA Mini and Blood Mini Handbook (Qiangen) was used for DNA extraction. In brief, tissues were cut into blocks (about 25 mg) and then placed in 1.5 ml-tubes, followed by addition of Buffer ATL (180 μL) and protein K (20 μL). Then, this mixture was incubated at 56°C until the blocks were absent. Centrifugation was done, and the supernatant was removed, followed by addition of Buffer AL (200 μL). After incubation at 70°C for 10 min, centrifugation was done, and the supernatant was removed. Following addition of absolute ethanol (200 μL), centrifugation was performed, and the mixed solution was transferred into 2-ml QMSC, followed by centrifugation at 8000 rpm for 1 min. Then, QMSC was placed into a 2-ml collector, followed by addition of Buffer AW1 (500 μL) and subsequent centrifugation at 8000 rpm for 1 min. QMSC was placed into a 2-ml collector, and centrifugation was done at 14000 rpm for 3 min following addition of Buffer AW2 (500 μL). QMSC was placed into a 1.5 ml-EP tube and allowed to stay at room temperature for 1 min following addition of Buffer AE (200 μL). Centrifugation was done for 1 min at 8000 rpm, and solution was harvested. Primers targeting the promoter I region of DNA was designed as follows: 5’CAGAGAATCTCAAAGGTGCA3’ (forward), 5’ACGACATCCCATAACTTC3’ (reverse). Pre-denaturation was done at 98°C for 4 min, 30 cycles of denaturation at 98°C for 15 s, annealing at 60°C for 15 s and extension at 72°C for 30 s, and a final extension at 72°C for 10 min. The products were subjected to gel electrophoresis, and the gel was retrieved for detection. Sequencing was done to detect the abnormal SNPs of promoter I region of GDNF gene.
Sequencing for BSP methylation
QIAamp DNA Mini and Blood Mini Handbook (Qiagen) was used for DNA extraction, and procedures were abovementioned above. Bisulfite treatment was done with EZDNA Methylation-GoldTM Kit. In brief, 1 μg of DNA (20 μL) was collected from each sample, and then mixed with 130 μL of CT Conversion Reagent, followed by centrifugation. The mixture was incubated at 98°C for 10 min, 64°C for 2.5 h and 4°C for 16 h. Then, 600 μl of M-Binding Buffer was added to Zymo-Spin IC Column and then placed in a collection tube. Products collected after centrifugation (150 μL) were added to 600 μL of above Binding Buffer, followed by vortexing. Centrifugation was done at >10000 rpm for 30 s. The supernatant was removed. Then, 200 μL of wash buffer was added, followed by centrifugation for 30 s at >10000 rpm. Following addition of desulphonation buffer (200 μL) into the column, incubation was done at room temperature (20°C-30°C) for 15-20 min, followed by centrifugation at >10000 rpm for 30 s. After addition of 200 μL of wash buffer, centrifugation was done for 30 s at >10000 rpm. The column was transferred into a new 1.5-ml tube, followed by addition of 20 μL of Elution Buffer into the column. The column was allowed to stay for 3-5 min, followed by centrifugation at >10000 rpm for DNA elution. Then, DNA was immediately used for expansion by PCR or stored at -20°C. Two pairs of primers were designed targeting the promoter I region of BSP as follows: Primer 1: 5’GAAGGGATTAGGGTTAGAATTTTT3’ (forward), 5’CCCAAACAAAAACRATATTTCT3’ (reverse); Primer 2: 5’ATGYGTTTGATTTTATTTTTAAAGA3’ (forward), CTACRCRAACAAACRAAA3’ (reverse). Reaction mixture was composed of 10× buffer (5 μL), 50 mM MgCl2 (2 μL), 10 mM dNTP (1 μL), forward primer (2 μL), reverse primer (2 μL), DNA (5 μL), Platinum Taq (Invitrogen; 0.3 μL) and ddH2O (32.7 μL). PCR was done at 95°C for 5 min, 42 cycles of 95°C for 30 s, 58°C for 30 s and 72°C for 40 s, and a final extension at 72°C for 10 min. Products after PCR were connected to pMD19-T (Takara) at room temperature for 2 h. Then, 10 μL of products after connection was used to infect competent DH5a cells which was then smeared on the Lysogeny Broth (LB) containing the antibiotic ampicillin (Amp), followed by incubation at 37°C over night. Eight colonies were selected from each plate for identification by PCR. Five positive colonies were seeded into 3 ml of liquid LB, followed by incubation at 37°C over night. Plasmid Miniprep Kit (Axygen) was used for plasmid extraction. Then, plasmids were subjected to sequencing. The extent of methylation was analyzed on the database (http://quma.cdb.riken.jp/).
Statistical analysis
Statistical analysis was done with SPSS version 16.0. Data were expressed as mean ± standard deviation (SD). Comparisons of data among groups were done with one way analysis of variance. A value of P<0.05 was considered statistically significant.
Results
Sequencing
There was no GDNF gene mutation in glioma and normal brain tissues.
PCR after bisulfite treatment
After expansion with primer 1 and primer 2, the promoter 1 region was identified at 469 bp and 507 bp, respectively (Figure 1).
Figure 1.
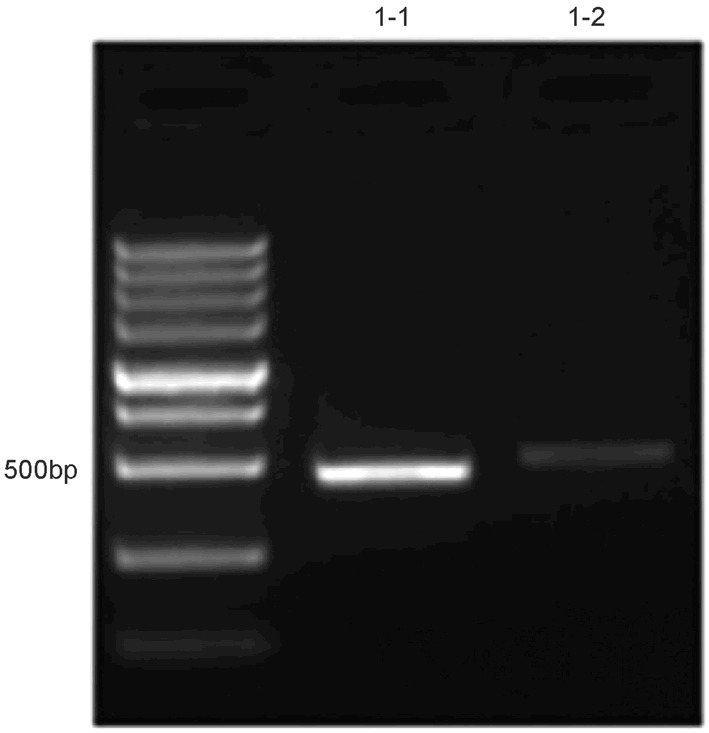
Products from PCR after bisulfite treatment.
Methylation status of promoter I
The promoter I region locates at about 1000 bp preceding the exon 2 of GDNF gene (Figure 2A). It was predicted that there were 40 CpG sites in the promoter I region (Figure 2B). After bisulfite treatment, sequencing was done (Figure 2C) (black: methylation; white: non-methylation). Statistical analysis showed the proportion of tissues with methylation of promoter I of GDNF gene was 72.25%, 86.25% and 86.75% in normal brain tissues, low grade glioma and high grade glioma. Significant difference was noted between normal brain tissues and low or high grade glioma, but there was no marked difference between low grade glioma and high grade glioma (Figure 3).
Figure 2.
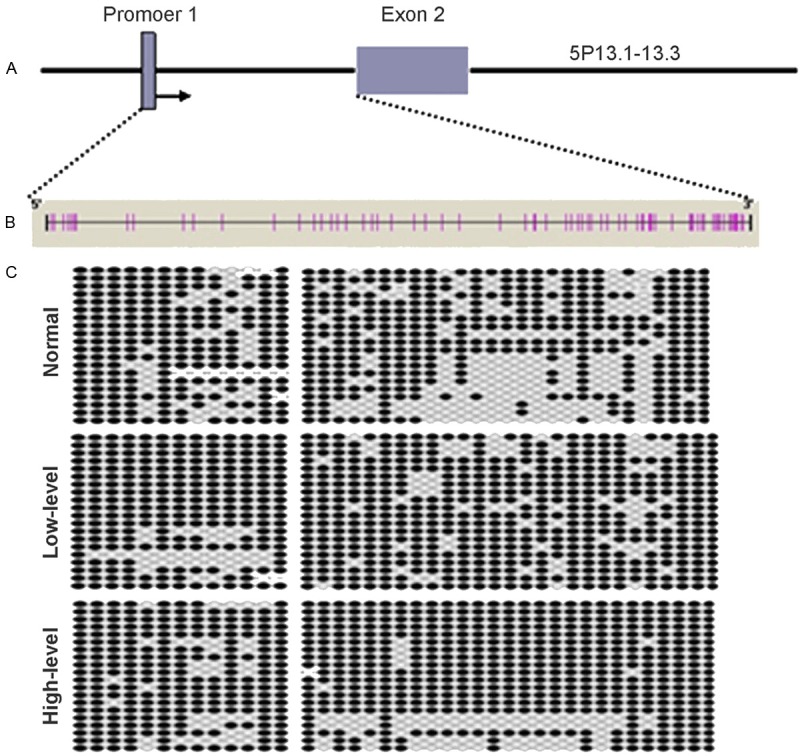
Sequencing of promoter 1 region of GDNF gene after bisulfite treatment. The promoter I region locates at about 1000 bp preceding the exon 2 of GDNF gene (A). There were 40 CpG sites in the promoter I region (B). After bisulfite treatment, sequencing was done (black: methylation; white: non-methylation) (C).
Figure 3.
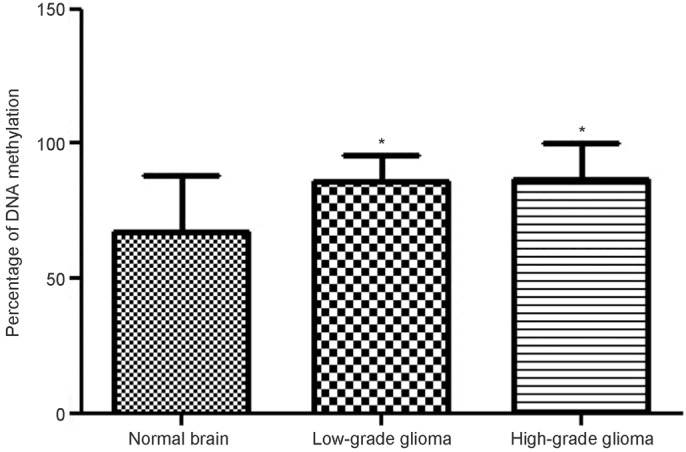
Methylation status of promoter 1 region of GDNF gene.
Discussion
GDNF belongs to the TGF-β superfamily and is a new neurotrophic factor first identified, purified and cloned from B49 glioma cell line in 1993 by Lin et al [6]. GDNF gene is mapped to 5P13.1-P13.3. It has neurotrophic effect on multiple types of cells and has been regarded an important bioactive neurotrophic factor [7,8]. In our study, results showed the primary GDNF sequence was identical between glioma and normal brain tissues, and the methylation of promoter I region of GDNF region increased markedly. This indicates that GDNF gene is not mutated and the highly methylated promoter I region of GDNF gene is likely an important factor causing silencing of promoter I region in the glioma.
Epigenetic modifications include DNA methylation, histone modification, nucleosome positioning and non-coding RNA. Of these modifications, DNA methylation has been deeply studied in mammalians [9,10]. DNA methylation induced gene silencing, together with histone modification and modifications of nuclear chromatin-associated proteins, is able to regulate the expression of genes. In cancer cells, the epigenetic modifications are characterized by whole genomic methylation and high CpG methylation of the promoter [11]. In cancer cells, high DNA methylation may cause gene silencing, which has been confirmed [12,13]. Since the CpG methylation was first identified at the promoter of Rb tumor suppressor gene, methylation is identified in a lot of genes. Studies have shown that DNA methylation of p16, MLH1 and BRCA1 may cause these gene silencing [14,15]. In our study, high methylation was found at the promoter I region of GDNF gene, which might cause promoter I region silencing in the glioma. However, a lot of findings suggest that GDNF expression is higher than that in normal brain tissues [16], which seem to be inconsistent with the fact of silencing of promoter I region due to its methylation.
GDNF gene is one with two promoters including promoter 1 and promoter 2. Suter-Crazzolara et al. [17] found that promoter 1 region locating at the up-stream of exon 2 had sites with high transcriptional activities. However, Baecker et al. [18] proposed promoter 2 played a major role in the transcription initiation. In different tissues and at different stages of development and diseases, the role of each promoter is distinct. In our study, results showed the promoter I region was highly methylated in GDNF. Some studies also reveal that the GDNF expression increases markedly in glioma [16]. Thus, we speculate that there might be low methylation of promoter 2 region of GDNF gene. In glioma, the methylation of promoter 1 region of GDNF causes the silencing of promoter 1 region, but that of promoter 2 region leads to the activation of promoter 2 region. In several studies on transcriptional factors of GDNF promoters, findings showed there were inhibitors targeting the promoters, which may cause low activity of these promoters, and the activity of theses transcriptional factors of promoters plays important roles in the transcriptional expression of GDNF gene.
In the present study, GDNF gene was sequenced, and results showed there were no mutations on GDNF gene. After bisulfite treatment, the promoter 1 region of GDNF gene was highly methylated, as shown by sequencing. Our findings provide evidence for further investigations on the epigenetic modifications in glioma.
Acknowledgements
The study was supported by Medical and Health Science Research Fund of Zhejiang Province and Nonprofit Applied Research Program of Zhejiang Province (2010C33015), and Medicine and Health Program of Zhejiang Province (2014RCA026).
Disclosure of conflict of interest
None.
References
- 1.Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109:93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- 2.Ping YF, Yao XH, Jiang JY, Zhao LT, Yu SC, Jiang T, Lin MC, Chen JH, Wang B, Zhang R, Cui YH, Qian C, Wang J, Bian XW. The chemokine CXCL12 and its receptor CXCR4 promote glioma stem cell-mediated VEGF production and tumour angiogenesis via PI3K/AKT signalling. J Pathol. 2011;224:344–354. doi: 10.1002/path.2908. [DOI] [PubMed] [Google Scholar]
- 3.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 4.Zhou FC. DNA methylation program during development. Front Biol (Beijing) 2012;7:485–494. doi: 10.1007/s11515-012-9246-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boumber Y, Issa JP. Epigenetics in cancer: what’s the future? Oncology (Williston Park) 2011;25:220–226. 228. [PubMed] [Google Scholar]
- 6.Lin LF, Doherty DH, Lile JD, Bektesh S, Collins F. GDNF: a glial cell line-derived neurotrophic factor for midbrain dopaminergic neurons. Science. 1993;260:1130–1132. doi: 10.1126/science.8493557. [DOI] [PubMed] [Google Scholar]
- 7.Jiang Y, Zeng SL, Lu YY, Zhu JB, Li T, Li FF, Xu CH. GDNF promotes to express Pitx3, Nurr1 and TH in transplanted midbrain-derived neural stem cells of Parkinson’s disease model of rats. Chin J Neuroanat. 2011;27:92–97. [Google Scholar]
- 8.Minnich JE, Mann SL, Stock M, Stolzenbach KA, Mortell BM, Soderstrom KE, Bohn MC, Kozlowski DA. Glial cell line-derived neurotrophic factor (GDNF) gene delivery protects cortical neurons from dying following a traumatic brain injury. Restor Neurol Neurosci. 2010;28:293–309. doi: 10.3233/RNN-2010-0528. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M. The necessity of a human epigenome project. Carcinogenesis. 2006;27:1121–1125. doi: 10.1093/carcin/bgl033. [DOI] [PubMed] [Google Scholar]
- 10.Veeck J, Esteller M. Breast cancer epigenetics: from DNA methylation to microRNAs. J Mammary Gland Biol Neoplasia. 2010;15:5–17. doi: 10.1007/s10911-010-9165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De S, Babu MM. Genomic neighbourhood and the regulation of gene expression. Curr Opin Cell Biol. 2010;22:326–333. doi: 10.1016/j.ceb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Ma X, Wang YW, Zhang MQ, Gazdar AF. DNA methylation data analysis and its application to cancer research. Epigenomics. 2013;5:301–316. doi: 10.2217/epi.13.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li KK, Li F, Li QS, Yang K, Jin B. DNA methylation as a target of epigenetic therapeutics in cancer. Anticancer Agents Med Chem. 2013;13:242–247. doi: 10.2174/1871520611313020009. [DOI] [PubMed] [Google Scholar]
- 14.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(Suppl 1):S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 16.Wiesenhofer B, Stockhammer G, Kostron H, Maier H, Hinterhuber H, Humpel C. Glial cell line-derived neurotrophic factor (GDNF) and its receptor (GFR-alpha 1) are strongly expressed in human gliomas. Acta Neuropathol. 2000;99:131–137. doi: 10.1007/pl00007416. [DOI] [PubMed] [Google Scholar]
- 17.Orth SR, Ritz E, Suter-Crazzolara C. Glial cell line-derived neurotrophic factor (GDNF) is expressed in the human kidney and is a growth factor for human mesangial cells. Nephrol Dial Transplant. 2000;15:589–595. doi: 10.1093/ndt/15.5.589. [DOI] [PubMed] [Google Scholar]
- 18.Morisseau C, Newman JW, Tsai HJ, Baecker PA, Hammock BD. Peptidyl-urea based inhibitors of soluble epoxide hydrolases. Bioorg Med Chem Lett. 2006;16:5439–5444. doi: 10.1016/j.bmcl.2006.07.073. [DOI] [PMC free article] [PubMed] [Google Scholar]


