Abstract
The sperm or eggs of sexual organisms go through a series of cell divisions from the fertilized egg; mutations can occur at each division. Mutations in the lineage of cells leading to the sperm or eggs are of particular importance because many such mutations may be shared by somatic tissues and also may be inherited, thus having a lasting consequence. For decades, little has been known about the pattern of the mutation rates along the germline development. Recently it was shown from a small portion of data that resulted from a large-scale mutation screening experiment that the rates of recessive lethal or nearly lethal mutations differ dramatically during the germline development of Drosophila melanogaster males. In this paper the full data set from the experiment and its analysis are reported by taking advantage of a recent methodologic advance. By analyzing the mutation patterns with different levels of recessive lethality, earlier published conclusions based on partial data are found to remain valid. Furthermore, it is found that for most nearly lethal mutations, the mutation rate at the first cell division is even greater than previous thought compared with those at other divisions. There is also some evidence that the mutation rate at the second division decreases rapidly but is still appreciably greater than those for the rest of the cleavage stage. The mutation rate at spermatogenesis is greater than late cleavage and stem-cell stages, but there is no evidence that rates are different among the five cell divisions of the spermatogenesis. We also found that a modestly biased sampling, leading to slightly more primordial germ cells after the eighth division than those reported in the literature, provides the best fit to the data. These findings provide conceptual and numerical basis for exploring the consequences of differential mutation rates during individual development.
Keywords: Drosophila melanogaster, cell coalescent, germline mutation rate, likelihood inference
Sperm from any organism results from a series of cell divisions during individual development, and each of these divisions may lead to mutations (Gilbert 2003). Because mutation is the ultimate cause of genetic variation, numerous studies have focused on understanding various aspects of mutation, including estimations of its rate. The overall mutation rate per generation is an essential quantity for various evolutionary/population genetic studies and is thus the focus of much research. However, a detailed understanding of mutation rates along individual development is also indispensable and an integral knowledge of biology and its importance has been widely recognized in medical genetics, particularly in the study of cancer/tumor development. A mutation that occurred early in the development will likely lead to more descendants (somatic or germ cells) than one that occurred later and thus will likely have more impact on the host as well as on its chance of survival in the population (Woodruff et al. 1996; Huai and Woodruff 1997; Woodruff and Thompson 2005). Differential mutation rates during development also provide new insights on male-driven evolution (Gao et al. 2011).
Until recently, there has been little progress on dissecting mutation rates during germline development at the level of individual cell divisions. Although next-generation sequencing may hold great promise for providing rich information for such purposes, well-developed classic experiments can still be a powerful and cost-effective approach, particularly for some model organisms. Gao et al. (2011) reported an analysis of the mutation patterns from a large-scale experiment for screening recessive lethal or nearly lethal mutations and found that mutation rates differ substantially in the germline lineage. In particular, the first cell division harbors the greatest mutation rate, followed by the divisions in spermatogenesis, whereas cell divisions in between have at least a magnitude smaller mutation rate. Gao et al. (2011) analyzed only those mutations of extreme recessive lethality, which are only a small fraction of the available data, due to a technical difficulty that also limited the analysis to families of exactly 20 offspring with at most two mutations per family. Fu (2013) extended the previous inference framework (Gao et al. 2011) with a new method for approximating the probability of a mutation pattern and a refined coalescent algorithm for simulating sample genealogies which are necessary for deriving coefficients used in the analysis. The refined inference framework not only removes the limitation of at most two mutations per families but also allows families of different sizes. This framework also established the confidence for conclusions derived from likelihood ratios based statistical tests for mutation screening data.
Taking advantage of the aforementioned methodologic progress, we report in this paper the analysis of the complete data set from the experiment, which consists of 9872 families of various sizes and which contains three times more mutations than previously reported. The greater resolution of the data as well as the contrast of results from analyzing mutations with different recessive lethalities allows us to obtain more accurate/stable estimates of mutation rates, to explore some aspects of the mutation process, and to test hypotheses that were unattainable previously. As a result, a deeper understanding of the mutation rate patterns along germline development is achieved. Furthermore, new hypotheses are presented and the male-driven hypothesis is re-evaluated in light of the new results from the current analysis.
Materials and Methods
Materials
Drosophila melanogaster stocks from Woodruff’s laboratory in Bowling Green State University were used. These flies were maintained by taking advantage of the balancer chromosomes that were pioneered by H. J. Muller (Muller 1928) for the purpose of maintaining newly isolated mutations, including recessive lethals, without selection (Muller and Oster 1963; Abrahamson and Lewis 1971; Ashburner 1989; Greenspan 1997). Balancers for each of the major chromosomes of these D. melanogaster contain multiple inversions and one or more dominant visible mutations. The inversions, which were mapped by using giant polytene chromosomes, act as crossover suppressors and the clearly visible dominant mutations allows for the identification of heterozygotes. With these chromosome stocks, new lethal or nearly lethal mutations are maintained in the heterozygous state against the balancer chromosomes without the concern of being lost due to recombination. The experiment employed three types of autosomal haploid chromosomes (genomes), which are denoted by β, γ, and z. More specifically they are as follows:
The β type balancer is homozygous lethal and is marked with the dominant visible, and recessive lethal mutations, Curly (Cy) wings, Lobe (L) eye, and Ultrabithorax (Ubx) enlarged halteres. This balancer segregates as a unit and suppresses crossing over on both the second and third chromosomes effectively (Lindsley and Zimm 1992; Thompson 1977). Similarly, the γ chromosome is also homozygous lethal and carries dominant visible markers. Type z represents a haploid genome with wild-type second and third chromosomes that are free of lethal mutations at the start of experiment. Recessive lethal or nearly lethal mutations in z are the screening target of the experiment.
Experiment
Graf et al.’s (1992) methods for culturing the flies were followed with some modifications. Drosophila medium containing water, glucose, agar, corn meal, and the antifungal agent methyl-p-hydroxybenzoate was cooked and dispensed into glass culture vials (10 cm in height and 3.5 cm in diameter) via a self-made dispenser. Also, self-made vial holders (designed to hold vials in a 10 × 10 array to match the dispenser) were used to facilitate the work. After drying and cooling the medium, a small piece of sterilized filter paper was folded and inserted into the medium with forceps to increase the surface and to regulate humidity within the vial, and then a small amount of live baker’s yeast was seeded into the vial. The culture vials, with cotton plugs added, were used to start the cultures. The flies were reared in a chamber, which allowed simultaneous culturing of more than 20,000 culture vials of flies under standard conditions (25°, approximately 60% relative humidity and 16-hr light:8-hr dark). The temperature in the culturing chamber was adjusted with air-conditioners and a self-regulating electric heating system, and humidity was manually controlled with a humidifier.
As briefly described in Gao et al. (2011), the mutation screening experiment, which is similar to protocols that have been used in various laboratories (Thompson and Woodruff 1980; Woodruff et al. 1984; Mason et al. 1985; Brodberg et al. 1987; Woodruff et al. 1996), consists of two parts. The first is to employ a three-generation assay to identify autosomal-recessive lethal or nearly lethal mutations in approximately 1200 genes on the second and third chromosomes in D. melanogaster. The second component of the experiment is known as the allelism test, which is to delineate the identity of mutations leading to different mutants.
The first component of the experiment was designed to screen β/z male offspring of crosses between single β/z males and multiple β/γ virgin females to see whether a new lethal or nearly lethal mutation occurred in the z chromosomes during germline development of the father. In essence, the mating scheme within each family derived from a single β/z male is as follows:
The intention here was to follow 20 offspring (lines) per family, but because some lines would not succeed for a variety of reasons, including death and failing quality control, up to 35 lines were initiated per family. As a result, the number of offspring in a family ranged from 2 to 35. Parental flies were removed from culture vials well before we examined the progeny and collected virgin females. The progeny adult flies were etherized using the Drosophila Fly Anesthetizer (Burco) and examined for phenotype and sex under a stereo microscope. Virgin β/γ or β/z females were collected within about 8 hr after we removed parental flies and then collection was repeated every 8 hr (usually at about 8:00 am, 4:00 pm, and midnight) during the eclosion.
Care was taken to ensure that β/γ females were virgin during the experiment. As a quality control measure, if β/γ offspring in the F3 were observed, the line was deemed disqualified because such an event can only result from nonvirgin β/z females. Also, to make sure the degree d of recessive lethality or simply lethality (Fu 2013), which is one minus the percent of z/z homozygote among all offspring, is adequately estimated, 40 offspring were set to be the minimum number of offspring examined in F3. A line is declared to be tentatively a mutant with lethality d if the percentage of its z/z homozygotes among all surviving offspring is not larger than d.
The β/z males used in the Parental stage were selected from families of F3 in which no mutation of detectable lethality was found (i.e., the percentage of z/z offspring is normal). In addition, the selection favored vigorous young males with distinct phenotypes. The process ensured that the chromosome z in the β/z male was devoid of recessive lethal mutation in general, but newly arisen recessive lethal mutation in F3 may escape such surveillance. The counting of F3 offspring was performed when there were a sufficient number of matured offspring. By the time a line was judged to be devoid of the targeted mutations, it would be typically 2−4 d after the minimum age for a matured adult.
The second component of the experiment is the allelism test, which is to delineate the identity of the mutation leading to each mutant. This was achieved by a series of crosses between mutant lines, with one line contributing β/z males and another virgin β/z females. There are usually many ways the crosses can be arranged, but each mutant line must be involved in at least one cross. The percentage of z/z individuals among offspring of a cross is expected to be similar to those of the parental lines if the mutations in the two different lines are the same and significantly higher when they are different (if mutations are indeed recessive). When there are multiple mutant lines in a family, a series of interconnected crosses between different mutant lines are necessary to resolve ambiguities.
Statistical method
Using the same notation as Fu (2013), the information conveyed by mutation(s) in a family can be represented by a mutation pattern
| (1) |
in which each element represents a mutation and its value is the number of offspring carrying the mutation, or simply the size of the mutation. After a line obtains a mutation of lethality ≥ d%, a further mutation will likely do nothing or increase the level of lethality, but the effect is usually difficult to distinguish from the first one under our experimental setting. This often led to masking or nondetection of the second mutation (Fu 2013). As a result, each identified mutant offspring is associated with one and only one mutation in the mutation pattern. Therefore, there is a natural constraint that i + j + k … is not larger than the family size. The aggregation of mutation patterns for families of size f can be concisely represented as
| (2) |
where n(κi) is the number of occurrences of pattern κi and for brevity can be omitted if its value is 1. The statistical analysis of the experimental results consists of determining the mutation pattern in each family, estimating mutation rates, and testing hypotheses.
Determination of the mutation pattern in a family:
The log-likelihood ratio test is used to test the null hypothesis that two mutant lines share the same causal mutation against the alternative that they result from independent mutations. Under the null hypothesis, the likelihood is the product of three binomial distributions all having the same frequency for z/z offspring, while under the alternative the binomial distribution for the cross has different frequencies for z/z offspring. Suppose zi and ni are the numbers of z/z and total offspring for parental line i(i = 1, 2) respectively, and z3 and n3 are the corresponding numbers for the cross. Then the test statistic is
| (3) |
where p123 = (z1 + z2 + z3)/(n1 + n2 + n3), p12 = (z1 + z2)/(n1 + n2) and p3 = z3/n3. The test statistic follows asymptotically a χ2 distribution with one degree of freedom under the null hypothesis that the cross results in the same z/z percentage as the two parental lines. Therefore, a significant test result indicates that the two lines result from different mutations. Often, multiple crosses are performed among lines within a family. If the overall significance level is α, and there are m crosses, then the level of significance for each test should be set to about α/m. In general, larger values of α will result in more significant tests and thus more independent mutations will be inferred, but the number of mutants for a given lethality does not change significantly, which are mostly determined by the percentage of z/z offspring in the F3. The results presented in this paper corresponds to α = 0.10.
Estimating mutation rate and hypothesis testing:
Suppose the development from the fertilized egg to the sperms are divided into I consecutive time intervals [ti − 1 + 1, ti] (i = 1,…I) (t0 = 0 and tI = maximum number of cell divisions), and let ui be the mutation rate per cell division within the ith interval. The estimate of μ = (u1, … uI) and statistical tests about them were performed through the maximum likelihood approach developed in Gao et al. (2011) and further refined in Fu (2013). The inference makes use of the information about population dynamics, intervals of cell divisions, and coalescent structure of the sample genealogy (Figure 1), but in essence, the inference framework is based on evaluating the likelihood function
| (4) |
where f represents family size, κ is a mutation pattern for a given family size f, nf(κ) is the number of occurrences of pattern κ in families of size f, and pf(κ) is the probability of mutation pattern κ, which is a function of μ and coefficients that are derived from the dynamics of the germline population. The probability can be computed through a combined approach of both analytic derivation and coalescent simulation of samples. The first product enumerates over all the different family sizes, while for each family size the second product enumerates over all observed patterns. The maximum likelihood estimate is the value of μthat maximizes the above likelihood function.
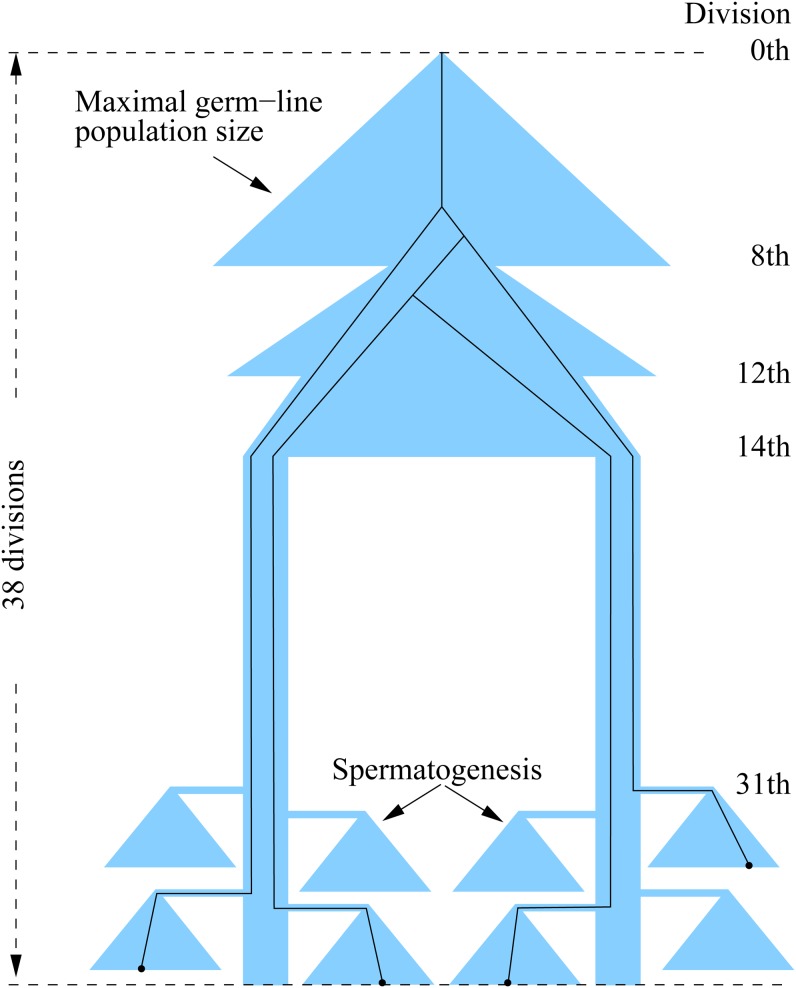
Figure 1.
Population dynamics and an example of the genealogy of four sperm sampled at the time at which maximal 38th cell division has occurred (adapted from Fu 2013).
The overall mutation rate μ is defined as the sum of mutation rates over all divisions in the development. Thus the maximum likelihood estimate of μ is
| (5) |
Alternatively, an unbiased estimate of μ can be obtained by a classical approach (Fu and Huai 2003), which is
| (6) |
This estimate has the advantage of being unbiased regardless of the assumptions made on the dynamics of the germline lineage development.
Results
Data and summary
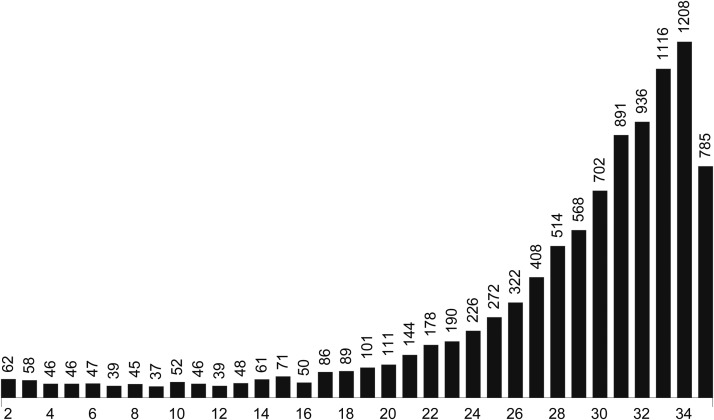
More than 10,000 families were screened in a 4-yr period (2004−2008), from which 9872 succeeded with at least one survival line at F3. To obtain positional information about observed mutation(s), we further require at least two completed lines for a family. To make sure β/γ females in F2 are virgin, we further require that no β/γ offspring among the F3 was observed. Furthermore, when the recessive lethality of a line exceeds a given threshold and the identity of the mutation cannot be determined, the line was also removed. This occurs when such a line was not used in crosses with other mutant lines (if they exist). After the cleanup, 9594 families passed the quality control, which resulted in a total of 271,794 lines, which is 90% of the 300,737 examined lines. The mean number of lines per families is 28.3 and the mean number of offspring in the qualified lines in F3 is 117.9. The distribution of the family sizes is given in Figure 2.
Figure 2.
Distribution of family sizes in 9594 screened families.
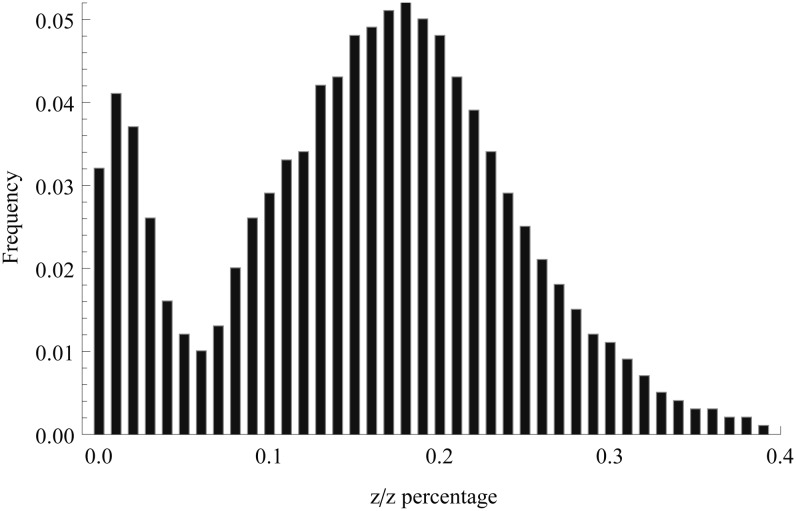
The histogram of the percentage of z/z offspring in the 271,794 lines is given in Figure 3. There are two obvious modes in the histogram, one at 1% and another at 18%. The first one corresponds to those with relatively high lethality mutations, and the latter corresponds to the normal z/z homozygotes. Note that because β/β is lethal, only β/z and z/z genotypes can potentially survive. If the genotypes are of the same fitness, their proportions will be 2/3 and 1/3, respectively. Figure 3 shows that z/z appears to be less fit than the β/z genotype, resulting in an average of approximately 17.5% only. One possible reason may be that the normal z chromosome used in the initial experiment contained some mildly deleterious recessive mutations. The reduced fitness of the z/z homozygotes does not affect significantly the identification of recessive mutations of high lethality but makes the dissection of those of lower lethality more challenging.
Figure 3.
Distribution of the percentage of z/z offspring among 271,794 lines in F3.
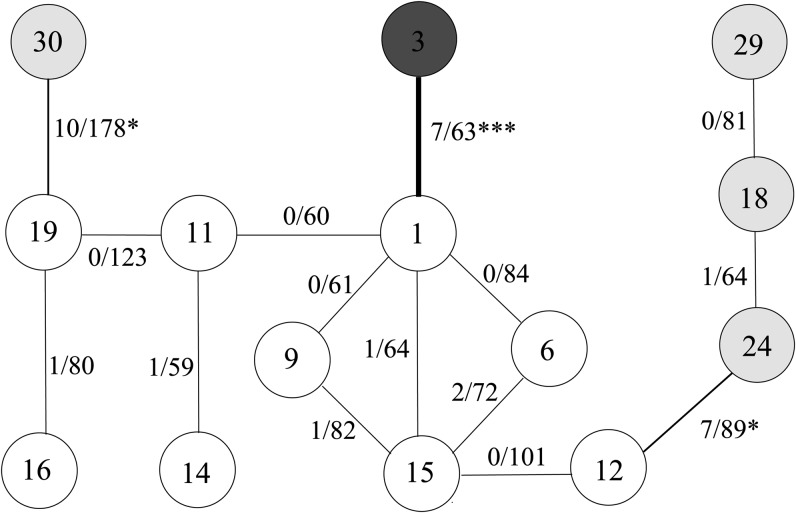
The identification of mutations starts with lines having a low percentage of z/z offspring in the F3 and proceeds with cross experiments and inference. One example is presented to illustrate the process. Table 1 shows an example of the F3 results of a family early in the experiment. The family starts with 30 lines but after initial quality control (one with β/γoffspring and two with too few total offspring and a few lines did not survive), a total of 17 lines are recorded.
Table 1. An example of F3 data for family 140.
| Line # | z/z | Total | z/z Percent |
|---|---|---|---|
| 1 | 0 | 110 | 0.000 |
| 2 | 3 | 90 | 0.033 |
| 3 | 0 | 56 | 0.000 |
| 5 | 29 | 112 | 0.259 |
| 6 | 2 | 72 | 0.028 |
| 9 | 3 | 127 | 0.024 |
| 11 | 0 | 120 | 0.000 |
| 12 | 1 | 64 | 0.016 |
| 14 | 1 | 101 | 0.010 |
| 15 | 0 | 70 | 0.000 |
| 16 | 2 | 63 | 0.032 |
| 18 | 3 | 71 | 0.042 |
| 19 | 1 | 54 | 0.019 |
| 24 | 2 | 103 | 0.019 |
| 28 | 0 | 62 | 0.000 |
| 29 | 1 | 125 | 0.008 |
| 30 | 1 | 118 | 0.008 |
It is clear that all lines, except for line 5, are suspects of new mutations of relatively high lethality. For a line to be declared as a mutant line for lethality level d, at minimum it should have a percentage of z/z offspring not larger than 1 − d and it is crossed with at least one other line if available. We were not sure how low the d can be so it was decided to perform as many crosses as feasible. As a result, all the lines except 5 and 2 are used for cross experiments. The exclusion of line 2 was not intentional but was due to the late completion of F3 for that line.
There are multiple ways to carry out allelism tests but the principle is that ambiguity needs to be resolved. Figure 4 shows the diagram of crosses used and their results. It is clear that there is no evidence that the six lines, represented by white circles, resulted from different mutations because the percentages of z/z offspring from crosses among them are similar to their parental lines. The crosses between lines 1 and 3, lines 19 and 30, and lines 12 and 24, resulted in significant test results, but after adjusting multiple tests as described in the Statistical Method section, only the cross between 1 and 3 is significant at the 10% level. Therefore, two mutations are identified, the first one includes line 3 with the z/z percentage being 0, and the second one includes the other 13 lines tested with an overall z/z percentage of 0.0171.
Figure 4.
Cross and results for family 140. a/b (e.g. 0/101) beside a line indicates the numbers of z/z and total offspring of the cross, respectively. Three crosses, between lines 19 and 30, between 12 and 24, and between lines 1 and 3 are individually significant (with * and *** representing, respectively, significance at 5 and 1% level). After adjusting for multiple tests, all three crosses are significant at the 50% significance level but only the cross between lines 1 and 3 remains significant at the 10% significance level.
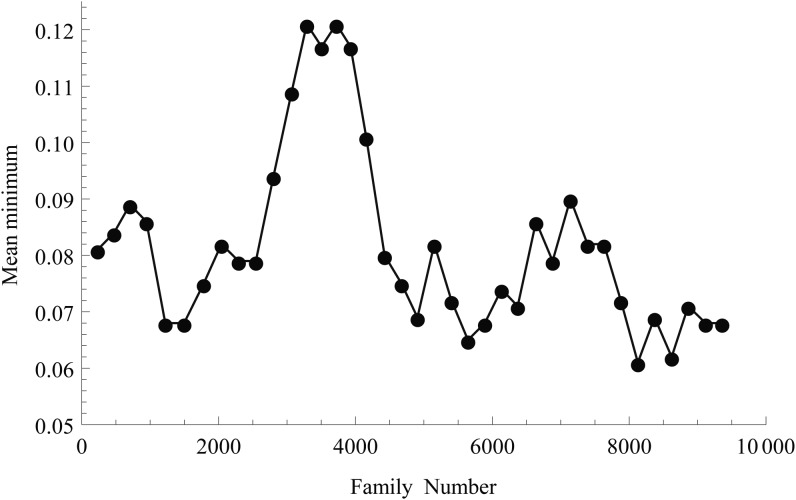
The number of identifiable mutations of relatively high lethality depends on the extensiveness of lines used in the allelism test. Although we were interested in determining as many mutations as feasible, it becomes too laborious when the number of lines to be crossed is large. Therefore, a compromise had to be made, which was achieved progressively. Figure 5 shows the mean minimum z/z percentage for lines in a family that are not subjected to the allelism crossing experiment. It can be seen that after the initial 3000 families, the minimum was raised from approximately 8% to approximately 12% for a short period of time. The practice was deemed impractical, and the minimum then dropped back to approximately 7%. The overall average is 8.2%.
Figure 5.
Mean minimum percentage of z/z offspring among nontested lines in windows of 200 families.
Despite the fact that lines with a recessive lethality of more than 92% were generally subjected to the allelism test, there is an increasing level of difficulties and ambiguity in determining the identity of mutations with decreasing lethality. As a result, we are cautious about those mutations identified with a recessive lethality lower than 95%. To be conservative, yet allowing for sufficient contrast, we will focus on those with lethality of 97% or greater. The mutation patterns identified through the allelism test depends on how stringent the criteria is, which is controlled by the value of α as described in the Materials and Methods section. Because α is the overall error rate, the larger it is, there will be more independent mutations declared. Table 2 provides the summary of mutations identified under different lethality intervals under two values of α. Both the number of mutations and the number of mutants are greatest in the lethality interval [97%, 98%), followed by the lethality interval [98%, 99%). Overall, the pattern appears to agree with that of the z/z percentages shown in Figure 3, where the second percentile is the most frequent mutant type and is followed by the third. As predicted previously, the number of mutations under α = 0.5 is larger in most cases, particularly for greater lethalities. However, the overall mutation rate does not change significantly in any of the cases. It turns out that subsequent analysis of mutations from different α values also leads to a relatively small difference. Therefore, we will focus on the presentation of analysis resulting from the mutations identified with α = 0.10.
Table 2. Distributions of 9594 families by mutation count under different lethality intervals.
| Mutations | [99–100%] | [98–99%) | [97–98%) |
|---|---|---|---|
| Allelism test with α = 0.1 | |||
| 0 | 8459 | 8391 | 8296 |
| 1 | 1052 | 1134 | 1240 |
| 2 | 81 | 66 | 56 |
| 3 | 2 | 3 | 2 |
| mt | 1220 | 1275 | 1358 |
| nm | 2673 | 10,766 | 17,342 |
| 0.0098 | 0.0396 | 0.0638 | |
| Allelism test with α = 0.5 | |||
| 0 | 8304 | 8273 | 8265 |
| 1 | 1174 | 1219 | 1232 |
| 2 | 112 | 95 | 93 |
| 3 | 4 | 7 | 4 |
| mt | 1410 | 1430 | 1430 |
| nm | 2897 | 11,328 | 16,574 |
| 0.0107 | 0.0417 | 0.0610 | |
mt, total number of mutations; nm, total number of mutants; , overall mutation rate estimated by Equation (6).
The data analyzed by Gao et al. (2011) is a subset of those with lethality [99%,100%] (or ≥99%). However, a 5% significance level without adjustment for multiple tests was used, so their data are not strictly a subset of column 1 of Table 2 from either α = 0.10 or α = 0.50. Because the number of crosses in a family increases with the number of likely mutants, the major effect of adjusting for multiple tests is that it makes the declaration of different mutations easier for families with a smaller number of likely mutants and harder for families with many likely mutants. Comparatively, because the mean number of crosses for families with a cross is approximately 17 (data not shown), this translates into about a 3% significance level on average per family, even α = 0.5 still appears slightly more stringent than the criteria used by Gao et al. (2011). It is reassuring that the subsequent inference is rather robust and as a result no qualitative conclusions made previously need to be revoked, which will be seen from the inference in a later section.
It is also useful to consider mutations for given minimum lethality levels. Table 3 provides the distribution under four minimum lethalities (notice that lethality ≥99% is the same as [99%, 100%]). It should be pointed out that mutation patterns for a given minimum lethality, say 97%, cannot be obtained as a simple aggregation of those in the lethality intervals [99–100%], [98–99%), and [97–98%). This occurs because sometimes mutations of lethalities falling into different intervals may occur in the same family, which can be seen from Table 3 in which there are families with four mutations, whereas there are none when considering lethality intervals separately.
Table 3. Distributions of 9594 families by mutation count under different minimum lethalities.
| Mutations | ≥99% | ≥98% | ≥97% |
|---|---|---|---|
| 0 | 8459 | 7397 | 6321 |
| 1 | 1052 | 1919 | 2743 |
| 2 | 81 | 258 | 481 |
| 3 | 2 | 20 | 48 |
| 4 | 0 | 0 | 1 |
| mt | 1220 | 2495 | 3853 |
| nm | 2673 | 13439 | 30,781 |
| 0.0098 | 0.0494 | 0.1133 |
mt, total number of mutations; nm, total number of mutants; , overall mutation rate estimated by Equation (6).
There are 111 families of size 20 (Figure 2) and under the lethality level [99%,100%] the collection of mutation patterns is
which indicates that 11 have one mutation of pattern <1>, one each for patterns of <2>, <3>, and <17>, respectively and two have a mutation of pattern <1, 1>. The number of mutant families is 11 + 1 + 1 + 1 + 2 = 16 and, thus, the number of nonmutant families is 111 − 16 = 95. Similarly, the mutation patterns for a family of size 20 under lethality level [98%, 99%) and [97%,98%) are, respectively,
and
The complete mutation patterns of the data corresponding to different lethality levels are given Table A1, Table A2, and Table A3 in the Appendix. For the sake of space, mutation patterns for given minimum lethalities will not be listed but can be obtained from the authors.
Inference about mutation rates
Although it is desirable to make an inference about the mutation rate for every cell division along germline development, the lack of sufficient resolution in the experimental data and the prohibiting computational burden limit our inference to six or fewer intervals. As described in the Statistical Method section, the intervals can be represented by a series of integers defining the boundary locations. For example, the sequence 1, 2, 14, and 31 means that there are five intervals: [1, 1], [2, 2], [3, 14], [15, 31], and [32, n] where n is the last cell division (e.g., 36). However, because different sperm may experience different numbers of divisions (but at least 36), while regardless of the number, the last five are spermatogenesis, it is more logical to put the last five divisions into the last interval and divisions from the 15th to just prior to spermatogenesis as the fourth. This interval definitions is conveniently represented by 1, 2, 14, −5.
The evaluation of the likelihood function makes use of coefficients computed from simulated samples from the germline population, which is dependent on the assumptions about the population dynamics. Let N(i) be the population size of germline lineage after the ith cell division. The assumptions about N(i) used in Gao et al. (2011) are provided in Table 1 in Fu (2013) and one assumption that has significant impact on the analysis is about N(8). Previous knowledge (Drost and Lee 1995, 1998) suggests that after the eighth division, about four to six cells move to the posterior region and become the primordial germ cells (PGCs). For a relatively small number of PGCs, are the PGCs a random sample out of the 256 cells available after the eighth division, or are they more closely related being descendants of an ancestral cell a few divisions earlier, or something in between? We will refer to this factor as sampling bias leading to N(8). This issue can be investigated effectively by examining the value of N(5), because after three divisions a cell in N(5) will have eight descendant cells in N(8), making it possible that all the PGCs are derived from a single cell in N(5). On the other hand, if N(5) is set to 32, it is equivalent to assuming that N(8) is a random sample out of the 356 cells.
The maximum number D of cell divisions is one factor that impacts the likelihood analysis. Because 36 cell divisions is the minimum and young mature males were used in the experiment, D was set to 36 as in Gao et al. (2011), although large values also were examined to some extent. As pointed out in the Experiment section, the likely range of cell divisions for sperm leading to F1 offspring is between 36 to about 40. Hence, we conducted likelihood analysis with D from 36 to 42, and found that D = 38 yields the overall best fit, which is significantly better than the case with D = 36, but goodness of fit declines gradually after D > 40. Even so, the pattern of likelihood estimates differ only marginally for D in the range between 36 and 42 and the impact on the interpretation of inference is relatively low. Because we are reasonably confident that the majority of sperm used in the experiments are from young males, we shall report results throughout the paper with D = 38.
Assumptions on the dynamics of the germline population
Although we are primarily interested in the distribution of mutation rates during germline development, it is important to evaluate the impact of the assumptions about the dynamics of the population on the inference. One such assumption concerns how the four to six cells are selected after the eighth cell division. The primary analysis in Gao et al. (2011) assumed that these cells are randomly drawn from 256 cells which resulted from the eighth cell division. Although this does seem consistent with the Drosophila embryonic development literature (Sonnenblick 1965), the impact of biased sampling were evaluated to some extent. We carried out more extensive analysis using the full data set.
We first investigated the impact of the values of N(5) and N(8) on the value of the likelihood function. To make the computation manageable, we divided the range (1−32) of N(5) into groups each containing two consecutive integers, and similarly the range (1−256) of N(8) into groups each containing three consecutive integers. Because the dynamics of the germline population are properties of the germline and thus should be more or less independent of types of mutations, one uses as much data as possible for judging its goodness of fit. Therefore, we examine the effect of gradually adding more data into the analysis on the strength of conclusions. Table 4 shows the log-likelihood values for a number of combinations of N(5) and N(8) for data under three lethality levels.
Table 4. Differential decrease to the maximum likelihood with intervals [1, 1], [2, 2], [3, 14], [15 − 6], [−5, 38].
| Recessive Lethality | ||||
|---|---|---|---|---|
| N(5) | N(8) | ≥99% | ≥98% | ≥97% |
| 1−2 | 1−3 | 80.3 | 622.9 | 1519.1 |
| 4−6 | 31.7 | 261.6 | 636.8 | |
| 7−9 | 42.5 | 329.3 | 809.6 | |
| 10−12 | 53.4 | 403.2 | 996.1 | |
| 13−15 | 59.6 | 443.3 | 1093.4 | |
| 3-4 | 1−3 | 53.6 | 396.8 | 958.7 |
| 4−6 | 5.3 | 48.9 | 114.9 | |
| 7−9 | 2.2 | 26.0 | 55.9 | |
| 10−12 | 2.8 | 27.3 | 62.6 | |
| 13−15 | 3.8 | 32.7 | 75.3 | |
| 5-6 | 1−3 | 54.2 | 401.8 | 970.8 |
| 4−6 | 2.1 | 20.8 | 47.5 | |
| 7−9 | 0.0 | 0.0 | 0.0 | |
| 10−12 | 1.0 | 3.3 | 13.6 | |
| 13−15 | 2.0 | 8.8 | 30.6 | |
| 7-8 | 1−3 | 53.3 | 401.8 | 963.3 |
| 4−6 | 2.3 | 17.7 | 39.3 | |
| 7−9 | 1.9 | 7.1 | 25.3 | |
| 10−12 | 4.0 | 17.3 | 56.6 | |
| 13−15 | 5.7 | 27.1 | 84.7 | |
| 9-10 | 1−3 | 52.5 | 398.1 | 953.3 |
| 4−6 | 2.8 | 18.3 | 45.4 | |
| 7−9 | 3.7 | 16.6 | 53.4 | |
| 10−12 | 6.7 | 32.8 | 100.3 | |
| 13−15 | 9.2 | 46.7 | 128.9 | |
Table 4 shows that regardless of recessive lethality the maximum likelihood is achieved at the combination of N(5) = 5 ~ 6 and N(8) = 7 ~ 9. Because twice the difference is the value of the log-likelihood ratio test (with one degree of freedom when one of the N(5) and N(8) is fixed), it follows that for 98% and 97% recessive lethality, any other combination of N(5) and N(8) can be rejected at the 1% significance level. For 99% recessive lethality, the resolution is less, but all other combinations except N(5) = 5 ~ 6 and N(8) = 10 ~ 12 can be rejected at the 5% level (including the random sampling). These results imply that N(5) is about 5−6, which represents a sampling for the eighth population that is far more restrictive than random sampling, which corresponds to N(5) = 32. Note that Gao et al. (2011) categorically defined N(5) = 4 as mild sampling bias largely due to mistaking the value as the number of ancestral sequences after the fifth cell division. Given the amount of reduction from 32 to 5−6, its classification as modest bias (at least) appears to be in order. These results also suggest that N(8) = 4 − 6, based on various experimental observations, appears to be conservative. Hence, a slightly larger number for N(8) may be more the norm.
Estimates of mutation rates
In the previous section, we established that N(5) = 5 ~ 6 and N(8) = 7 ~ 9 is the best assumption for the germline population dynamics; therefore, we will use this assumption for subsequent analysis. Because we investigate the mutation rates for different lethality levels, it is desirable to know whether a slight deviation of the optimal parameters will lead to a significant difference in the subsequent mutation rate estimation and hypothesis testing. We performed full likelihood analysis for several lethalities around N(5) = 5 − 6 and N(8) = 7 − 9 and found that indeed the impact is rather marginal unless the deviation leads to a substantially smaller likelihood value. Table 5 lists, as examples, the maximum likelihood estimates of mutation rates for combinations of N(5) and N(8) that differs no more than one step from the optimal. As can be seen, overall the estimates are rather similar to those under the optimal, but there are some notable differences in u2 for cases two steps away from the optimal; but then the difference of likelihood to that of the optimal is substantial. Therefore, we are confident that conclusions based on the optimal N(5) and N(8) will be robust.
Table 5. Full maximum likelihood estimates of u × 103 for lethality ≥ 97% and N(5) = 5 ~ 6 and N(8) = 7 ~ 9 with intervals [1,1], [2,2],[3,3], [4 14], [15 -6], and [-5 38].
| N(5) | N(8) | u1 | u2 | u3 | u4 | u5 | u6 | −ln(L) |
|---|---|---|---|---|---|---|---|---|
| 3−4 | 4−6 | 68.666 | 5.685 | 0.000 | 0.001 | 0.189 | 1.955 | 15272.36 |
| 3−4 | 7−9 | 69.001 | 3.028 | 0.000 | 0.001 | 0.190 | 1.951 | 15216.65 |
| 3−4 | 10−12 | 68.801 | 2.388 | 0.000 | 0.001 | 0.193 | 1.951 | 15226.09 |
| 5−6 | 4−6 | 68.801 | 2.393 | 0.001 | 0.001 | 0.190 | 1.951 | 15211.19 |
| 5-6 | 7−9 | 67.082 | 1.635 | 0.000 | 0.002 | 0.187 | 1.951 | 15164.41 |
| 5−6 | 10−12 | 66.437 | 1.374 | 0.013 | 0.002 | 0.188 | 1.951 | 15175.13 |
| 7−8 | 4−6 | 67.866 | 1.876 | 0.000 | 0.001 | 0.191 | 1.951 | 15205.74 |
| 7−8 | 7−9 | 65.919 | 1.316 | 0.013 | 0.002 | 0.189 | 1.951 | 15185.64 |
| 7−8 | 10−12 | 65.216 | 1.167 | 0.000 | 0.002 | 0.190 | 1.951 | 15213.69 |
Table 6 gives the estimates of u for three recessive lethalities for 38 cell divisions and the optimal combination of N(5) and N(8). The most striking pattern is that the mutation rate for the first division varies considerably, but regardless of how the data are examined, it is markedly larger than those for subsequent divisions. Similar to the pattern observed in Gao et al. (2011), the spermatogenesis has a relatively higher mutation rate than the interval divisions. With data aggregation, there appears to be a trend of appreciable mutation rate for the second division, and a similar pattern is observed for the stem cell stage although the magnitude is less appreciable. The cleavage stage, excluding the first (and perhaps also the second), harbors the smallest mutation rate. The ratio of the first division mutation rate to the mean mutation rate of the internal divisions are all larger than 100, with the highest ratio of 620 for lethality [97%, 98%).
Table 6. Full maximum likelihood estimates of u × 103 for several lethalities.
| Lethality | u1 | u2 | u3 | u4 | u5 | u6 | Ratio | |
|---|---|---|---|---|---|---|---|---|
| [99%, 10%) | 4.054 | 0.000 | 0.000 | 0.001 | 0.028 | 0.850 | 0.0089 | 206 |
| [98%, 99%) | 24.460 | 0.337 | 0.006 | 0.010 | 0.067 | 0.551 | 0.0289 | 469 |
| [97%, 98%) | 39.577 | 0.605 | 0.002 | 0.002 | 0.077 | 0.398 | 0.0437 | 620 |
| ≥ 98% | 28.660 | 0.272 | 0.006 | 0.018 | 0.089 | 1.443 | 0.0380 | 439 |
| ≥ 97% | 67.371 | 1.258 | 0.021 | 0.031 | 0.177 | 1.954 | 0.0821 | 445 |
Ratio: the ratio of the mutation rate of the first cell division and the mean rate for the interval cell divisions, computed as u1/[(u2 + u3 + 11u4 + 18u5)/32].
Mutation rate hypotheses testing
As defined in Materials and Methods, μ = (u1, u2, … uI) with ui being the mutation rate per cell division in the ith interval. Then different constraints on the rates will affect the estimates of μ and the associated log-likelihood values are used as the basis for testing the hypotheses about the mutation rates. The following nine hypotheses will be considered:
All the hypotheses except H4b are labeled the same as those in Gao et al. (2011) and Fu (2013). Table 7 gives the values of these tests against the H8.
Table 7. The values of the log-likelihood ratio test for various hypotheses Hi against H8.
| i | ||||||||
|---|---|---|---|---|---|---|---|---|
| Lethality | 1 | 2 | 3 | 4 | 4b | 5 | 6 | 7 |
| [98%, 99%) | 2304.9 | 1.0 | 176.3 | 0.5 | 0.4 | 0.7 | 236.1 | 455.9 |
| [97%, 98%) | 4197.6 | 3.6 | 345.7 | 1.0 | 0.0 | 3.4 | 142.7 | 893.0 |
| ≥99% | 1004.3 | 3.1 | 31.9 | 0.0 | 0.0 | 2.5 | 654.5 | 52.2 |
| ≥98% | 2953.9 | 1.2 | 193.0 | 0.2 | 0.2 | 0.6 | 855.6 | 492.3 |
| ≥97% | 6613.4 | 1.6 | 489.7 | 0.9 | 1.1 | 1.3 | 934.3 | 1287.6 |
Asymptotic χ2 distribution for Hi against H8 has 4, 2, 1,1,1,1 and 1 degree of freedom, respectively.
As in Gao et al. (2011), the assumption of equal mutation rate during germline development is soundly rejected and the evidence is stronger with data aggregation. As mentioned previously, with data aggregation there appears to be a trend that the second division and the stem-cell stage also have appreciable mutations (Table 6), but the log-likelihood ratio tests in Table 7 does not provide significant support for the trend. In fact, there is no significant evidence to reject the hypothesis that all internal divisions, that is, from the second until right before gametogenesis, share the same mutation rate.
To investigate the sensitivity of our inference with regard to possible sporadic inclusions of families with pre-existing mutations, we can exclude a certain fraction of families with a high percentages of mutants among offspring. However, it is not easy to decide the proper fractions to use, so a conservative approach was taken by removing all families with a percentage of mutants exceeding a given threshold value. Table 8 shows the results of the likelihood ratio tests for two different thresholds. At a 90% threshold value, for example, families of size 20 with 18 or more mutants were excluded, and families of size 30 with 27 or more mutants were excluded. As expected, the number of families excluded increases with the width of the lethality interval for a given threshold value, and decreasing the threshold from 90% to 85% results in doubling the number of families being removed. For the latter threshold value, as high as 20% of mutant families were removed. Even with such extreme exclusions, all the cases with significant test results shown in Table 7 remain the same.
Table 8. The values of the log-likelihood ratio test for various hypotheses Hi against H8 excluding families with high percentage of mutants.
| i | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Lethality | No. Excluded Families | 1 | 2 | 3 | 4 | 4b | 5 | 6 | 7 |
| Mutants % ≥ 90 | |||||||||
| ≥99% | 22 | 1020.2 | 2.3 | 12.4 | 0.0 | 0.1 | 0.7 | 658.8 | 15.3 |
| ≥98% | 125 | 2155.2 | 0.7 | 94.0 | 1.0 | 0.0 | 1.9 | 843.5 | 228.3 |
| ≥97% | 324 | 4322.4 | 0.0 | 219.6 | 7.8 | 3.2 | 15.3 | 934.3 | 613.4 |
| Mutants% ≥ 85 | |||||||||
| ≥99% | 43 | 1007.5 | 0.6 | 6.2 | 0.0 | 0.0 | 0.6 | 660.7 | 5.3 |
| ≥98% | 272 | 1792.2 | 0.6 | 55.9 | 1.2 | 0.0 | 1.5 | 835.9 | 130.7 |
| ≥97% | 665 | 3283.6 | 4.5 | 125.7 | 7.9 | 0.0 | 8.2 | 921.8 | 339.7 |
Asymptotic χ2 distribution for Hi against H8 has 4, 2, 1, 1, 1, 1, and 1 degree of freedom, respectively.
So far we have grouped the five cell divisions in the spermatogenesis as one interval and thus assumed that mutation rates are constant within the interval. This hypothesis can be investigated by dividing the gametogenesis into two intervals. Table 9 lists the maximum likelihood estimates for different partitions of the five cell divisions in gametogenesis (to reduce the amount of computation, the third cell division and the remaining cleavage divisions are combined into one interval, since there is little evidence to suggest that their rates are different). It follows that the maximum difference in the log-likelihood value to that for the case of equal rate (i = 0) is 0.7. Therefore, there is little evidence to suggest different mutation rates in the process of gametogenesis, despite the trend that early cell divisions have a greater estimated mutation rate than the later divisions.
Table 9. Likelihood estimates of mutation rates with lethality ≥97% with gametogenesis split into two intervals with [1, 1], [2, 2], [3, 14], [15 − 6][−5, −(i + 1)], [−i, 38].
| i | u1 | u2 | u3 | u4 | u5 | u6 | −ln(L) |
|---|---|---|---|---|---|---|---|
| 0 | 67.082 | 1.457 | 0.016 | 0.183 | 1.936 | 1.936 | 15,164.7 |
| 1 | 67.082 | 1.425 | 0.023 | 0.165 | 2.282 | 0.825 | 15,164.0 |
| 2 | 67.082 | 1.379 | 0.026 | 0.167 | 2.279 | 1.548 | 15,164.1 |
| 3 | 67.082 | 1.289 | 0.034 | 0.162 | 2.429 | 1.713 | 15,164.3 |
| 4 | 67.082 | 1.240 | 0.039 | 0.159 | 2.814 | 1.797 | 15,164.1 |
i/(5 − i): first i divisions of gametogenesis and last 5 − i divisions as two intervals.
Discussion
The full data set from our large-scale mutation screening experiment provides rich information for exploration in much more detail than previously possible for various aspects of the mutation process during germline development. The inference, taking advantage of the improved framework, leads to the following conclusions: (1) mutation rates during germline development are not equal, (2) the first division harbors the greatest mutation rate, (3) gametogenesis has mutation rates greater than other divisions except the first (and perhaps the second as well), (4) after the first cell division, the rate drops rapidly, and after the second division the rate becomes flat throughout the cleavage stage, (5) no evidence of rate difference during the gametogenesis, and (6) the number of PGCs after the eighth division is likely greater than that reported in the literature and they are not derived at random from the 256 cells at that stage nor from one or two ancestral cells at the fifth division.
Because of reduced DNA repair efficiency during gametogenesis, greater mutation rate at gametogenesis is expected. Although it is generally known that zygotic control starts toward the end of the cleavage stage which for Drosophila is around the 10−14th divisions, current knowledge of Drosophila development does not provide an adequate explanation of why the first division (or with the second division) harbors a much elevated mutation rate compared with the rest of the cell divisions during the cleavage stage, which was pointed out earlier by Gao et al. (2011). However, it is now known that some mutations in the sperm may be repaired in the early cleavage after fertilization (Rathke et al. 2014), so a portion of observed mutations in the early cleavage may be the result of incomplete repairing of pre-existing mutations. If all early cleavage mutations are derived as such, one would expect that the mutation rate for the first cell division would be similar to or smaller than that of gametogenesis; therefore, the much elevated mutation rate for the first cell division remains to be illuminated biologically.
To safeguard our conclusions against artifacts in both the experiment and inference, we also carried out analysis with combinations of parameters deviated from the reported optimal set. Examining the impact of assumptions on the dynamics of population size is one such effort, another is to examine the impact of sporadic pre-existing mutations of high lethality that had managed to escape our surveillance. Such mutations would be identified by the experiment as ones that lead to 100% mutant offspring (if the experiment was perfect) or close to 100% mutant offspring if the z/z percentages in a few lines fluctuate upward to escape both the initial screening and subsequent allelism tests. Our analysis (Table 8) shows that the aggressive removal of families with a high percentage of mutants does not change the main conclusions. Therefore, the possibility of some sporadic pre-existing mutations escaping our surveillance cannot be the primary cause of the sharp contrast of mutation rates along the germline development. The experiment and subsequent dissection of mutations was not perfect, which led to our investigation of the impact of the overall significance level on assigning mutants into mutational groups and subsequent inference. Again, all the main conclusions remains intact. One can conclude that the consistency of conclusions under various adjustments come from overwhelming information (both quantity and quality) in the data.
The greater resolutions in data from 98% and 97% recessive lethality have resolved some ambiguity previously encountered but also reveals a striking pattern. Although the rate for gametogenesis increases from 1 to 1.5 and 1.9, the rate increase of the first cell division is much more profound, from 4 to 30 for 98% lethality and 60 for 97% lethality. Although some bias might have been introduced in the delineation of mutations from cross data, the impact of such factors is likely modest at best. This is because the same analysis was conducted for mutation patterns generated by using two different α values (0.5 and 0.95) and the conclusions remain mostly intact except for some relatively small changes in numerical values. Therefore, we are confident that the elevated mutation rate for the first cleavage with decreasing lethality is beyond reasonable doubt. However, there is an increasing discrepancy between the overall mutation rate estimated from likelihood analysis and that from the classical estimator when the rate is significantly higher than 1%. We are not certain about the cause of this discrepancy but again different criteria for delineating cross data are not the major cause. This is an issue that deserves further investigation and one possible reason may be due to increasing inaccuracy of approximating the larger probabilities of mutation patterns used in the likelihood analysis. If this is true, it implies that the mutation rate for the first cell division might have been underestimated. Nevertheless, the qualitative conclusions from the current analysis are unlikely to be altered significantly.
The pattern of the ratio of the mutation rate of the first cell division and the mean rate of the interval divisions also deserve to be examined carefully. The lower ratio for greater lethality level (≥99%) than those of lower lethality, say [97%,98%) for example, is apparent. One possible cause may be that a significant number of mutations, which are completely lethal or nearly lethal, have a dominant effect and the degree of dominance increases with the lethality in general. When the lethality is reduced, the second division starts to harbor a greater mutation rate, which also helps to lower the ratio. Another way to look at the phenomenon is to examine the average cluster size of mutations for different levels of lethality. Table 2 shows that on average (i.e., nm/mt), mutations of lethality ≥ 99 is 2.2 whereas the average for lethalities of level [98%, 99%) and [97%, 98%) are 8.4 and 12.8, respectively. In general, mutations leading to smaller clusters tend to occur later than those to larger clusters and thus the selective disadvantage of mutations of high lethality may be one reason which prevents them from reaching a high frequency in the cell population within a host.
It is clear that our experimental data have the resolution to test the validity of some assumptions about the dynamics of the population. The number of cell divisions for the sperm in the parental stage was initially thought to be 36, but after examining the experimental procedure carefully, we realized that by the time the β/z males were introduced into the parental stage, additional two to three stem cell divisions might have occurred because on average it takes 32 hr for a stem-cell cycle (Wallenfang et al. 2006). Furthermore, each male can only mate a limited number of times during 24 hr; thus, the offspring in the F1 may result from sperm of an even wider range of ages. Therefore, sperm in the Parental stage likely range from a minimal of 36 cell divisions to approximately 40 divisions. Indeed, the likelihood analysis suggests that 38 cell divisions provide the best overall fit. The population size after the eighth division was thought to be in the range of four to six as reported in the literature (Drost and Lee 1995, 1998), but the best fit suggests that may be a slight underestimate, and a more appropriate range should be 7−9. Although the aforementioned two quantities have previous knowledge to judge their validity, little can be found from the literature about how the PGCs after the eighth division are derived or selected among the 256 available cells. Constraints of physical space and the tendency that recently derived cells tend to cluster near each other suggests that the PGCs may not be a random sample from the 256 cells as assumed by default. This analysis provides strong evidence that they are derived from a relatively small number of ancestral cells at the fifth division, but the assumption that their common ancestor is a cell at the fifth generation can be soundly rejected.
Regardless of the mechanism leading to the increased mutation rate ratio with decreasing lethality between the first division and gametogenesis, the trend is clear, and one can envisage that for neutral or nearly neutral mutations, the ratio may be even greater. This prospect lends stronger support to the explanation in Gao et al. (2011) concerning a lower ratio of male vs. female mutation rate than expected due to the difference in the number of cell divisions between sperm and eggs. This study further suggests that the rate differential between the first one or two with the remaining cell divisions can differ for mutations of different nature (here different lethalities). It is conceivable that this may be true for different genes/regions in the genome as well. Human genetic disease cases (Vogel and Motulsky 1997) as well as DNA sequence data (Miyata et al. 1987; Shimmin et al. 1993; Li 1997; Crow 2006) has led to the conclusion known as male-driven evolution, i.e., males dominate females in generating inheritable mutations in evolution. For 30-yr-old human males and females, there are roughly 400 and 30 cell divisions leading to sperm and eggs, respectively (Drost and Lee 1995), so the expected ratio of male-to-female mutation rate is larger than 10; however, estimates from sequence data vary and in general are lower than this ratio. Several possible causes have been put forward for this apparent discrepancy, but if the first (or first and second) cell division has a mutation rate many-fold larger than the average mutation rates for the remaining cell division, the phenomenon becomes easily explainable. For example, suppose the ratio of the mutation rate of the first cell division and the mean of the internal divisions is 100, then the male to female ratio would be expected to be (100 + 399)/(100 + 29) ≈ 3.9 and if the ratio is 500, then the male to female mutation ratio would be about 1.7. Because different mutation types or regions may exhibit rather different ratios between the first division and the average, the ratio between male-to-female mutation rates can be expected to be quite variable.
Acknowledgments
We thank those who performed part of the experiment, including Ji-fen Li, Zhen Xie, Fang Chen, Jian-rui Zhou, Qun-li Wang, Lei-hua He, Qian-qian Zhao, Zong-jun Luo, Rui-lin Zhang, Ji Yao, Rui-hong Zhang, Xing-qin Yin, Xiao-qing Yang, Ji-qin Liu, Ji-xin Yang, Xing-yun Wang, Tian-fen Zhang, Yong-ping Meng, Qiu-qi Li, Yan-hong Du, Shu-li Xiong, Mei Zhang, Mei-lan Huo, Li-xian Lou, Xiao-yun Jiang, and Ya-ting Liu. This work was partly support by grants from the Chinese NSF (30570248 and 91231120 YF) and the Betty Wheless Trotter Endowment Fund from The University of Texas Health Science Center at Houston, TX.
Appendix
Mutation patterns for different recessive lethalities
Table A1. Mutation patterns for recessive lethality [99–100%).
| 2: <2> 3: <1>6 4: <1>4 < 3><1, 1> 5: <1>9 < 3> 6: <1>9 < 2> <1, 1>2 7: <1>4 < 6> 8: <1>9 < 3> 9: <1>6 < 2><3> 10: <1>12 < 2>4 < 4> 11: <1>8 < 3><2, 5> 12: <1>3 < 3><1, 6> 13: <1>5 < 2>2 < 3><1, 1> 14: <1>8 < 1, 3> 15: <1>7 < 1, 1> 16: <1>5 < 3> 17: <1>7 < 1, 1> 18: <1>9 19: <1>10 20: <1>11 < 2><3><17><1, 1>2 21: <1>19 < 2><1, 1> 22: <1>21 < 2>3 < 1, 1><1, 2> 23: <1>22 24: <1>24 < 4><1, 1>3 25: <1>21 < 1, 1>2 26: <1>26 < 2><3>2 < 8><1, 1> 27: <1>36 < 2>4 < 1, 1>3 < 1, 4, 19> 28: <1>38 < 2><3><1, 1><1, 2><2, 5> 29: <1>32 < 2><1, 1><1, 23> 30: <1>55 < 2>3 < 25><27><29><1, 1>2 < 1, 3><1, 4><2, 3> 31: <1>76 < 2>3 < 3><26>2 < 28><29><31><1, 1>2 < 1, 3> 32: <1>90 < 2>4 < 3><26><27><29>2 < 30>4 < 1, 1>7 < 1, 16><1, 24><1, 26><1, 27><1, 1, 1> 33: <1>134 < 2>4 < 3>2 < 21><25><26><27><30><32><33>2 < 1, 1>12 < 1, 2><1, 27><1, 28><3, 27><4, 25> 34: <1>131 < 2>4 < 3>5 < 5><22><25>2 < 29>2 < 31>2 < 33><1, 1>7 < 1, 2><1, 3>2 35: <1>86 < 2><3><4><28><30>2 < 31><32><33><1, 1>6 < 1, 2> |
Table A2. Mutation patterns for recessive lethality [98–99%).
| 2: <1>3 3: <1> 4: <1>4 < 2> 5: <1>4 6: <1>6 7: <1>5 < 2>2 8: <1>2 < 2> 9: <1>4 < 2> 10: <1>8 < 3><4> 11: <1>7 < 2>2 12: <1>5 < 2><7> 13: <1>5 < 3><4><7> 14: <1>6 < 2>2 < 6> 15: <1>8 < 2><3>2 < 5> 16: <1>6 < 2><3><1, 1> 17: <1>5 < 2>3 < 3><5><11><13><1, 1><1, 9> 18: <1>10 < 2>2 < 4> 19: <1>6 < 2>4 < 4><13> 20: <1>6 < 2>2 < 3>2 21: <1>11 < 2><4> 22: <1>10 < 2>3 < 3><10><11><1, 1> 23: <1>15 < 2><3><9><17><21> 24: <1>22 < 2>2 < 3><5><10><16><3, 17> 25: <1>16 < 2><6><22> 26: <1>21 < 2>6 < 9><18> 27: <1>18 < 4><9><12><20><21> 28: <1>23 < 2>3 < 3>2 < 6><7><18><21><1, 1>3 29: <1>42 < 2>8 < 14><16><20><21>2 < 23><25>4 < 1, 20><7, 10><11, 14><1, 1, 2> 30: <1>31 < 2>3 < 3><4><6>2 < 7><15><17>2 < 19><20><22><25>2 < 26>3 < 27>3 < 28><29><1, 1>4 < 1, 2><1, 27><2, 5><4, 20><1, 1, 5> 31: <1>39 < 2>6 < 3>3 < 4>2 < 5><9><10>2 < 19><22>2 < 23>6 < 24><25>4 < 26>4 < 27>3 < 28>5 < 29><30><1, 1>2 < 1, 25><6, 22><8, 9><2, 4, 15> 32: <1>64 < 2>8 < 3><4>2 < 5><6><7><10><12><13><19><22><23>2 < 24>2 < 25>6 < 26>9 < 27>4 < 28>3 < 29>8 < 30>3 < 31>4 < 32><1, 1>2 < 1, 2>2 < 1, 13><1, 26><1, 28><2, 17><4, 18> 33: <1>69 < 2>15 < 3>4 < 4><5>2 < 7><9><11><13>2 < 14><17><18><20><21><22>3 < 23><24>4 < 25>5 < 26>9 < 27>7 < 28>10 < 29>9 < 30>12 < 31>6 < 32>6 < 33>2 < 1, 1>7 < 1, 2><1, 3><1, 22><1, 27><1, 29><3, 23><3, 25><6, 19><6, 20><6, 23> 34: <1>93 < 2>5 < 3><4><6><8><10><16><18><20><22>3 < 23>3 < 24>3 < 25><26>3 < 27>5 < 28>12 < 29>9 < 30>12 < 31>10 < 32>7 < 33>3 < 34>4 < 1, 1>3 < 1, 3><1, 20><1, 22><1, 25>2 < 1, 31><2, 26><2, 29><7, 21> 35: <1>67 < 2>8 < 3>2 < 4><5>2 < 15>2 < 20><21><22>3 < 23>2 < 24>2 < 25>2 < 26>6 < 28>3 < 29>5 < 30>6 < 31>10 < 32>2 < 33>3 < 34>3 < 35><1, 1><1, 27><2, 2><3, 30> |
Table A3. Mutation patterns for recessive lethality [97–98%).
| 2: <1> 3: <1>2 4: <1>2 5: <1>2 6: <1>4 7: <1><4><6><1, 2> 8: <1>7 < 2>3 < 2, 2> 9: <2><3> 10: <2>2 < 3> 11: <1>4 < 2>2 < 3>2 < 6><9> 12: <1> 13: <1>3 < 2> 14: <1>2 < 2>2 < 3> 15: <1>4 < 3><7>2 < 12><13> 16: <1>4 < 4><5> 17: <1>5 < 2>3 < 3>2 < 13>2 < 17> 18: <1>4 < 2><15> 19: <1>6 < 2>2 < 3><4><11>2 < 2, 7> 20: <1>8 < 2><3>2 < 14><19><2, 10> 21: <1>6 < 3><8><12> 22: <1>13 < 2>5 < 4><6><8><14> 23: <1>10 < 2>4 < 4>2 < 11><12> 24: <1>12 < 2>4 < 3><12><16><1, 1> 25: <1>16 < 2><5>2 < 10><18> 26: <1>12 < 2>2 < 4>2 < 11><16><17><21><23><1, 1><1, 2> 27: <1>19 < 2>3 < 3><4><9><10><14><16><20><21><23>2 < 1, 6><2, 2> 28: <1>16 < 2>5 < 3>2 < 4><6><9><10><20><21>2 < 25><27><1, 2> 29: <1>22 < 2>5 < 3><7><20>2 < 21>2 < 22>3 < 23>3 < 25><27><28>2 < 1, 19><1, 24><2, 22> 30: <1>25 < 2>4 < 4>2 < 6><16><17><18>2 < 19><20><23><24>2 < 25><26>5 < 27>2 < 28>2 < 29>2 < 1, 1><1, 9> 31: <1>35 < 2>10 < 3>2 < 4><5>3 < 10><11><16><17>2 < 19>3 < 20>2 < 21>3 < 22>3 < 23>3 < 24>4 < 25>2 < 26>2 < 27>8 < 28>6 < 29>6 < 30>5 < 31><1, 1>3 < 1, 2><2, 23><2, 25><3, 17> 32: <1>61 < 2>10 < 3>6 < 4><5><6><9><12><15><17><18><19><20>2 < 21>4 < 22>3 < 23>3 < 24>4 < 25>7 < 26>11 < 27>4 < 28>7 < 29>12 < 30>6 < 31>7 < 32>3 < 1, 1><1, 27><1, 29><3, 26><7, 24> 33: <1>59 < 2>9 < 3>5 < 4>3 < 6><7><11>3 < 13><14>2 < 17><18><19>3 < 20>5 < 21>5 < 22>2 < 23>10 < 24>7 < 25>8 < 26>8 < 27>17 < 28>15 < 29>12 < 30>13 < 31>16 < 32>18 < 33>6 < 1, 1>2 < 1, 18><1, 25><1, 27>2 < 2, 20><3, 16>2 < 4, 25><6, 23> 34: <1>59 < 2>9 < 3>5 < 5><8><10><12><14><16><17><19>2 < 20>3 < 21>2 < 22><23>4 < 24>3 < 25>7 < 26>18 < 27>14 < 28>11 < 29>16 < 30>16 < 31>16 < 32>18 < 33>4 < 34>9 < 1, 1>2 < 1, 2>2 < 1, 24>2 < 2, 27><3, 27><3, 29><6, 21><9, 18><10, 15>2 < 11, 13><1, 1, 1> 35: <1>51 < 2>4 < 3>5 < 4>2 < 6><15><20><23><24>3 < 25>2 < 26>4 < 27>5 < 28>7 < 29>9 < 30>11 < 31>7 < 32>12 < 33>8 < 34>4 < 35>3 < 1, 2><1, 25><3, 28><9, 19><1, 1, 1> |
Footnotes
Communicating editor: K. S. McKim
Literature Cited
- Abrahamson S., Lewis E. B., 1971. The detection of mutations in Drosophila melanogaster, pp. 461–484 in Chemical Mutagens, edited by Hollaender A. Plenum Press, New York [Google Scholar]
- Ashburner M., 1989. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- Brodberg R. K., Mitchell M. J., Smith S. L., Woodruff R. C., 1987. Specific reduction of N-dimethylnitro-samine mutagenicity in Drosophila melanogaster by dimethyl sulfoxide. Environ. Mol. Mutagen. 10: 425–432 [DOI] [PubMed] [Google Scholar]
- Crow J. F., 2006. Age and sex effects on human mutation rates: an old problem with new complexities. J. Radiat. Res. 47 Suppl B: B75–82 [DOI] [PubMed] [Google Scholar]
- Drost J. B., Lee W. R., 1995. Biological basis of germline mutation: Comparisons of spontaneous germline mutation rates among Drosophila, mouse and human. Environ. Mol. Mutagen. 25(Suppl 26): 48–64 [DOI] [PubMed] [Google Scholar]
- Drost J. B., Lee W. R., 1998. The developmental basis for the germline mosaicism in mouse and Drosophila melanogaster. Genetica 102/103: 421–443 [PubMed] [Google Scholar]
- Fu Y. X., 2013. Statistical methods for analyzing Drosophila germline mutation rates. Genetics 194: 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. X., Huai H., 2003. Estimating mutation rate: how to count mutations? Genetics 164: 797–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J. J., Pan X. R., Hu J., Ma L., Wu J. M., et al. , 2011. Highly variable recessive lethal or nearly lethal mutation rates during germline development of male Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 108: 15914–15919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert S. F., 2003. Developmental Biology. Ed. 7 Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- Graf U., van Schaik N., Wurgler F. E., 1992. Drosophila Genetics: A Practical Course. Springer-Verlag, Berlin, Heidelberg [Google Scholar]
- Greenspan S. F., 1997. Fly Pushing: The Theory and Practice of Drosophila Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- Huai H., Woodruff R. C., 1997. Clusters of identical new mutations can account for the overdispersed molecular clock. Genetics 147: 339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. H., 1997. Molecular Evolution. Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- Lindsley D. L., Zimm G. G., 1992. The Genome of Drosophila melanogaster. Academic Press, Inc., New York [Google Scholar]
- Mason J. M., Valencia R., Woodruff R. C., Zimmering S., 1985. Genetic drift and seasonal variation in spontaneous mutation frequencies in Drosophila. Environ. Mutagen. 7: 663–676 [DOI] [PubMed] [Google Scholar]
- Miyata T., Hayashida H., Kuma K., Mitsuyasa K., Yasunaga T., 1987. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harb. Symp. Quant. Biol. 52: 863–867 [DOI] [PubMed] [Google Scholar]
- Muller H. J., 1928. The measurement of gene mutation rate in Drosophila, its high variability, and its dependence upon temperature. Genetics 13: 279–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., Oster I. I., 1963. Some mutational techniques in Drosophila, pp. 249–278 in Methodology in Basic Genetics, edited by Burdette W. J. Holden-Day, Inc., San Francisco, CA [Google Scholar]
- Rathke C., Baarends W. R., Awe S., Renkawitz-Pohl R., 2014. Chromatin dynamics during spermiogenesis. Biochim. Biophys. Acta 1839: 155–168 [DOI] [PubMed] [Google Scholar]
- Shimmin L., Chang B. H.-J., Li W. H., 1993. Male-driven evolution of DNA sequences. Nature 362: 745–747 [DOI] [PubMed] [Google Scholar]
- Sonnenblick B. P., 1965. The early embryology of Drosophila melanogaster, pp. 62–167 in Biology of Drosophila, edited by Demerec M. Hafner Publishing Company, New York [Google Scholar]
- Thompson J. N., Jr., Woodruff R. C., 1980. Increased mutation in crosses between geographically separated strains of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 77: 1059–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson V., 1977. Recombination and response to selection in Drosophila melanogaster. Genetics 85: 125–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel F., Motulsky A. G., 1997. Human Genetics: Problems and Approaches. Ed. 3 Springer-Verlag, New York [Google Scholar]
- Wallenfang M. R., Nyak R., DiNardo S., 2006. Dynamics of the male germline stem cell population during aging of Drosophila melanogaster. Aging Cell 5: 297–304 [DOI] [PubMed] [Google Scholar]
- Woodruff R. C., Thompson J. N. J., 2005. The fundamental theorem of neutral evolution: Rates of substitution and mutation should factor in premeiotic clusters. Genetica 125: 333–339 [DOI] [PubMed] [Google Scholar]
- Woodruff R. C., Thompson J. J. N., Seeger M. A., Spivey W. E., 1984. Variation in spontaneous mutation and repair in natural population lines of Drosophila melanogaster. Heredity 58: 223–234 [Google Scholar]
- Woodruff R. C., Huai H., Thompson J. N., Jr, 1996. Clusters of identical new mutation in the evolutionary landscape. Genetica 98: 149–160 [DOI] [PubMed] [Google Scholar]