Abstract
When treated with 17β-estradiol, female ACI rats (Rattus norvegicus) rapidly develop mammary cancers that share multiple phenotypes with luminal breast cancers. Seven distinct quantitative trait loci that harbor genetic determinants of susceptibility to 17β-estradiol−induced mammary cancer have been mapped in reciprocal intercrosses between susceptible ACI rats and resistant Brown Norway (BN) rats. A panel of unique congenic rat strains has now been generated and characterized to confirm the existence of these quantitative trait loci, designated Emca3 through Emca9, and to quantify their individual effects on susceptibility to 17β-estradiol−induced mammary cancer. Each congenic strain carries BN alleles spanning an individual Emca locus, introgressed onto the ACI genetic background. Data presented herein indicate that BN alleles at Emca3, Emca4, Emca5, Emca6, and Emca9 reduce susceptibility to 17β-estradiol−induced mammary cancer, whereas BN alleles at Emca7 increase susceptibility, thereby confirming the previous interval mapping data. All of these Emca loci are orthologous to regions of the human genome that have been demonstrated in genome-wide association studies to harbor genetic variants that influence breast cancer risk. Moreover, four of the Emca loci are orthologous to loci in humans that have been associated with mammographic breast density, a biomarker of breast cancer risk. This study further establishes the relevance of the ACI and derived congenic rat models of 17β-estradiol−induced mammary cancer for defining the genetic bases of breast cancer susceptibility and elucidating the mechanisms through which 17β-estradiol contributes to breast cancer development.
Keywords: ACI rat, Brown Norway rat, quantitative trait locus, estradiol, breast cancer
Breast cancer remains the second-leading cause of cancer-related mortality for women in the United States despite significant improvements in prevention, diagnosis, and treatment over the past two decades. The etiology of breast cancer is complex and incompletely understood. A family history of breast cancer and/or ovarian cancer is among the strongest known risk factors. Approximately 5–10% of breast cancers result from inheritance of rare, but highly to moderately penetrant, mutations in a small number of well-studied tumor suppressor genes, including BRCA1 and BRCA2 (Ponder et al. 2005). More recently, genome-wide association studies (GWAS) have localized within the human genome more than 70 common genetic variants that act as low-penetrance determinants of breast cancer risk, and together these variants are estimated to explain approximately 15% of heritable risk (Easton et al. 2007; Stacey et al. 2008; Thomas et al. 2009; Turnbull et al. 2010; Siddiq et al. 2012; Garcia-Closas et al. 2013; Ghoussaini et al. 2013; Michailidou et al. 2013). The identities as well as the sites and mechanisms of action of the causal genetic variants mapped in GWAS have not been defined.
Numerous laboratory, clinical, and population-based studies implicate endogenous and exogenous estrogens in breast cancer etiology. For example, use of selective estrogen receptor modulators, such as tamoxifen and raloxifene, has been shown to reduce dramatically the incidence of breast cancer in women who are at a high risk for developing the disease (Umar et al. 2012; Den Hollander et al. 2013). Similarly, aromatase inhibitors, which block estrogen production by inhibiting the aromatization of androgen precursors, also have been demonstrated to reduce the incidence of breast cancer in high-risk populations (Umar et al. 2012; Den Hollander et al. 2013). Conversely, the use of hormone-replacement regimens by postmenopausal women has been strongly associated with an increased risk of breast cancer (Narod 2011).
We are using the ACI rat model of 17β-estradiol (E2)-induced mammary cancer in genetic studies to map and identify genetic variants that determine breast cancer risk as well as to define more fully the mechanisms through which estrogens contribute to breast cancer development. Female ACI rats are uniquely susceptible to mammary cancer when treated with physiological levels of E2 (Shull et al. 1997; Spady et al. 1998; Shull et al. 2001). The mammary cancers that develop in E2-treated ACI rats express estrogen receptor-alpha and progesterone receptor, are dependent upon estrogens for survival and growth, and exhibit genome instability (Harvell et al. 2000; Adamovic et al. 2007; Ruhlen et al. 2009). Each of these tumor phenotypes is also a feature of luminal type breast cancers in humans. Interval mapping studies that use F2 progeny generated in intercrosses between susceptible ACI rats and resistant Copenhagen (COP) or Brown Norway (BN) rats revealed the locations of nine quantitative trait loci (QTL), designated Emca1 (Estrogen-induced mammary cancer) through Emca9, that influence susceptibility to E2-induced mammary cancer (Gould et al. 2004; Schaffer et al. 2006; Shull 2007; B. Schaffer and J. Shull, unpublished data). As data from GWAS continue to emerge, it is becoming increasingly clear that this rat model of estrogen-induced mammary cancer and humans share multiple genetic determinants of breast cancer risk. The objectives of this study were to develop and characterize congenic rat strains to confirm the existence of Emca3, Emca4, Emca5, Emca6, Emca7, and Emca9, to quantify the effect of each of these QTL on mammary cancer development and to evaluate the relevance of these QTL to risk loci identified in GWAS. The animal models characterized in this study will serve as valuable resources for fine mapping and identifying the causal genetic variants that reside within each QTL as well as for defining the sites and mechanisms of action of these variants on mammary cancer development.
Materials and Methods
Care, treatment, and phenotypic characterization of animals
Studies presented herein were performed at two different institutions. All procedures involving live animals were approved by the respective Institutional Animal Care and Use Committees. ACI/SegHsd and BN/SsNHsd were obtained from Harlan Sprague Dawley, Inc. (Indianapolis, IN). The congenic rat strains used in this study were generated in our laboratory as described herein. All rats were housed under controlled temperature, humidity, and 12-hr light/12-hr dark conditions in facilities that were accredited by the American Association for Accreditation of Laboratory Animal Care and operated in accordance with The Guide for the Care and Use of Laboratory Animals. All procedures related to the care, propagation, genotyping, treatment with E2 and evaluation for presence of mammary cancer have been previously described (Shull et al. 1997; Spady et al. 1998; Harvell et al. 2000; Shull et al. 2001; Harvell et al. 2002; Gould et al. 2004; Gould et al. 2005; Schaffer et al. 2006; Kurz et al. 2008; Ding et al. 2013; Schaffer et al. 2013). The animals were generally killed after196 ± 5 days of treatment or earlier if necessitated because of tumor burden. However, 13 ACI rats were treated for up to 282 days. Raw latency and tumor number data for each rat strain are compiled in Supporting Information, Table S1.
Generation and characterization of congenic rat strains
The congenic strains described herein were generated using a marker assisted selective breeding protocol as described previously (Schaffer et al. 2013). The markers used for positive and negative selection during backcrossing are listed in Table S2. Once a male rat was obtained that was heterozygous for BN alleles across a desired Emca locus and homozygous for ACI alleles at all background markers, that male was backcrossed to ACI females and sibling progeny carrying the same recombinant chromosome were intercrossed to produce rats that were homozygous for BN alleles across the specific Emca locus of interest (Table 1).
Table 1. Genetic characteristics of congenic strains.
| Strain Designation | Strain Abbreviation | Chr Length, Mb | Proximal ACI | Proximal BN | Distal BN | Distal ACI | RGD ID |
|---|---|---|---|---|---|---|---|
| ACI.BN-(D2Rat251-D2Mgh3)/Shul | ACI.BN-Emca3 or Emca3 | 258.2 | D2Mit29 5.60 Mb | D2Rat251 6.33 Mb | D2Mgh3 63.00 Mb | D2Rat22 69.30 Mb | 5687969 |
| ACI.BN-(D7Rat36-D7Rat11)/Shul | ACI.BN-Emca4 or Emca4 | 143 | Telomere | D7Rat36 1.53 Mb | D7Rat11 118.83 Mb | D7Rat9 125.40 Mb | 6482654 |
| ACI.BN-(D3Rat80-D3Rat3)/Shul | ACI.BN-Emca5 or Emca5 | 171 | D3Rat52 14.05 Mb | D3Rat80 32.05 Mb | D3Rat3 162.58 Mb | D3Rat59 163.44 Mb | 5688396 |
| ACI.BN-(D4Rat5-D4Got131)/Shul | ACI.BN-Emca6 or Emca6 | 187.1 | D4Mgh22 4.41 Mb | D4Rat5 9.63 Mb | D4Got131 162.28 Mb | D4Mgh11 171.20 Mb | 5688400 |
| ACI.BN-(D6Rat148-D6Rat109)/Shul | ACI.BN-Emca7 or Emca7 | 147.6 | D6Rat105 18.62 Mb | D6Rat148 20.82 Mb | D6Rat109 146.16 Mb | Telomere | 5688402 |
| ACI.BN-(D18Rat30-D18Rat89)/Shul | ACI.BN-Emca9 or Emca9 | 87.3 | Telomere | D18Rat133 3.55 Mb | D18Rat89 63.69 Mb | D18Rat5 77.45 Mb | 5687973 |
BN, Brown Norway.
Statistical analyses of data
MSTAT Version 5.4 was used to perform all statistical analyses (Drinkwater 2010). P values ≤ 0.05 were considered to be statistically significant. Latency was defined as the number of days separating initiation of E2 treatment and the first detection of a palpable mammary tumor. Median latency was derived from Kaplan-Meier analyses. The log rank test was used to compare latencies between strains and to calculate the relative risk of each set of congenic rats in comparison to ACI rats. The Wilcoxon rank sum test was used to compare the numbers of mammary tumors observed at necropsy. The susceptibilities of the different congenic rat strains to E2-induced mammary cancer were compared with ACI and BN rats evaluated contemporaneously at the same institution.
Sources of genomic data
The locations of all genetic markers described herein were obtained from the Rat Genome Database and are based on Rat Genome Assembly, version 3.4 (Laulederkind et al. 2013; Nigam et al. 2013). Sequence variants between ACI and BN rats were obtained using the Variant Visualizer Tool available at Rat Genome Database. The lists of genes residing within each Emca locus were generated using the Biomart Tool at Ensembl and are based on Rat Genome Assembly version 3.4, Ensembl release 69, October 2012 (Kinsella et al., 2011; Flicek et al. 2014). All references to genome coordinates in humans are based on genome assembly GRCh37.p10.
Results and Discussion
Validation and characterization of Emca3
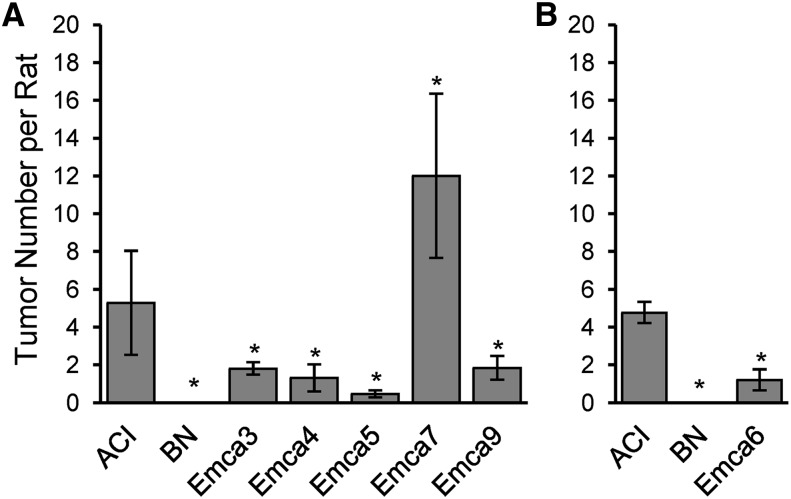
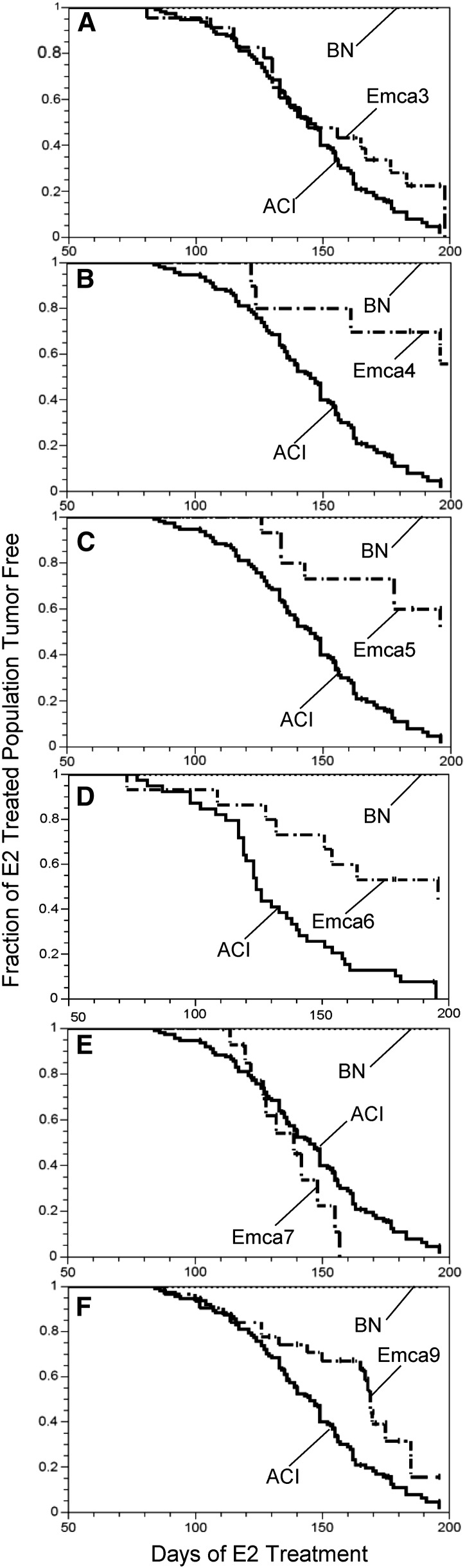
Emca3 was localized to RNO2 in an interval mapping study of F2 progeny generated in an intercross between ACI females and BN males (B. Schaffer and J. Shull, unpublished data). The peak LOD (logarithm of odds ratio) region for Emca3 was nearest D2Rat16 (43.4 Mb), although the LOD peak for this QTL did not achieve genome-wide significance (Table 3). To evaluate this putative QTL further, the ACI.BN-Emca3 congenic rat strain was generated by introgressing BN alleles from D2Rat251 (6.3Mb) to D2Mgh3 (63.0Mb) onto the ACI genetic background (Table 1). Based on the interval mapping data, it was hypothesized that ACI.BN-Emca3 congenic rats would exhibit reduced susceptibility to E2-induced mammary cancer when compared with ACI rats. ACI rats developed an average of 5.3 ± 4.5 (mean ± SD) mammary cancers when treated with E2 (Figure 1A and Table 2 illustrate data on group sizes). By contrast, treated ACI.BN-Emca3 rats developed only 1.8 ± 1.5 cancers per rat (P < 0.0001 compared with E2-treated ACI rats). Latency to appearance of palpable mammary cancer did not differ between E2-treated ACI and ACI.BN-Emca3 rats (Figure 2A and Table 2). Data from E2-treated BN rats, which are highly resistant to E2-induced mammary cancer and did not develop mammary cancers within the 196-day course of treatment, are illustrated for comparison. These data confirm the existence of Emca3 on RNO2.
Table 3. Orthologous relationships between Emca susceptibility loci in rat and breast cancer risk loci in humans.
| Locus | Peak LOD Marker | Proximal 95% Confidence Interval Marker | Distal 95% Confidence Interval Marker | Human Locus | SNP | Chr. Position of Human SNP | Gene | Orthologous Rat Position, Mb | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Emca3 | D2Rat16 | − | − | 5q11 | rs1353747 | 58337481 | PDE4D | 41.3 | Michailidou et al. 2013 |
| 43.4 Mb | 5q11 | rs10472076 | 58184061 | RAB3C | 41.4 | Michailidou et al. 2013 | |||
| 5q11 | rs889312 | 56031884 | MAP3K1, MIER3 | 43.2 | Easton et al. 2007; Turnbull et al. 2010 | ||||
| 5q11 | rs16886165 | 56023083 | MAP3K1 | 43.2 | Thomas et al. 2009 | ||||
| 5q11 | rs30099 | 52418582 | − | 46.9 | Easton et al. 2007 | ||||
| 5p12 | rs981782 | 45285718 | − | 49.9 | Easton et al. 2007 | ||||
| 5p12 | rs2067980 | 44982317 | MRPS30 | 50.3 | Thomas et al. 2009 | ||||
| 5p12 | rs7716600 | 44875005 | MRPS30 | 50.5 | Thomas et al. 2009 | ||||
| 5p12 | rs10941679 | 44706498 | MRPS30, HCN1 | 50.6 | Stacey et al. 2008; Thomas et al. 2009 | ||||
| Emca4 | D7Rat19 | D7Rat44 | D7Rat15 | 8q24 | rs13281615 | 128355618 | MYC | 98.4 | Easton et al. 2007 |
| 98.6 Mb | 66.2 Mb | 107.4 Mb | 8q24 | rs1562430 | 128387852 | MYC | 98.5 | Thomas et al. 2009; Turnbull et al. 2010 | |
| 8q24 | rs11780156 | 129194641 | MIR1208 | 99.3 | Michailidou et al. 2013 | ||||
| Emca5 | D3Rat114 | D3Rat227 | D3Rat210 | 2q31 | rs2016394 | 172972971 | METAP1D | 53.9 | Michailidou et al. 2013 |
| 149.5 Mb | 41.1 Mb | 156.6 Mb | 2q31 | rs1550623 | 174212894 | CDCA7 | 54.9 | Michailidou et al. 2013 | |
| 8p12 | rs9693444 | 29509616 | − | 63.2 | Michailidou et al. 2013 | ||||
| 20q11 | rs2284378 | 32588095 | RALY | 145.3 | Siddiq et al. 2012 | ||||
| Emca6 | D4Rat103 | D4Rat14 | D4Rat202 | 7q35 | rs720475 | 144074929 | ARHGEF5, NOBOX | 71.1 | Michailidou et al. 2013 |
| 82.7Mb | 41.7Mb | 159.1 Mb | 3p26 | rs6762644 | 4742276 | ITPR1, EGOT | 143.9 | Michailidou et al. 2013 | |
| Emca7 | D6Rat22 | D6Rat68 | D6Rat81 | 2p24 | rs4666451 | 19286943 | − | 33.3 | Easton et al. 2007 |
| 75.5 Mb | 2.8 Mb | 112.0 Mb | 2p24 | rs12710696 | 19320803 | − | 33.3 | Garcia-Closas et al. 2013 | |
| 14q13 | rs2236007 | 37132769 | PAX9, SLC25A21 | 77.1 | Michailidou et al. 2013 | ||||
| 14q24 | rs2588809 | 68660428 | RAD51L1 | 102.5 | Michailidou et al. 2013 | ||||
| 14q24 | rs999737 | 69034382 | RAD51B | 103.9 | Thomas et al. 2009 | ||||
| 14q32 | rs941764 | 91841069 | CCDC88C | 125.3 | Michailidou et al. 2013 | ||||
| Emca8 | D5Rat95 | D5Rat134 | D5Rat37 | 9q31 | rs10759243 | 110306115 | − | 73.5 | Michailidou et al. 2013 |
| 130.6 Mb | 52.43 Mb | 148.5 Mb | 9p21 | rs1011970 | 22062134 | CDKN2A, CDKN2B | 109.0 | Turnbull et al. 2010 | |
| 1p36 | rs616488 | 10566215 | PEX14 | 166.1 | Michailidou et al. 2013 | ||||
| Emca9 | D18Rat30 | − | − | 18q11 | rs527616 | 24337424 | − | 6.5 | Michailidou et al. 2013 |
| 5.7 Mb | 18q11 | rs1436904 | 24570667 | CHST9 | 6.8 | Michailidou et al. 2013 |
LOD, logarithm of odds ratio; SNP, single-nucleotide polymorphism.
Figure 1.
Effects of Emca loci on mammary cancer multiplicity. Beginning at 9 wk of age, female rats of each of the indicated rat strains were treated with E2 released continuously from subcutaneous Silastic implants. Female rats from each congenic strain were evaluated contemporaneously with batches of ACI and BN rats. Because the mammary cancer phenotypes exhibited by susceptible ACI and resistant BN rats at each of the two institutions were highly reproducible, the individual batches of ACI and BN rats evaluated at each institution were pooled for comparison with congenic rats evaluated at the same institution. (A) Animals evaluated at the University of Nebraska Medical Center. (B) Animals evaluated at the University of Wisconsin-Madison. Each data bar indicates the mean tumor number (± SEM) observed at necropsy for rats treated for at least 160 days and no more than 201 days; n = 10−83 animals per group. Asterisks indicate statistical significance (P < 0.05) compared with ACI females evaluated at the same institution.
Table 2. Mammary cancer phenotypes.
| Strain | Median Latency, da | Latency P* vs. ACIb | Hazard Ratiob | Mean Latency, dc | Incidence, %d | Mean Tumor Number per Rate | Tumor Number P* vs. ACIf | Ng |
|---|---|---|---|---|---|---|---|---|
| ACI (UNMC) | 147 | − | 1.000 | 140 ± 27 | 94 | 5.3 ± 4.5 | − | 126 (83) |
| BN (UNMC) | − | 2.66e-6 | 0.000 | − | 0 | 0 | 1.08e-6 | 10 (10) |
| ACI.BN-Emca3 | 144 | 0.2280 | 0.7358 | 141 ± 29 | 100 | 1.8 ± 1.5 | 0.0001 | 23 (20) |
| ACI.BN-Emca4 | NAh | 0.0017 | 0.2322 | 151 ± 35 | 43 | 1.3 ± 2.3 | 0.0004 | 11 (10) |
| ACI.BN-Emca5 | NAh | 0.0003 | 0.2704 | 156 ± 28 | 48 | 0.5 ± 0.7 | 6.17e-7 | 15 (13) |
| ACI.BN-Emca7 | 139 | 0.1153 | 1.640 | 135 ± 14 | 100 | 12 ± 5.9 | 0.0002 | 15 (11) |
| ACI.BN-Emca9 | 169 | 0.2251 | 0.7327 | 142 ± 30 | 84 | 1.8 ± 3.0 | 5.13e-6 | 32 (23) |
| ACI (UW) | 126 | − | 1.000 | 128 ± 25 | 100 | 4.8 ± 3.6 | − | 45 (42) |
| BN (UW) | − | 8.39e-6 | 0.000 | − | 0 | 0 | 1.19e-5 | 10 (9) |
| ACI.BN-Emca6 | 196 | 0.0010 | 0.3027 | 138 ± 37 | 56 | 1.2 ± 2.1 | 8.82e-5 | 15 (15) |
UNMC, University of Nebraska Medical Center; BN, Brown Norway; NA, not applicable; UW, University of Wisconsin.
Calculated from Kaplan-Meier analysis.
Calculated using log rank test.
Calculated for tumor positive rats (mean ± SD).
Calculated for population at risk.
Calculated for all rats that were treated with E2 ≥160 days but ≤200 days (mean ± SD).
Calculated using Wilcoxon rank sum test.
Total number of rats treated with E2 (total number of rats treated ≥160 days but ≤200 days.)
Not applicable. Median latency exceeds 196 days.
P ≤ 0.05 indicates statistical significance.
Figure 2.
Effects of Emca loci on mammary cancer latency and incidence. Female rats of each of the indicated rat strains were treated with E2 as described in the section Materials and Methods and Figure 1 and were evaluated by palpation once or twice weekly to detect the presence of mammary cancer. Each data panel illustrates the number of days separating initiation of E2 treatment and detection of palpable mammary cancer or grossly apparent mammary cancer at necropsy. (A) ACI.BN-Emca3, (B) ACI.BN-Emca4, (C) ACI.BN-Emca5, (D) ACI.BN-Emca6, (E) ACI.BN-Emca7, and (F) ACI.BN-Emca9. Congenic rat strains were compared with populations of E2-treated ACI and BN rats evaluated at the same institution. Sample sizes ranged from 11 to 126 rats per strain.
The Emca3 locus is orthologous to two distinct regions of human chromosome (Chr) 5, 5p12 and 5q11.2, and multiple single-nucleotide polymorphisms (SNPs) residing within these regions of Chr5 have been associated with breast cancer (Table 3). Positional candidate genes residing near the Chr5 breast cancer associated SNPs and/or suggested to mediate the actions of the causal genetic variants include MRPS30, HCN1, MAP3K1, MIER3, RAB3C, and PDE4D (Easton et al. 2007; Stacey et al. 2008; Thomas et al. 2009; Turnbull et al. 2010; Michailidou et al. 2013). Moreover, three studies suggest that mammographic breast density, a well-established biomarker of breast cancer risk, is influenced by genetic variants that reside within this same region of the human genome, near MRPS30, HCN1, and MAP3K1 (Table 4) (Woolcott et al. 2009; Odefrey et al. 2010; Fernandez-Navarro et al. 2013). Each of these genes that have been associated with breast cancer risk and/or mammographic density resides within the Emca3 peak LOD region on RNO2. Taken together, the data from the rat models and humans suggest that the causal variants responsible for the actions of Emca3 on mammary cancer susceptibility may reside on RNO2 within the maximal interval defined by the individual GWAS; i.e., Pde4d (40.1 Mb) to Mrps30 (50.6 Mb). This region contains 55 known protein coding genes, including the six noted above (Table S3). Evaluation of whole-genome sequences for the ACI and BN rat strains revealed nonsynonymous coding region variants within several genes residing within Emca3, including Map3k1 (2 nonsynonymous variants), Ndufs4, Mocs2, Hcn1, and Mrps30 (Table S4). The impact of these variants on the functions of their respective proteins and mammary cancer susceptibility is not known. Emca3 also harbors several genes that encode small RNAs, including miR449a, miR449c, and miR582 (Table S3). Mcs1, a complex QTL that determines susceptibility to mammary cancers induced by the polycyclic aromatic hydrocarbon dimethylbenz[a]anthracene in backcross progeny generated in crosses between susceptible Wistar-Furth rats and resistant COP rats, has been mapped to the same region of RNO2 as Emca3 (Hsu et al. 1994; Shepel et al. 1998; Haag et al. 2003; DenDekker et al. 2012). Together, these data suggest that genetic variants residing within the Emca3 and Mcs1 loci on RNO2 may influence development of mammary cancer in response to both estrogens and the genotoxic carcinogen dimethylbenz[a]anthracene.
Table 4. Orthologous relationship between Emca susceptibility loci in rat and breast density loci in humans.
| Locus | Peak Marker | Proximal 95% Confidence Interval Marker | Distal 95% Confidence Interval Marker | Human Locus | SNP | Chr. Position of Human SNP | Gene | Orthologous Rat Position, Mb | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Emca3 | D2Rat16 | − | − | 5q11 | rs889312 | 56031884 | MAP3K1, MIER3 | 43.2 | Odefrey et al. 2010 |
| 43.4 Mb | 5p12 | rs10941679 | 44706498 | HCN1, MRPS30 | 50.6 | Woolcott et al. 2009 | |||
| 5p12 | rs4415084 | 44662515 | MRPS30 | 69.5 | Fernandez-Navarro et al. 2013 | ||||
| Emca4 | D7Rat19 98.6 Mb | D7Rat44 66.2 Mb | D7Rat15 107.4 Mb | 8q24 | rs13281615 | 128355618 | MYC | 98.4 | Odefrey et al. 2010 |
| Emca6 | D4Rat103 82.7 Mb | D4Rat14 41.7Mb | D4Rat202 159.1 Mb | 7q34 | rs4728251 | 131858336 | RAB19 | 59.3 | Greenwood et al. 2011 |
| Emca7 | D6Rat22 75.5 Mb | D6Rat68 2.8 Mb | D6Rat81 112.0 Mb | 14q24.1 | rs10483813 | 69031284 | RAD51L1 | 102.9 | Vachon et al. 2012 |
SNP, single-nucleotide polymorphism.
Validation and characterization of Emca4
Emca4 was mapped to RNO7 in an interval mapping study of F2 progeny generated in a BN × ACI intercross (Schaffer et al. 2006). The LOD peak for this QTL was near D7Rat19 (98.6 Mb) and the 95% confidence interval extended from D7Rat44 (66.2Mb) to D7Rat15 (107.4Mb) (Table 3). The ACI.BN-Emca4 congenic strain was generated by introgressing BN alleles from D7Rat36 (1.53Mb) to D7Rat11 (118.83Mb) onto the ACI genetic background (Table 1). ACI.BN-Emca4 rats exhibited significantly reduced susceptibility to E2-induced mammary cancer relative to ACI rats, thereby confirming the published mapping data (Table 2). Tumor number at termination of the study was lower in ACI.BN-Emca4 rats than in ACI rats; 1.3 ± 2.3 tumors per rat vs. 5.3 ± 4.5, P = 0.0004 (Figure 1A and Table 2). Moreover, latency to appearance of palpable mammary cancer was prolonged in ACI.BN-Emca4 rats, relative to ACI rats, and only 43% of ACI.BN-Emca4 rats at risk developed mammary cancer within the 196 day course of E2 treatment, compared to 94% for E2 treated ACI rats (Figure 2B and Table 2).
The peak LOD region of Emca4 is orthologous to the Chr8q24 breast cancer risk locus in humans identified in multiple GWAS (Easton et al. 2007; Thomas et al. 2009; Turnbull et al. 2010; Michailidou et al. 2013). A genetic determinant of mammographic breast density has also been mapped to the 8q24 locus (Odefrey et al. 2010). The 8q24 breast cancer risk locus is defined by a gene desert that surrounds the MYC proto-oncogene. Myc similarly resides at the Emca4 LOD peak (Schaffer et al. 2006). It has been hypothesized that genetic variants in the 8q24 risk locus reside within cis-acting regulators of transcription, impact MYC expression in a cell type specific or temporal manner, and thereby modify breast cancer risk (Ahmadiyeh et al. 2010). Together, the data from the rat models and humans suggest an evolutionarily conserved determinant of breast cancer risk resides within Emca4 near Myc. Table S5 and Table S6 list the genes and variants, respectively, residing within the 10-Mb region of RNO7 centered on Myc. The 8q24 locus is pleiotropic with respect to its influence on disease susceptibility and has been associated with multiple cancer types in addition to breast cancer, including ovarian, prostate, colorectal, and bladder cancers (Braem et al. 2011; Ishak and Giri 2011; Theodoratou et al. 2012; Ma et al. 2013; Sur et al. 2013). Ept7, a QTL that impacts development of E2-induced pituitary tumors in (BNxACI)F2 rats, has been mapped to the same region of RNO7 as Emca4 (Shull et al. 2007; Kurz et al. 2014). Together, these data suggest the possibility that the causal variant(s) residing within the overlapping Emca4 and Ept7 loci may exert pleiotropic effects on tumor development in multiple estrogen responsive tissues. We are fine mapping Emca4 and Ept7 to identify the causal genetic variants that determine susceptibility to mammary cancers and pituitary tumors and to define the sites and mechanisms of action of these causal variants.
Validation and characterization of Emca5
Interval mapping analyses of (BNxACI)F2 progeny localized Emca5 to RNO3 (Schaffer et al. 2006). The LOD peak for this QTL was near D3Rat114 (149.5 Mb) and the 95% confidence interval extended from D3Rat227 (41.1 Mb) to near D3Rat210 (156.6 Mb) (Table 3). The ACI.BN-Emca5 congenic strain was generated by introgressing BN alleles from D3Rat80 (32.1 Mb) to D3Rat3 (162.6 Mb) onto the ACI genetic background (Table 1). ACI.BN-Emca5 rats exhibited dramatically reduced susceptibility to E2-induced mammary cancer, relative to ACI rats (Table 2). Tumor number at the termination of the experiment was significantly reduced (Figure 1A, Table 2), latency to appearance of palpable mammary cancer was significantly prolonged, and mammary cancer incidence was reduced in E2-treated ACI.BN-Emca5 rats compared with treated ACI rats (Figure 2C, Table 2).
The peak LOD region of Emca5 is orthologous to a segment of the long arm of Chr20q11 that has been associated with breast cancer risk (Siddiq et al. 2012). RALY has been identified as a candidate gene for this risk locus, and the rat ortholog of Raly resides at 145.3 Mb on RNO3, the peak LOD region for Emca5. The 20-Mb interval of RNO3 that extends from 135 to 155 Mb and flanks the peak LOD region of Emca5 harbors more than 250 annotated genes (Table S7). No functionally significant variants were identified in Raly upon comparison of available ACI and BN genome sequences, although nonsynonymous variants were identified in other genes residing within the Emca5 region (Table S8). It is also noteworthy that the proximal shoulder of Emca5 is orthologous to two additional regions of the human genome that have been linked to breast cancer risk in humans, Chr2q31 and Chr8p12 (Michailidou et al. 2013). Together, these data confirm the existence of Emca5 as a genetic determinant of susceptibility to E2-induced mammary cancer in the rat and suggest that Emca5 may harbor one or more genetic determinants of mammary cancer susceptibility that are shared with humans. Identification of Emca5 candidates for further evaluation will require this locus to be mapped more precisely using additional congenic rat strains.
Validation and characterization of Emca6
Emca6 was defined in (BNxACI)F2 rats as a QTL with a LOD peak located proximal to D4Rat103 (82.7 Mb) and a 95% confidence interval spanning from D4Rat14 (41.7 Mb) to D4Rat202 (159.1 Mb) (Table 3) (Schaffer et al. 2006). The ACI.BN-Emca6 congenic rat strain harbors BN alleles spanning from D4Rat5 (9.6 Mb) to D4Got131 (162.3 Mb) introgressed onto the ACI genetic background (Table 1). As predicted from the published interval mapping data, ACI.BN-Emca6 rats were less susceptible than ACI rats to E2-induced mammary cancer. Mammary tumor number was lower (Figure 1B and Table 2), latency to appearance of mammary cancer was prolonged and incidence of mammary cancer was reduced (Figure 2D and Table 2) in E2 treated ACI.BN-Emca6 rats when compared to treated ACI rats. These data confirm the existence of the Emca6 mammary cancer QTL.
The peak LOD region of Emca6 is orthologous to a human breast cancer risk locus at Chr7q35, tagged by SNP rs720475 and harboring ARHGEF5 and NOBOX (Michailidou et al. 2013). In the rat, these genes reside at 71.1 Mb on RNO4 (Table 3 and Table S9). Comparison of genome sequences for ACI and BN rats revealed multiple variants within Arhgef5 and Nobox; however, none of these occur in conserved positions suggesting they may not be of functional significance (Table S10). The peak LOD region of Emca6 is also orthologous to an interval on Chr7q34 containing RAB19 that has been associated with mammographic density in premenopausal women (Greenwood et al. 2011) (Table 4). Rab19 in rat resides at 66.9 Mb on RNO4. The 20 Mb interval centered on the Emca6 LOD peak contains 221 annotated protein coding genes and 21 genes that encode various classes of RNA (Table S9). Genes within this interval that harbor nonsynonymous variants include Trim24, which encodes an estrogen receptor-alpha interacting protein that has been suggested to contribute to breast cancer development (Tsai et al. 2010) and Hoxa7, which encodes a transcription factor that regulates expression of estrogen receptor-alpha (Zhang et al. 2013). In addition, a more distal segment of Emca6 is orthologous to a second human breast cancer risk locus at Chr3p26 that harbors ITPR1 and EGOT (Michailidou et al. 2013). Itpr1 resides at position 143.7 Mb on RNO4. Egot, a non-protein encoding gene, is not annotated in rat.
Validation and characterization of Emca7
Emca7 was mapped to RNO6 in a BN × ACI intercross (Schaffer et al. 2006). The LOD peak for this QTL resided proximal to D6Rat22 (75.5 Mb) and the 95% confidence interval extended from D6Rat68 (2.8 Mb) to D6Rat81 (112.0 Mb) (Table 3). Emca7 was unique among the Emca loci mapped in reciprocal intercrosses between susceptible ACI rats and resistant BN rats in that BN alleles at Emca7 were associated with increased susceptibility to E2-induced mammary cancer. ACI.BN-Emca7 congenic rats harbor BN alleles from D6Rat148 (20.8 Mb) to D6Rat109 (146.2 Mb) introgressed on the ACI genetic background (Table 1). When treated with E2, ACI.BN-Emca7 rats exhibited increased susceptibility to mammary cancer, relative to ACI rats. The number of mammary tumors observed at termination of the experiment was significantly higher in E2-treated ACI.BN-Emca7 rats compared with that observed in treated ACI rats (Figure 1A and Table 2). However, latency to appearance of palpable mammary cancer did not differ significantly upon comparison of ACI.BN-Emca7 and ACI rats (Figure 2E and Table 2).
Three distinct regions within Emca7 are orthologous to breast cancer risk loci identified in GWAS (Table 3). The region of RNO6 at 33.3 Mb is orthologous to a risk locus at Chr2p24 tagged by SNPs rs4666451 and rs12710696 (Easton et al. 2007; Garcia-Closas et al. 2013). The region of RNO6 near D6Rat22 (75.5 Mb), the marker nearest the Emca7 LOD peak, is orthologous to a breast cancer risk locus at Chr14q13, which harbors two suggested candidate genes PAX9 and SLC25A21 (77.1 Mb on RNO6) (Michailidou et al. 2013). Finally, the distal segment of the Emca7 confidence interval is orthologous to a risk locus at Chr14q24, which harbors two candidate genes RAD51B and RAD51L1 (102.1 Mb on RNO6) (Thomas et al. 2009; Michailidou et al. 2013). This region of Chr14q24 has also been associated with mammographic breast density (Vachon et al. 2012) (Table 4). The genes and variants that reside within 20 Mb of the LOD peak for Emca7 are listed in Table S11 and Table S12, respectively.
Validation and characterization of Emca9
Emca9 was identified in an intercross between ACI and BN rats as a suggestive QTL on RNO18 with a LOD peak at D18Rat30 (5.7 Mb) (Table 3) (B. Schaffer and J. Shull, unpublished data). To confirm the existence of this QTL, the ACI.BN-Emca9 congenic strain was generated by introgressing onto the ACI genetic background BN alleles from D18Rat133 (3.6 Mb) to D18Rat89 (63.7 Mb) (Table 1). Compared with ACI rats, ACI.BN-Emca9 congenic rats exhibited reduced susceptibility to E2-induced mammary cancer as evidenced by fewer mammary tumors and a lower mammary cancer incidence upon termination of the experiment (Figure 1A and Table 2). Latency to appearance of palpable mammary cancer did not differ significantly between E2-treated ACI and ACI.BN-Emca9 rats (Figure 2F and Table 2).
The peak LOD region of Emca9 is orthologous to Chr18q11, which harbors two distinct breast cancer risk loci, the latter of which contains the candidate gene CHST9 (Table 3) (Michailidou et al. 2013). The proximal 15 Mb of RNO18, which constitutes the peak LOD region for Emca9, harbors 71 annotated protein coding genes (Table S13). Comparison of whole-genome sequences for ACI and BN rats revealed three nonsynonymous variants in Chst9 (Table S14). These data confirm the existence of Emca9 as a genetic determinant of susceptibility to E2-induced mammary cancer and suggest that this QTL may be orthologous to a breast cancer risk locus in humans. Finally, it is noted that Emca9 overlaps with Emca2, a genetic determinant of susceptibility to E2-induced mammary cancer that was mapped to RNO18 in an intercross between susceptible ACI rats and resistant COP rats (Gould et al. 2004).
Sham-treated control rats do not develop mammary cancer
Small groups of ACI, BN, and ACI.BN-Emca congenic rats (n = 4−7 rats per strain) received empty Silastic implants and were evaluated as sham-treated contemporaneous controls. No mammary cancers were observed in sham-treated control ACI or BN rats or in control female rats from any of the ACI.BN-Emca congenic strains described herein. These data indicate that development of mammary cancers in the ACI and the different strains of ACI.BN-Emca congenic rats requires treatment with E2, and are consistent with previous studies that indicate that ACI and congenic rat strains established on the ACI genetic background are resistant to spontaneously arising mammary cancer (Maekawa and Odashima 1975; Shull et al. 1997; Shull et al. 2001; Gould et al. 2004; Schaffer et al. 2006; Kurz et al. 2008; Schaffer et al. 2013).
Six unique congenic rat strains were generated and evaluated in this study to define further the genetic bases of susceptibility to mammary cancer in a rat model in which tumor development is induced by treatment with E2, a naturally occurring steroid hormone that has been inextricably implicated in breast cancer development. The data presented herein provide the first published evidence of the existence of Emca3 on RNO2 and Emca9 on RNO18 and confirm the existence of Emca4 on RNO7, Emca5 on RNO3, Emca6 on RNO4, and Emca7 on RNO6. A similar congenic rat based approach was used recently to confirm the existence of Emca8 on RNO5 (Schaffer et al. 2013). Therefore, the existence of all seven of the Emca QTL mapped previously in intercrosses between susceptible ACI rats and resistant BN rats has now been independently verified.
Data presented herein also indicate that the peak LOD region for each of the 6 Emca loci characterized in this study is genetically orthologous to one or more genetic determinants of breast cancer risk mapped in GWAS. The orthologous relationship between the peak LOD region of Emca8 and the Chr9p21 breast cancer risk locus in humans was noted previously (Schaffer et al. 2013). Moreover, a recently published GWAS identified a breast cancer risk locus at Chr9q31 that is orthologous to a second local LOD peak within the complex Emca8 locus (Michailidou et al. 2013; Schaffer et al. 2013). Together, these data strongly suggest that the rat and human share multiple genetic determinants of mammary cancer susceptibility. By confirming the existence of seven distinct Emca loci and illustrating their genetic relatedness to breast cancer risk loci in humans, the present and recently published studies further establish the relevance of the ACI and ACI.BN-Emca congenic rat models of E2-induced mammary cancer to breast cancer in humans.
Interestingly, the peak LOD regions of Emca3, Emca4, Emca6 and the distal segment of Emca7 are orthologous to regions of the human genome that have been associated with mammographic breast density, a biomarker of breast cancer risk (Woolcott et al. 2009; Odefrey et al. 2010; Greenwood et al. 2011; Vachon et al. 2012; Fernandez-Navarro et al. 2013). Mammographic density is determined by the relative amounts of radiodense epithelium and stroma relative to the amount of radiolucent adipose tissue. Multiple cellular and molecular phenotypes have been identified that differ quantitatively and/or qualitatively between mammary glands of E2-treated ACI and BN rats, including epithelial density, which is greater in susceptible ACI rats, and luminal ectasia and associated collagenous stroma, which are more prominent in mammary glands of resistant BN rats (Ding et al. 2013). These data suggest that some radiodense features, e.g., epithelial density, may segregate with susceptibility conferring alleles, whereas others, e.g., luminal ectasia and associated collagenous stroma, may segregate with resistance conferring alleles. If true, then development of imaging technologies that can discriminate these specific phenotypes may enhance the value of breast imaging for breast cancer risk prediction.
The data emerging from studies focused on identification of low penetrance determinants of breast cancer risk strongly suggest that the causal genetic variants, once identified, will likely prove to be noncoding nucleotide or structural variants that influence expression of specific genes in a cell type specific and/or temporal manner. Moreover, the actions of the causal genetic variants on breast cancer risk may be extrinsic to the mammary epithelial cell population in which breast cancers arise. These considerations impose limits upon the use of breast cancer cell lines and human subjects for identifying the causal variants and establishing their functional roles in breast cancer etiology. The ACI.BN-Emca congenic rat strains described herein will serve as valuable models for identifying the causal genetic variants that influence susceptibility to E2-induced mammary cancer as well as for identifying the cell type(s) in which these variants exert their effects and defining the molecular mechanisms through which these actions are conferred. The knowledge to be generated by further characterization of these rat models should significantly enhance our understanding of the genetic bases underlying breast cancer risk as well as the mechanisms through which estrogens contribute to breast cancer development.
Supplementary Material
Acknowledgments
We thank Jason Huerta and Martin Tochacek for their contributions to this study. This work was supported by grant R01-CA77876 from the National Institutes of Health and grant KG081343 from the Susan G. Komen for the Cure Foundation. Shared research resources at the University of Nebraska Medical Center were supported by Cancer Center Support Grant P30-CA036727, whereas those at the University of Wisconsin-Madison were supported by Cancer Center Support Grant P30-CA014520. John Colletti was supported by National Institutes of Health training grant T32-CA009135. Beverly Schaffer was supported by grants DAMD17-03-1-0477 and DAMD17-00-1-0361 from the Department of Defense Breast Cancer Training Program.
Footnotes
Communicating editor: D. W. Threadgill
Literature Cited
- Adamovic T., Roshani L., Chen L., Schaffer B. S., Helou K., et al. , 2007. Nonrandom pattern of chromosome aberrations in 17beta-estradiol-induced rat mammary tumors: indications of distinct pathways for tumor development. Genes Chromosomes Cancer 46: 459–469 [DOI] [PubMed] [Google Scholar]
- Ahmadiyeh N., Pomerantz M. M., Grisanzio C., Herman P., Jia L., et al. , 2010. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc. Natl. Acad. Sci. USA 107: 9742–9746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braem M. G., Schouten L. J., Peeters P. H., van den Brandt P. A., Onland-Moret N. C., 2011. Genetic susceptibility to sporadic ovarian cancer: a systematic review. Biochim. Biophys. Acta 1816: 132–146 [DOI] [PubMed] [Google Scholar]
- denDekker A. D., Xu X., Vaughn M. D., Puckett A. H., Gardner L. L., et al. , 2012. Rat Mcs1b is concordant to the genome-wide association-identified breast cancer risk locus at human 5q11.2 and MIER3 is a candidate cancer susceptibility gene. Cancer Res. 72: 6002–6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander P., Savage M. I., Brown P. H., 2013. Targeted therapy for breast cancer prevention. Front Oncol 3: 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Zhao Y., Warren C. L., Sullivan R., Eliceiri K. W., et al. , 2013. Association of cellular and molecular responses in the rat mammary gland to 17beta-estradiol with susceptibility to mammary cancer. BMC Cancer 13: 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinkwater, N. R., 2010 MSTAT Version 5.4.1. Available at: http://mcardle.wisc.edu/mstat/version/ Accessed June 3, 2014.
- Easton D. F., Pooley K. A., Dunning A. M., Pharoah P. D., Thompson D., et al. , 2007. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447: 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Navarro P., Pita G., Santamarina C., Moreno M. P., Vidal C., et al. , 2013. Association analysis between breast cancer genetic variants and mammographic density in a large population-based study (Determinants of Density in Mammographies in Spain) identifies susceptibility loci in TOX3 gene. Eur. J. Cancer 49: 474–481 [DOI] [PubMed] [Google Scholar]
- Flicek P., Amode M. R., Burrell D., Beal K., Billis K., et al. , 2014. Ensembl 2014. Nucleic Acids Res. 42: D749–D755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Closas M., Couch F. J., Lindstrom S., Michailidou K., Schmidt M. K., et al. , 2013. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat. Genet. 45: 392–398, 398e391–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoussaini M., Pharoah P. D., Easton D. F., 2013. Inherited genetic susceptibility to breast cancer: the beginning of the end or the end of the beginning? Am. J. Pathol. 183: 1038–1051 [DOI] [PubMed] [Google Scholar]
- Gould K. A., Tochacek M., Schaffer B. S., Reindl T. M., Murrin C. R., et al. , 2004. Genetic determination of susceptibility to estrogen-induced mammary cancer in the ACI rat: mapping of Emca1 and Emca2 to chromosomes 5 and 18. Genetics 168: 2113–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould K. A., Pandey J., Lachel C. M., Murrin C. R., Flood L. A., et al. , 2005. Genetic mapping of Eutr1, a locus controlling E2-induced pyometritis in the Brown Norway rat, to RNO5. Mamm. Genome 16: 854–864 [DOI] [PubMed] [Google Scholar]
- Greenwood C. M., Paterson A. D., Linton L., Andrulis I. L., Apicella C., et al. , 2011. A genome-wide linkage study of mammographic density, a risk factor for breast cancer. Breast Cancer Res. 13: R132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag J. D., Shepel L. A., Kolman B. D., Monson D. M., Benton M. E., et al. , 2003. Congenic rats reveal three independent Copenhagen alleles within the Mcs1 quantitative trait locus that confer resistance to mammary cancer. Cancer Res. 63: 5808–5812 [PubMed] [Google Scholar]
- Harvell D. M., Strecker T. E., Tochacek M., Xie B., Pennington K. L., et al. , 2000. Rat strain-specific actions of 17beta-estradiol in the mammary gland: correlation between estrogen-induced lobuloalveolar hyperplasia and susceptibility to estrogen-induced mammary cancers. Proc. Natl. Acad. Sci. USA 97: 2779–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvell D. M., Strecker T. E., Xie B., Pennington K. L., McComb R. D., et al. , 2002. Dietary energy restriction inhibits estrogen-induced mammary, but not pituitary, tumorigenesis in the ACI rat. Carcinogenesis 23: 161–169 [DOI] [PubMed] [Google Scholar]
- Hsu L. C., Kennan W. S., Shepel L. A., Jacob H. J., Szpirer C., et al. , 1994. Genetic identification of Mcs-1, a rat mammary carcinoma suppressor gene. Cancer Res. 54: 2765–2770 [PubMed] [Google Scholar]
- Ishak M. B., Giri V. N., 2011. A systematic review of replication studies of prostate cancer susceptibility genetic variants in high-risk men originally identified from genome-wide association studies. Cancer Epidemiol. Biomarkers Prev. 20: 1599–1610 [DOI] [PubMed] [Google Scholar]
- Kinsella R. J., Kahari A., Haider S., Zamora J., Proctor G., et al. , 2011. Ensembl biomarts: a hub for data retrieval across taxonomic space. Database 2011: bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz S. G., Hansen K. K., McLaughlin M. T., Shivaswamy V., Schaffer B. S., et al. , 2008. Tissue-specific actions of the Ept1, Ept2, Ept6, and Ept9 genetic determinants of responsiveness to estrogens in the female rat. Endocrinology 149: 3850–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz S. G., Dennison K. L., Samanas N. B., Hickman M. P., Eckert Q. A., et al. , 2014. Ept7 influences estrogen action in the pituitary gland and body weight of rats. Mamm. Genome 25: 244–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laulederkind S. J., Hayman G. T., Wang S. J., Smith J. R., Lowry T. F., et al. , 2013. The Rat Genome Database 2013–data, tools and users. Brief. Bioinform. 14: 520–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Hu Q., Chen Z., Tao S., Macnamara L., et al. , 2013. Systematic evaluation of bladder cancer risk-associated single-nucleotide polymorphisms in a Chinese population. Mol. Carcinog. 52: 916–921 [DOI] [PubMed] [Google Scholar]
- Maekawa A., Odashima S., 1975. Spontaneous tumors in ACI/N rats. J. Natl. Cancer Inst. 55: 1437–1445 [DOI] [PubMed] [Google Scholar]
- Michailidou K., Hall P., Gonzalez-Neira A., Ghoussaini M., Dennis J., et al. , 2013. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 45: 353–361, 361e351–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narod S. A., 2011. Hormone replacement therapy and the risk of breast cancer. Nat. Rev. Clin. Oncol. 8: 669–676 [DOI] [PubMed] [Google Scholar]
- Nigam R., Laulederkind S. J., Hayman G. T., Smith J. R., Wang S. J., et al. , 2013. Rat Genome Database: a unique resource for rat, human, and mouse quantitative trait locus data. Physiol. Genomics 45: 809–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odefrey F., Stone J., Gurrin L. C., Byrnes G. B., Apicella C., et al. , 2010. Common genetic variants associated with breast cancer and mammographic density measures that predict disease. Cancer Res. 70: 1449–1458 [DOI] [PubMed] [Google Scholar]
- Ponder B. A., Antoniou A., Dunning A., Easton D. F., Pharoah P. D., 2005. Polygenic inherited predisposition to breast cancer. Cold Spring Harb. Symp. Quant. Biol. 70: 35–41 [DOI] [PubMed] [Google Scholar]
- Ruhlen R. L., Willbrand D. M., Besch-Williford C. L., Ma L., Shull J. D., et al. , 2009. Tamoxifen induces regression of estradiol-induced mammary cancer in the ACI.COP-Ept2 rat model. Breast Cancer Res. Treat. 117: 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer B. S., Lachel C. M., Pennington K. L., Murrin C. R., Strecker T. E., et al. , 2006. Genetic bases of estrogen-induced tumorigenesis in the rat: mapping of loci controlling susceptibility to mammary cancer in a Brown Norway x ACI intercross. Cancer Res. 66: 7793–7800 [DOI] [PubMed] [Google Scholar]
- Schaffer B. S., Leland-Wavrin K. M., Kurz S. G., Colletti J. A., Seiler N. L., et al. , 2013. Mapping of three genetic determinants of susceptibility to estrogen-induced mammary cancer within the Emca8 locus on rat chromosome 5. Cancer Prev. Res. (Phila.) 6: 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepel L. A., Lan H., Haag J. D., Brasic G. M., Gheen M. E., et al. , 1998. Genetic identification of multiple loci that control breast cancer susceptibility in the rat. Genetics 149: 289–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull J. D., 2007. The rat oncogenome: comparative genetics and genomics of rat models of mammary carcinogenesis. Breast Dis. 28: 69–86 [DOI] [PubMed] [Google Scholar]
- Shull J. D., Spady T. J., Snyder M. C., Johansson S. L., Pennington K. L., 1997. Ovary-intact, but not ovariectomized female ACI rats treated with 17beta-estradiol rapidly develop mammary carcinoma. Carcinogenesis 18: 1595–1601 [DOI] [PubMed] [Google Scholar]
- Shull J. D., Pennington K. L., Reindl T. M., Snyder M. C., Strecker T. E., et al. , 2001. Susceptibility to estrogen-induced mammary cancer segregates as an incompletely dominant phenotype in reciprocal crosses between the ACI and Copenhagen rat strains. Endocrinology 142: 5124–5130 [DOI] [PubMed] [Google Scholar]
- Shull J. D., Lachel C. M., Murrin C. R., Pennington K. L., Schaffer B. S., et al. , 2007. Genetic control of estrogen action in the rat: mapping of QTLs that impact pituitary lactotroph hyperplasia in a BN x ACI intercross. Mamm. Genome 18: 657–669 [DOI] [PubMed] [Google Scholar]
- Siddiq A., Couch F. J., Chen G. K., Lindstrom S., Eccles D., et al. , 2012. A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum. Mol. Genet. 21: 5373–5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spady T. J., Harvell D. M., Snyder M. C., Pennington K. L., McComb R. D., et al. , 1998. Estrogen-induced tumorigenesis in the Copenhagen rat: disparate susceptibilities to development of prolactin-producing pituitary tumors and mammary carcinomas. Cancer Lett. 124: 95–103 [DOI] [PubMed] [Google Scholar]
- Stacey S. N., Manolescu A., Sulem P., Thorlacius S., Gudjonsson S. A., et al. , 2008. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 40: 703–706 [DOI] [PubMed] [Google Scholar]
- Sur I., Tuupanen S., Whitington T., Aaltonen L. A., Taipale J., 2013. Lessons from functional analysis of genome-wide association studies. Cancer Res. 73: 4180–4184 [DOI] [PubMed] [Google Scholar]
- Theodoratou E., Montazeri Z., Hawken S., Allum G. C., Gong J., et al. , 2012. Systematic meta-analyses and field synopsis of genetic association studies in colorectal cancer. J. Natl. Cancer Inst. 104: 1433–1457 [DOI] [PubMed] [Google Scholar]
- Thomas G., Jacobs K. B., Kraft P., Yeager M., Wacholder S., et al. , 2009. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat. Genet. 41: 579–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W. W., Wang Z., Yiu T. T., Akdemir K. C., Xia W., et al. , 2010. TRIM24 links a non-canonical histone signature to breast cancer. Nature 468: 927–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbull C., Ahmed S., Morrison J., Pernet D., Renwick A., et al. , 2010. Genome-wide association study identifies five new breast cancer susceptibility loci. Nat. Genet. 42: 504–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar A., Dunn B. K., Greenwald P., 2012. Future directions in cancer prevention. Nat. Rev. Cancer 12: 835–848 [DOI] [PubMed] [Google Scholar]
- Vachon C. M., Scott C. G., Fasching P. A., Hall P., Tamimi R. M., et al. , 2012. Common breast cancer susceptibility variants in LSP1 and RAD51L1 are associated with mammographic density measures that predict breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 21: 1156–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolcott C. G., Maskarinec G., Haiman C. A., Verheus M., Pagano I. S., et al. , 2009. Association between breast cancer susceptibility loci and mammographic density: the Multiethnic Cohort. Breast Cancer Res. 11: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Cheng J. C., Huang H. F., Leung P. C., 2013. Homeobox A7 stimulates breast cancer cell proliferation by up-regulating estrogen receptor-alpha. Biochem. Biophys. Res. Commun. 440: 652–657 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.