Abstract
Transposable elements (TEs) are DNA sequences able to be mobilized in host genomes. They are currently recognized as the major mutation inducers because of their insertion in the target, their effect on neighboring regions, or their ectopic recombination. A large number of factors including chemical and physical factors as well as intraspecific crosses have traditionally been identified as inducers of transposition. Besides environmental factors, interspecific crosses have also been proposed as promoters of transposition of particular TEs in plants and different animals. Our previous published work includes a genome-wide survey with the set of genomic TEs and shows that interspecific hybridization between the species Drosophila buzzatii and Drosophila koepferae induces genomic instability by transposition bursts. A high percentage of this instability corresponds to TEs belonging to classes I and II. The detailed study of three TEs (Osvaldo, Helena, and Galileo), representative of the different TE families, shows an increase of transposition in hybrids compared with parental species, that varies depending on the element. This study suggests ample variation in TE regulation mechanisms and the question is why this variation occurs. Interspecific hybridization is a genomic stressor that disrupts the stability of TEs probably contributing to a relaxation of the mechanisms controlling TEs in the Drosophila genome. In this commentary paper we will discuss these results and the molecular mechanisms that could explain these increases of transposition rates observed in interspecific Drosophila hybrids.
Keywords: Drosophila, transposable elements, interspecific hybridization, transposition, genomic instability, epigenetic factors
D. buzzatii, D. koepferae, and their Hybrids
Drosophila buzzatii and Drosophila koepferae are two cactophilic sibling species, belonging to the repleta group,1 morphologically undistinguishable except by the male genitalia. They have an overlapping distribution in the arid zones of southern South America, although D. buzzatii has a wider geographical distribution compared with D. koepferae, which is mainly restricted to the oriental side of the Andes Mountains.2 Moreover, they present a partial ecological isolation because D. buzzatii breeds and feeds on cacti from genus Opuntia whereas D. koepferae prefers columnar cacti from Cereus and Trichocereus genus.2 Although recent studies have provided evidences of recent gene flow between both species,3 we know they present reproductive isolation. Hybrids have not been found in the wild yet, due probably to the absence of a molecular tool allowing their quick identification. In laboratory, interspecific crosses are possible as D. buzzatii males can mate D. koepferae females and female hybrid offspring can be backcrossed to D. buzzatii males.4 This potential of natural hybridization makes these species an excellent model for speciation studies by transposition, particularly because very early studies had already showed increases of transposition rates of Osvaldo, the best characterized retrotransposon in D. buzzati,5 in hybrids compared with parental species.6
TE Mobilization in Drosophila Interspecific Hybrids
The most numerous and best documented cases of TE mobilization after interspecific hybridization were reported in plants,7-9 where hybrids are easier to obtain and study than in animals in which the number of offspring is low making the crosses very difficult. However, now some examples begin to be available in animals: one of the best known cases is the kangaroo where increases of transposition and centromere expansion were observed in hybrids.10,11 The first direct observation of increases of transposition in hybrids between D. buzzatii and D. koepferae was reported by Labrador and Fontdevila12 with the retrotransposon Osvaldo. These results were confirmed by a second experiment6 that included a larger sample size and a robust quantitative test. The next published work13 in Drosophila referred to hybrids between D. melanogaster and D. simulans and showed a widespread de-repression of different TE families in F1 hybrids. Our recent published work,14 on D. buzzzatii and D. koepferae, analyzes by the first time the set of genomic elements by a genome-wide approach. The hybrids were followed by four successive generations (F1 hybrids and three backcrosses) and stocks used did not contain mutations that rescued sterility in F1, reproducing the natural conditions of both species. We observed that a total of 33 TEs belonging to 14 TE families of class I and II were mobilized in hybrid genomes compared with parental species, where only one TE seemed to be mobilized. Likely the number of mobilized elements is greater than observed, due to the low size of AFLP bands and the lack of a reference sequenced genome, some elements could have been unnoticed. However, the number of elements in this study constitutes a representative part of the parental species genome. The detailed study of Osvaldo, Helena, and Galileo, representative of LTR-retrotransposons, non-LTR-retrotransposons and transposons respectively, showed an increase of transposition rates in hybrids compared with parental species, reaching in some cases one order of magnitude greater.
It is noteworthy that in these experiments differences were observed in the number of TEs mobilized as well as in the transposition rates across de different generations and hybrid families (hybrid crosses replicas). These results point first to the different mechanisms of regulation between elements and, second, that the portion of genome introgressed in hybrids could play an important role in TE activation. It is important to note that the percentage of D. buzzatii genome introgressed in hybrids is increasing in each backcross with D. buzzatii parental males, and the region of genome is different between families. Differences in hybrid instability or gene misexpression between generations of hybrids are not rare and have already been reported in plants15 and mouse.16
Another important point is the trend to a higher increase of transposition rates in males than in females, even if the differences were only significant in one family. We ignore how the elements are awakened in hybrids or why that is especially relevant in males, as seen mainly for Osvaldo retrotransposon.14 We think that most transposition events occurring in hybrids happened in female germline because a higher number of insertions are segregating along hybrid generations and it is hybrid females which are repeatedly crossed with D. buzzatii males. Transpositions observed in hybrid males could correspond to events in somatic cells or/and in hybrid male germinal tissues that do not contribute to the next generation because hybrid males are sterile. Out of the hybrid context, cases of copia17,18 transpositions or expression of 412,19 micropia,20 and 173121 have been reported in Drosophila males suggesting regulation mechanisms different from females.
Why TE Activity Increases in Drosophila Hybrids?
The mechanism that induces transposition in hybrids is largely unknown and to date only an experimental study has addressed this issue in Drosophila.13 But, the similarities between events (sterility and transposition) occurring during interspecific hybridization and hybrid dysgenesis22 suggest a possible parallelism. During the Drosophila hybrid dysgenesis, by crosses between wild and laboratory strains, sterility and increases of transposition were observed in the F1 offspring as a consequence of the activation of male-derived TEs. This happened because maternally contributed Piwi-interacting RNAs (piRNA) were unable to repress TEs paternally derived. In the same way, during interspecific hybridization between Arabidopsis species, the parental copies of Athila TE, normally silenced in heterochromatin, were expressed in hybrids.23 The authors hypothesize that the activation of the element could be due to the insufficient production of small interference RNAs (siRNAs) in females or their low specificity because of the sequence divergence between the two Arabidopsis species used in crosses. It is proposed that hybrid genetic dysfunctions occurred not only by the sequence divergence of genes in the two species but also by a large variety of mechanisms related with integrity and maintenance of chromatin.24 It is well known that the epigenetic control can affect gene expression and TE mobilization, a phenomenon extensively studied in Drosophila especially in the case of genes of the piwi pathway responsible for defending germline from TE proliferation.25 Kelleher et al.13found both TE misregulation and aberrant piRNA production in interspecific hybrids between D. melanogaster and D. simulans, suggesting an adaptive divergence of piRNA pathway genes rather than differences in piRNAs derived from TEs in the species under study.
Increases of transposition were observed in our D. buzzatii/D. koepfere hybrids and raises de question about the involvement of TE mobilization in hybrid sterility and the molecular causes. Previous genetic work, with these same hybrids, proposes that male hybrid sterility results from the cumulative action of many interacting genes of minor effects.26 We ignore the direct impact of TE derepression in fecundity of D. buzzatii/D. koepferae hybrids but the divergence between alleles of piRNA pathway could contribute both to sterility and TE mobilization. In Drosophila, proteins encoded by these genes participate in piRNA biogenesis, transposon silencing,27 and germline stem cell (GSC) self-renewal in both males an females28 and are important in the maintaining of fertility.29,30 Mutations in Aubergine gene (a member of piRNA pathway) lead to accumulation of retrotransposons on ovaries and testes, and Stellate transcripts in testes that are associated to male sterility.29 In view of the importance of these mentioned genes I suggest that the hybrid sterility is not directly caused by transposition but that could be a consequence of adaptive divergence of piwi pathway genes. However, we cannot completely discard the possibility that TE mobilization in hybrids affect sterility because of their capability of neighboring genes misregulation.31
On the other hand to explain TE mobilization, it is known that piRNAs silencing transposons come from heterochomatic TE-rich regions named piRNA clusters.32,33 For many TEs the maternal deposition of TE specific piRNAs is critical for their propagation and, in the case of interspecific hybrid, if they do not inherited the piRNA clusters, or the elements inserted inside have diverged between the two species, the amount of piRNAs would not be sufficient to repress TEs. This point could be the cause of differences in the amount of TE mobilized through different hybridization generations and/or families observed in our work.
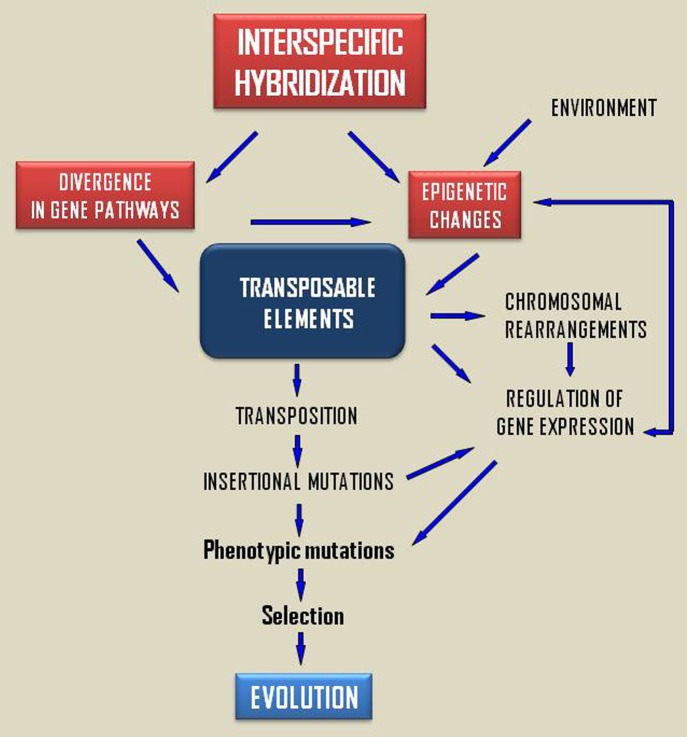
To summarize, the Figure 1 depicts putative scenarios leading to transposition in hybrids and their implications in speciation processes. The genomic “shock” by interspecific hybridization leads to an epigenetic reprogramming24 and a TE release associated to mutations and new gene regulatory ways. Some of these mutations could be fixed by selection in a novel environment contributing to speciation processes. There is increasing evidences suggesting that TEs had and have a promising role in evolutionary processes. Transposition bursts ensuing hybridization have been suggested as makers of rapid genome reorganizations and source of evolutionary innovations,34 notwithstanding more empirical work will be necessary to address these questions conclusively. The recent advances in the knowledge of TE silencing associated to interference RNAs (RNAi and piRNA) could open new ways to the understanding of TE activation in hybrids. Provided that the genomic context could affect epigenetic regulation, a way to disentangle the mechanisms implicated in TEs deregulation in hybrids could consist of comparing the small RNA content of hybrids and parental species. This study could be completed with the analysis of the expression of piRNA pathway genes in order to detected putative expression patterns altered in hybrids compared with parental species.
Figure 1. Ways of TE activation in hybrids and the putative role in speciation events.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
I want to thank O Cuba for reviewing the English and A Fontdevila, D Vela, and C Vieira as authors of the Plos One previous published work that thanks to their results, this Commentary has been possible. This work was supported by research grants CGL2009-12912-C03-01 and CGL2010-15395 from the Ministerio de Ciencia e Innovación (Spain), and grants 2009SGR 636 from Generalitat de Catalunya to the Grup de Biologia Evolutiva.
Glossary
Abbreviations:
- TEs
transposable elements
- AFLP
amplified fragments length polymorphism
- piRNAs
Piwi-interacting RNAs
- siRNAs
small interfering RNAs
References
- 1.Vilela C. A revision of the Drosophila repleta species group (Diptera, Drosophilidae) Rev Bras Entomol. 1983;1983:1–114. [Google Scholar]
- 2.Fontdevila A, Pla C, Hasson E, Wasserman M, Sanchez A, Naveira H, Ruiz A. Drosophila koepferae new member of the Drosophila serido (Diptera, Drosophilidae) superspecies taxon. Ann Entomol Soc Am. 1988;81:380–5. [Google Scholar]
- 3.Piccinali R, Aguadé M, Hasson E. Comparative molecular population genetics of the Xdh locus in the cactophilic sibling species Drosophila buzzatii and D. koepferae. Mol Biol Evol. 2004;21:141–52. doi: 10.1093/molbev/msh006. [DOI] [PubMed] [Google Scholar]
- 4.Marín I, Fontdevila A. Stable Drosophila buzzatii-Drosophila koepferae hybrids. J Hered. 1998;89:336–9. doi: 10.1093/jhered/89.4.336. [DOI] [PubMed] [Google Scholar]
- 5.Pantazidis A, Labrador M, Fontdevila A. The retrotransposon Osvaldo from Drosophila buzzatii displays all structural features of a functional retrovirus. Mol Biol Evol. 1999;16:909–21. doi: 10.1093/oxfordjournals.molbev.a026180. [DOI] [PubMed] [Google Scholar]
- 6.Labrador M, Utzet F, Fontdevila A. Interspecific Hybridization Increases Transposition Rates of Osvaldo. 1998:931–7. doi: 10.1093/oxfordjournals.molbev.a026182. [DOI] [PubMed] [Google Scholar]
- 7.Michalak P. An eruption of mobile elements in genomes of hybrid sunflowers. Heredity (Edinb) 2010;104:329–30. doi: 10.1038/hdy.2009.187. [DOI] [PubMed] [Google Scholar]
- 8.Ungerer MC, Strakosh SC, Zhen Y. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. 2006 doi: 10.1016/j.cub.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 9.Josefsson C, Dilkes B, Comai L. Parent-dependent loss of gene silencing during interspecies hybridization. Curr Biol. 2006;16:1322–8. doi: 10.1016/j.cub.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 10.Metcalfe CJ, Bulazel KV, Ferreri GC, Schroeder-Reiter E, Wanner G, Rens W, Obergfell C, Eldridge MDB, O’Neill RJ. Genomic instability within centromeres of interspecific marsupial hybrids. Genetics. 2007;177:2507–17. doi: 10.1534/genetics.107.082313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Neill RJW, O’Neill MJ, Graves JAM. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature. 1998;393:68–72. doi: 10.1038/29985. [DOI] [PubMed] [Google Scholar]
- 12.Labrador M, Fontdevila A. High transposition rates of Osvaldo, a new Drosophila buzzatii retrotransposon. Mol Gen Genet. 1994;245:661–74. doi: 10.1007/BF00297273. [DOI] [PubMed] [Google Scholar]
- 13.Kelleher ES, Edelman NB, Barbash DA. Drosophila interspecific hybrids phenocopy piRNA-pathway mutants. PLoS Biol. 2012;10:e1001428. doi: 10.1371/journal.pbio.1001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vela D, Fontdevila A, Vieira C, García Guerreiro MP. A genome-wide survey of genetic instability by transposition in Drosophila hybrids. PLoS One. 2014;9:e88992. doi: 10.1371/journal.pone.0088992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madlung A, Tyagi AP, Watson B, Jiang H, Kagochi T, Doerge RW, Martienssen R, Comai L. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 2005;41:221–30. doi: 10.1111/j.1365-313X.2004.02297.x. [DOI] [PubMed] [Google Scholar]
- 16.Turner LM, White MA, Tautz D, Payseur BA. Genomic networks of hybrid sterility. PLoS Genet. 2014;10:e1004162. doi: 10.1371/journal.pgen.1004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filatov DA, Morozova TV, Pasyukova EG. Age dependence of the copia transposition rate is positively associated with copia transcript abundance in a Drosophila melanogaster isogenic line. Mol Gen Genet. 1998;258:646–54. doi: 10.1007/s004380050778. [DOI] [PubMed] [Google Scholar]
- 18.Pasyukova E, Nuzhdin S, Li W, Flavell AJ. Germ line transposition of the copia retrotransposon in Drosophila melanogaster is restricted to males by tissue-specific control of copia RNA levels. Mol Gen Genet. 1997;255:115–24. doi: 10.1007/s004380050479. [DOI] [PubMed] [Google Scholar]
- 19.Borie N, Maisonhaute C, Sarrazin S, Loevenbruck C, Biémont C. Tissue-specificity of 412 retrotransposon expression in Drosophila simulans and D. melanogaster. Heredity (Edinb) 2002;89:247–52. doi: 10.1038/sj.hdy.6800135. [DOI] [PubMed] [Google Scholar]
- 20.Lankenau S, Corces VG, Lankenau DH. The Drosophila micropia retrotransposon encodes a testis-specific antisense RNA complementary to reverse transcriptase. Mol Cell Biol. 1994;14:1764–75. doi: 10.1128/mcb.14.3.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haoudi A, Rachidi M, Kim M-H, Champion S, Best-Belpomme M, Maisonhaute C. Developmental expression analysis of the 1731 retrotransposon reveals an enhancement of Gag-Pol frameshifting in males of Drosophila melanogaster. Gene. 1997;196:83–93. doi: 10.1016/S0378-1119(97)00203-5. [DOI] [PubMed] [Google Scholar]
- 22.Bregliano JC, Picard G, Bucheton A, Pelisson A, Lavige JM, L’Heritier P. Hybrid dysgenesis in Drosophila melanogaster. Science. 1980;207:606–11. doi: 10.1126/science.6766221. [DOI] [PubMed] [Google Scholar]
- 23.Josefsson C, Dilkes B, Comai L. Parent-dependent loss of gene silencing during interspecies hybridization. Curr Biol. 2006;16:1322–8. doi: 10.1016/j.cub.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 24.Michalak P. Epigenetic, transposon and small RNA determinants of hybrid dysfunctions. Heredity (Edinb) 2009;102:45–50. doi: 10.1038/hdy.2008.48. [DOI] [PubMed] [Google Scholar]
- 25.O’Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morán T, Fontdevila A. Genome-wide dissection of hybrid sterility in Drosophila confirms a polygenic threshold architecture. J Hered. 2014;105:381–96. doi: 10.1093/jhered/esu003. [DOI] [PubMed] [Google Scholar]
- 27.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 28.Szakmary A, Cox DN, Wang Z, Lin H. Regulatory relationship among piwi, pumilio, and bag-of-marbles in Drosophila germline stem cell self-renewal and differentiation. Curr Biol. 2005;15:171–8. doi: 10.1016/j.cub.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA. 2007;13:1911–22. doi: 10.1261/rna.744307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darricarrère N, Liu N, Watanabe T, Lin H. Function of Piwi, a nuclear Piwi/Argonaute protein, is independent of its slicer activity. Proc Natl Acad Sci U S A. 2013;110:1297–302. doi: 10.1073/pnas.1213283110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–35. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khurana JS, Wang J, Xu J, Koppetsch BS, Thomson TC, Nowosielska A, Li C, Zamore PD, Weng Z, Theurkauf WE. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell. 2011;147:1551–63. doi: 10.1016/j.cell.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fontdevila A. Hybrid genome evolution by transposition. Cytogenet Genome Res. 2005;110:49–55. doi: 10.1159/000084937. [DOI] [PubMed] [Google Scholar]