Abstract
Plant cell walls are composite structures mainly composed of polysaccharides, also containing a large set of proteins involved in diverse functions such as growth, environmental sensing, signaling, and defense. Research on cell wall proteins (CWPs) is a challenging field since present knowledge of their role into the structure and function of cell walls is very incomplete. Among CWPs, hydroxyproline (Hyp)-rich O-glycoproteins (HRGPs) were classified into three categories: (i) moderately glycosylated extensins (EXTs) able to form covalent scaffolds; (ii) hyperglycosylated arabinogalactan proteins (AGPs); and (iii) Hyp/proline (Pro)-Rich proteins (H/PRPs) that may be non-, weakly- or highly-glycosylated. In this review, we provide a description of the main features of their post-translational modifications (PTMs), biosynthesis, structure, and function. We propose a new model integrating HRGPs and their partners in cell walls. Altogether, they could form a continuous glyco-network with non-cellulosic polysaccharides via covalent bonds or non-covalent interactions, thus strongly contributing to cell wall architecture.
Keywords: arabinogalactan protein, extensin, hydroxyproline, O-glycosylation, proline-rich protein
Introduction
Plant cell walls are composite structures mainly composed of polysaccharides, namely cellulose, hemicelluloses and pectins, containing also a large set of proteins involved in the cell dynamics through diverse functions such as growth, environmental sensing, signaling, and defense (Fry, 2004). Research on cell wall components emerged in the nineteen sixties (Lamport and Northcote, 1960; Rees and Wight, 1969) and is still a very active field with continuous advances on the nature, structure and functions of polysaccharides (Carpita and Gibeaut, 1993; Willats et al., 2006; Scheller and Ulvskov, 2010) and of proteins (Rose and Lee, 2010; Albenne et al., 2013). However, the question of how these components are connected to make a functional matrix is still a matter of debate (Keegstra et al., 1973; Park and Cosgrove, 2012; Wang et al., 2012).
Among cell wall proteins (CWPs), hydroxyproline (Hyp)-rich O-glycoproteins (HRGPs) are complex macromolecules with various structures and functions. Identified several decades ago, HRGPs were classified into three categories: (i) moderately glycosylated extensins (EXTs); (ii) hyperglycosylated arabinogalactan proteins (AGPs); and (iii) Hyp/Pro-rich proteins (H/PRPs) that may be non-, weakly- or highly-glycosylated. Each HRGP sub-family is characterized by repetitive consensus sequences which determine the way they are glycosylated according to the so-called Hyp-O-glycosylation code (Kieliszewski, 2001; Tan et al., 2004; Estevez et al., 2006). From a functional point of view, HRGPs are also very diverse. AGPs are implicated in a variety of physiological processes including cell expansion, reproductive development, embryogenesis, signaling, and defense (Seifert and Roberts, 2007). EXTs are mostly described as structural proteins able to form covalent scaffolds (Qi et al., 1995; Brady et al., 1996; Cannon et al., 2008; Velasquez et al., 2012). Finally, H/PRPs are the less documented HRGPs and little is known about their structure and function. They seem to be associated to development and defense against biotic and abiotic stresses (Bradley et al., 1992; Bernhardt and Tierney, 2000; Battaglia et al., 2007). Hybrid and chimeric HRGPs also exist, enlarging the diversity of this superfamily. As previously defined, hybrid HRGPs are composed of HRGP modules from different families, and chimeric HRGPs are composed of one or more HRGP modules within a non-HRGP protein (Showalter et al., 2010). An expert bioinformatics analysis of the Arabidopsis thaliana genome identified 166 HRGPs classified in 85 AGPs, 59 EXTs, 18 H/PRPs, and 4 AGP/EXT hybrid proteins (Showalter et al., 2010). Besides, related to HRGPs but not classified in any of its three sub-families, some allergen proteins containing Hyp residues substituted by arabinogalactans (AGs) were identified in Artemisia vulgaris and Ambrosia artemisiifolia (Léonard et al., 2005, 2010).
Despite the great interest that plant biologist have had in HRGPs for more than 50 years, many questions about their mode of action in cell walls are still unanswered and HRGP research is still very challenging. In this review, we provide an update on (i) their post-translational modifications (PTMs) which consist in Pro-hydroxylation and O-glycosylation on serine (Ser) and Hyp residues and (ii) their roles in cell walls. We also focus on new insights into HRGP supramolecular assembly and propose a model including most recent data on covalent and non-covalent networks connecting HRGPs and polysaccharides.
Extensins (EXTs)
EXTs as structural molecules in plant cell walls
EXTs are modular, highly repetitive HRGPs showing similar features as collagen that contain Tyr cross-linking motifs. Unlike collagen, EXTs also undergo plant specific post-translational O-glycosylation on Ser-(Hyp)n ≥ 2 motifs. EXTs are represented in the A. thaliana genome by 59 members, some are classical EXTs while others are EXT-like chimeras and hybrid-EXTs that also contain other domains. Despite the high number of proteins with EXT domains in plant cell walls (Lamport et al., 2011), we know little about their exact functions and how this protein diversity is coordinated during plant development. There are several reasons that may explain our current lack of understanding of the EXT biology: (i) a high similarity in their protein sequences that make their characterization at the molecular level very difficult; (ii) the highly repetitive nature of their sequences since they are modular proteins, large in size and with complex chemical structures that carry several PTMs. Consequently, the biochemical characterization of a single EXT protein is still today very challenging; (iii) large number of EXTs and EXTs-related proteins encoded in known plant genomes; and (iv) several EXT genes are expressed at the same time in the same plant tissues (see Genevestigator database, https://www.genevestigator.com). In addition, most of the available EXT mutants analyzed until now show no clear phenotype. Few exceptions are the mutants atext3 (embryo lethal), atext6, 7, 10, 12 (shorter root hairs) and lrx1, 2 (root hair morphogenesis) that showed clear phenotypes (see Table 1).
Table 1.
Examples of EXTs and EXT-related proteins characterized in the last years.
| Protein/Gene name | Tissue or sub-cellular localization | Assumed function /Phenotype of mutants | References |
|---|---|---|---|
| EXTs | |||
| AtEXT1 (At1g76930) | Roots and Inflorescences | Cell wall formation/Induction in response to mechanical wounding, pathogen infection, senescence and at abscission zones, and treatment with hormones (methyl jasmonate, salicylic acid, auxin, brassinosteroids) | Merkouropoulos and Shirsat, 2003 |
| AtEXT3 (At1g21310) | Embryo | Cell wall formation/Embryo-lethal mutant. Incomplete cross wall assembly | Hall and Cannon, 2002; Cannon et al., 2008 |
| AtEXT6 (At2g24980) | Root hairs | Cell wall formation/Short root hair | Velasquez et al., 2011 |
| AtEXT7 (At4g08400) | |||
| AtEXT10 (At5g06640) | |||
| AtEXT11 (At5g49080) | |||
| AtEXT12 (At4g13390) | |||
| AtMOP10 (At5g05500) | Root hairs | Cell wall formation/Short root hair | Velasquez et al., 2011 |
| AtEXT-LIKE (At4g26750) | Root hairs | Cell wall formation/Short root hair | Velasquez et al., 2011 |
| SlEXT1 | Trichoblasts | -/Induced by ethylene | Bucher et al., 2002 |
| BnExtA | External and internal phloem of the main stem | -/Greatest expression in regions where a maximum tensile stress is exerted | Shirsat et al., 1996 |
| NtEXT1.4 | Stems, Roots and Carpels | -/Cells under mechanical stress: emergence of lateral roots, junction stem/petiole, fusion of carpels. Induction by mechanical stress in roots and stems | Hirsinger et al., 1999; Salvà and Jamet, 2001 |
| NsEXT1.2A | Stems and Roots | -/Expression in the root transition zone, in stem inner and outer phloem and in cortical cells at the stem/petiole junction. Induced by wounding | Guzzardi et al., 2004 |
| LRXs | |||
| AtLRR-EXT (At4g29240) | Root hairs | Cell wall formation /Short root hair | Velasquez et al., 2011 |
| AtLRX1 (At1g12040) | Root hairs | Cell wall formation/Morphogenesis of root hair | Baumberger et al., 2001 |
| AtLRX2 (At1g62440) | Root hairs | atlrx2 acts synergistically with atlrx1. atlrx1/atlrx2 show osmophilic aggregates and local disintegration of the cell wall | Baumberger et al., 2003 |
| VcISG (Inversion-Specific Glycoprotein) | Extracellular matrix | – | Ertl et al., 1992 |
| ZmPex1/ZmPex2/SlPEx (Pollen extensin-like) | Callose portion of the pollen tube cell wall | – | Rubinstein et al., 1995; Stratford et al., 2001 |
Dc, Daucus carota; Dca, Dianthus caryophyllus; La, Lupines albus; Ns, Nicotiana sylvestris; Nt, N. tabacum; Sl, Solanum lycopersicon; Vc, Volvox carteri; Zm, Zea mays.
PTMs of EXTs and the enzymes involved
Structural O-glycoproteins containing EXT domains that are ultimately secreted into plant cell walls are first shaped by several and complex PTMs that include: (i) signal peptide processing (in the ER), (ii) hydroxylation of Pro into Hyp residues, (iii) O-glycosylation on Hyp and Ser residues (in the ER-Golgi apparatus) and finally, (iv) Tyr cross-linking to promote the formation of a covalent network (in the cell wall). In the last few years, great progress has been made in our knowledge of the molecular players that act on the EXT biosynthetic pathway with the identification of several enzymes involved in their PTMs (summarized in Table A1). Hydroxylation of peptidyl-Pro is catalyzed by prolyl 4-hydroxylases (P4Hs) providing reactive hydroxyl groups for further O-glycosylation. Plant P4Hs are membrane-bound enzymes that belong to a family of 2-oxoglutarate-dependent dioxygenases (Hieta and Myllyharju, 2002; Koski et al., 2007, 2009). Partial in vitro and in vivo characterization of plant P4Hs (see Table A1) has been carried out in several plant model systems (Hieta and Myllyharju, 2002; Tiainen et al., 2005; Yuasa et al., 2005; Keskiaho et al., 2007; Vlad et al., 2007, 2010; Asif et al., 2009; Velasquez et al., 2011, 2012, in revision; Parsons et al., 2013). Most P4Hs are able to hydroxylate with different affinities several types of substrates containing collagen-like, polyproline EXT-type as well as AGP-like sequences. On the other hand, structural information on plant P4Hs is scarce since only one P4H from Chlamydomonas reinhardtii (CrP4H1) has been crystallized (Koski et al., 2007, 2009) and few P4Hs were characterized in vivo (Velasquez et al., in revision). Recent evidence showed that in A. thaliana, P4H5 forms homo-/hetero-dimers with P4H2 and P4H13 in the Golgi, suggesting the existence of P4H complexes required for proper Pro hydroxylation. It is plausible that more than one type of P4H complex would be formed in the ER-Golgi compartment, and in the case of the hetero-complexes, the presence of specific P4Hs (e.g., AtP4H5) may be required either for the correct recruitment or the scaffolding of the other P4Hs (e.g., AtP4H2) (Velasquez et al., in revision).
Hydroxylated EXTs are usually O-glycosylated with chains of up to four linear Ara residues on each Hyp (Velasquez et al., 2011; Ogawa-Ohnishi et al., 2013). The usual arabinoside structure linked to each Hyp unit is composed of β-L-Araf-(1,2)-β-L-Araf-(1,2)-β-L-Araf-(1,3)-α-L-tAraf. A fifth arabinose unit was reported in some tissues (Lamport, 1973). Specifically, three groups of arabinosyltransferases (AraTs) HPAT1-HPAT3 (GT8 CAZy family), RRA1-RRA3 (GT77 family), and XEG113 (GT77 family) have recently been implicated in the sequential addition of the innermost three L-Ara residues (Egelund et al., 2007; Gille et al., 2009; Velasquez et al., 2011; Ogawa-Ohnishi et al., 2013). The AraT that would transfer the fourth (1,3)-α-L-Araf moiety was identified very recently as Extensin Arabinose Deficient transferase (ExAD) within the GT47 family (Petersen et al. in preparation). In addition, one novel peptidyl-Ser galactosyltransferase named as SGT1 has been reported to add a single α-Galp residue to each Ser residue in Ser-(Hyp)4 motifs of EXTs and it would belong to a new family of CAZy (Saito et al., 2014). Glycosylated EXTs are cross-linked, at least in vitro, by putative type-III peroxidases (PERs) at the Tyr residues (Schnabelrauch et al., 1996; Jackson et al., 2001; Price et al., 2003) forming intra- and inter-EXT linkages (Cannon et al., 2008; Lamport et al., 2011). Thus, EXTs are able to form a three-dimensional glycoprotein network that possibly interacts with other cell wall components like pectins (Nuñez et al., 2009; Dick-Perez et al., 2011). Although the in vivo molecular mechanism of the covalent cross-link is unknown, there is evidence of PER-catalyzed oxidative coupling of Tyr residues in vitro that mediates the insolubilization of the proteins (Schnabelrauch et al., 1996; Jackson et al., 2001; Price et al., 2003). Recently, six apoplastic type-III PERs were identified as putative candidates for the cross-linking of EXTs specifically in the root hairs of A. thaliana (Velasquez et al., in revision). Structural proteins with polyproline sequences like collagen can also be Tyr-cross-linked by the action of a PER not only in vitro but also in vivo (Edens et al., 2001) suggesting that EXTs and collagen, as extracellular building blocks, would share structural features and functions.
Root hair as models to study EXT functions and related GTs
Root hairs have been used as a single-cell model to study cell wall biosynthesis in general and specifically EXTs during tip-growth (Park et al., 2011; Velasquez et al., 2011). Mutants deficient in the synthesis of a single wall polymer specifically in the root hair are generally impaired in growth because their cell wall structure is severely compromised (Diet et al., 2006; Cavalier et al., 2008; Ringli, 2010; Park et al., 2011; Pena et al., 2012; Velasquez et al., 2012). In this framework of interconnected wall polymers (Cosgrove, 2005; Dick-Perez et al., 2011), cross-linked EXTs have a key role during cell expansion and growth (Cannon et al., 2008; Ringli, 2010; Lamport et al., 2011; Velasquez et al., 2011). EXT domains seem to be important during polarized cell expansion since several EXT-related mutants have shorter root hairs such as classical ext6, ext7, ext10 and ext12 (Velasquez et al., 2011, 2012) and lrx1 and lrx2 mutants (Baumberger et al., 2001, 2003; Ringli, 2010).
Impact of O-glycosylation on EXT function
It is accepted that O-glycans increase HRGP solubility, resistance to proteolytic degradation and thermal stability (Kieliszewski et al., 1989; Ferris et al., 2001; Shpak et al., 2001; Kieliszewski et al., 2011; Lamport et al., 2011). Most of the mutants that correspond to glycosyltransferases (GTs) known to glycosylate EXTs (Table A1) have been related to root hair drastic phenotypes, highlighting that even minor changes in the O-glycosylation status of EXTs affect EXT function during polarized cell expansion (Velasquez et al., in revision). In addition, it was found that both O-glycosylation types present in EXTs (Hyp-O-arabinosylation and Ser-O-galactosylation) are required and have additive effects for correct EXT function in root hair growth (Velasquez et al., in revision). The known roles of EXTs in cell wall assembly, cell shape and growth raise the question about the function of each individual EXT molecule (Hall and Cannon, 2002; Cannon et al., 2008; Velasquez et al., 2011). Some examples of already characterized EXT or EXT-related genes are presented in Table 1. Recently, it was reported that EXTs can form, at least in vitro, a tridimensional covalent network through Tyr-linkages mediated by EXT PERs between individual EXT molecules and also via self-recognition and alignment of hydrophilic O-glycosylated Ser-(Hyp)3−4 repeats and hydrophobic peptide-cross-linking modules (Cannon et al., 2008). Thus, the ordered EXT monomer assembly in plant cell walls would involve a zipper-like endwise association via cross-linking at the ends of the molecules (Kieliszewski et al., 2011; Lamport et al., 2011). Recently, molecular dynamics and homology modeling experiments suggested that classical EXTs would be able to form a putative triple helix structure by lateral staggered alignment (Cannon et al., 2008) and Tyr cross-linking, similar to that present in collagen (Velasquez et al., in revision). It is also proposed that EXTs interact with pectins by a simple acid-base reaction forming a supramolecular ionic structure in the nascent cell wall (Valentin et al., 2010), which would serve as a template for further cell wall deposition (Cannon et al., 2008; Lamport et al., 2011). In addition, covalent EXT-pectin cross-links were also suggested (Qi et al., 1995; Nuñez et al., 2009). However, it is unclear how EXT monomers are secreted and assembled into the glyco-network and how EXT-pectin interactions are controlled in a coordinated way during new cell wall formation. In addition, pectin methyl esterases de-esterify galacturonic acid residues in homogalaturonans and liberate acidic charges for ionic interactions (Micheli, 2001) with positively-charged domains in molecules like EXTs.
Arabinogalactan proteins (AGPs)
Many articles reporting the state of the art concerning AGP structure, function and biosynthesis have been published recently (Seifert and Roberts, 2007; Ellis et al., 2010; Tan et al., 2012; Lamport and Várnai, 2013; Nguema-Ona et al., 2013; Knoch et al., 2014). AGPs are HRGPs containing a high proportion of sugars, up to 90%. They are characterized by repetitive X(Pro)n motifs in which X is mostly alanine (Ala) or Ser. In this review, we focus on specific aspects concerning (i) the characterization of their O-glycan moiety and (ii) their interactions with cell wall polysaccharides.
Structure of O-glycans of AGPs
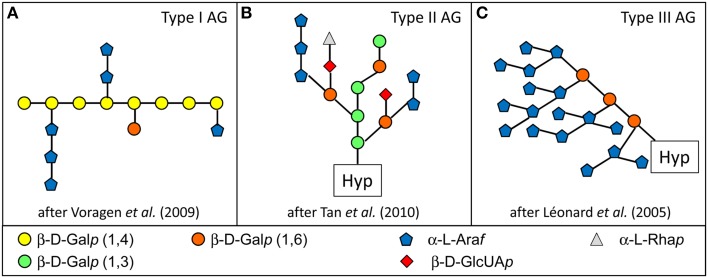
A remarkable work performed on proteins deriving from synthetic genes and produced in cell suspension cultures has allowed to characterize AGP O-glycans covalently linked to [Ser(Hyp)]n and [Ala(Hyp)]n motifs (Tan et al., 2010). It has been possible to precisely define the structure of type II AGs by combining monosaccharide and linkage analyses to mass spectrometry and NMR. An example of type II AG is given in Figure 1B. Type II AGs contain a β-D-Galp backbone formed by a succession of three β-1,3 linked D-Galp interrupted by a β-1,6 linkage causing a reverse turn. Gal residues of side chains can be substituted with α-L-Araf, α-L-Rhap or Me-GlcpA (Tan et al., 2010; Tryfona et al., 2012). The chelation of Ca2+ ions could occur at the level of GlcA residues located in close proximity. It should be noted that different variants of this basic structure exist, for example β-1,6 side chains can vary in length from 1 over 20 Gal residues (Tryfona et al., 2012). Type II AGs react positively with the β-glucosyl and β-galactosyl Yariv reagents, but not with the α-glucosyl and α-galactosyl Yariv reagents (Kitazawa et al., 2013). The β-galactosyl Yariv reagent has been shown to recognize the β-1,3-Gal main chains of type II AGs. They are different from previously described type I AGs which constitute lateral branches of RGI (Voragen et al., 2009). Type I AGs are formed by a linear chain of β-D-Galp (1,4) with lateral chains of α-L-Araf (1,5 attached to Gal O-3) and β-D-Galp (attached to Gal-O-6) (Figure 1A). Type II AGs also differ from type III AGs found on allergens like the Artemisia vulgaris Art v 1 (Léonard et al., 2005). The structure of type III AGs has been determined by combining the results of linkage analysis, NMR and enzymatic degradation. They are formed by a short linear chain of β-D-Galp (1,6). They only contain Gal and Ara residues, and they have large branched Ara chains. The linkage analysis indicates the presence of terminal Araf, 5-Araf, 3,5-Araf, 2,5-Araf, 2,3,5-Araf and 3,6-Galp. This Hyp O-glycan was shown to consist of Hyp1Gal3Ara5−28 series by MALDI-TOF MS. Type III AGs react with the β-glucosyl Yariv reagent suggesting that Art v 1 is an AGP. As for type II AGs, type III AGs probably exist in various forms and only a consensus model can be proposed (Figure 1C). Another kind of type III AGs has been later described for Amb a 4, an allergen of Ambrosia artemisiifolia (Léonard et al., 2010). It differs from the that of Art v 1 by the presence of different Hyp1Gal1Ara5−20series with a lower amount of Gal, the presence of more α-Araf (1,5) and less α-Araf (1,3).
Figure 1.
The three main types of AGs. One of the main differences between these AG types consist in the type of linkages between Gal residues of the main chain: β-1,4 in type I AG (A); β-1,3 and β-1,6 in type II AG (B); β-1,6 in type III AG (C). These differences have been highlighted on the figure by using different colors for Gal residues. Other differences are described in the text.
The existence of different types of AGs linked to AGPs (types II and III) raises the questions of (i) the role of the amino acid sequence and (ii) the presence of different types of GTs in plants to ensure the appropriate O-glycosylation of HRGPs (Léonard et al., 2005).
Interactions of AGPs with polysaccharides
The question of how AGPs are connected to other cell wall components and the involvement of their carbohydrate moieties in the interactions is of great importance, but still poorly documented. It has been assumed that AGPs could act as covalent cross-linkers in polysaccharide networks. Several lines of evidence suggested associations between AGPs and pectins. More than 40 years ago, it was hypothesized that Rha residues on type II AG side chains could be attached to RGI (Keegstra et al., 1973). Since then, several studies have reported the existence of strong associations between AGPs and pectins from different plant tissues, including grape (Pellerin et al., 1995), carrot (Immerzeel et al., 2006) or sugar beet (McKenna et al., 2006). Pectins were shown to co-localize with AGPs in pollen tubes (Li et al., 1995; Jauh and Lord, 1996; Mollet et al., 2002). Besides, enzymatic treatment of cell wall fractions with pectin-degrading enzymes allowed for an increased release of AGPs (Immerzeel et al., 2006; Lamport et al., 2006). One study also suggested the existence of AGP/xylan complexes (Kwan and Morvan, 1995). However, all these AGP/polysaccharide complex analyses were either indirect or achieved on preparations containing a mixture of AGPs, thus preventing a detailed characterization of the interactions. The first in depth structural study of an AGP polysaccharide complex involving pure AGP was only recently reported (Tan et al., 2013). It was shown that two isoforms of a purified A. thaliana AGP, At3g45230, are covalently attached to pectins and hemicelluloses. Linkages have been demonstrated between: (i) RGI/homogalacturonan and the Rha residue in the AGP type II AG domain and (ii) arabinoxylan and either a Rha residue of RGI or an Ara residue in the type II AG domain. A model was proposed for this complex called Arabinoxylan Pectin Arabinogalactan Protein1 (APAP1). The apap1 mutant showed an increased extractability of pectin and xylan, suggesting a structural role for APAP1 (Tan et al., 2013). However, since APAP1 was isolated from suspension culture media, it could correspond to a simplified structure with pectin and xylan domains smaller than expected in plant cell walls. Larger APAP1 complexes may exist in cell walls, but their extraction is undoubtedly the bottleneck preventing their characterization. Large AGP/pectin/xylan complexes should also be found in other plants, corroborating all prior studies indirectly suggesting their existence (Tan et al., 2013).
Present knowledge on AGP/polysaccharide interactions indicates that some AGPs may serve as cross-linker in cell walls and act as polysaccharide plasticizers as previously assumed (Lamport, 2001; Lamport et al., 2006). Chimeric proteins containing AGP domains were also suggested to interact with polysaccharides. In particular, SOS5 (SALT-OVERLY SENSITIVE 5), a Fasciclin-AGP, was assumed to interact with pectins, thus mediating mucilage adherence (Griffiths et al., 2014). SOS5 interacting partners were not identified. Further efforts will be necessary to highlight the contribution of AGPs to cell wall architecture and to give more insight into its molecular basis.
Hyp/Pro-rich proteins
Like EXTs and AGPs, H/PRPs belong to the HRGP superfamily and some of them are chimeric proteins. As mentioned above, little is known about the O-glycosylation of H/PRPs and their interactions with polysaccharides. With regard to O-glycosylation, information is only available for H/PRPs having X(Pro/Hyp)n ≥ 2X motifs. This type of domain can be associated with a short N-terminal AGP domain, a histidine (His)-stretch and a C-terminal PAC (Proline-rich protein and AGP, containing Cys) domain like in the A. thaliana AtAGP31 (Liu and Mehdy, 2007; Hijazi et al., 2012). Up to now, twelve such proteins have been identified in A. thaliana, Daucus carota, Gossypium hirsutum, Nicotiana alata, N. tabacum, Phaseolus vulgaris, Capsicum annuum and Petunia hybrida (Hijazi et al., 2014).
Structure of the O-glycans of H/PRPs of the AtAGP31 type
O-glycosylated amino acid motifs of the H/PRP domain of AtAGP31 have been characterized by mass spectrometry: Lys(Ala/Ser)HypVal, Lys(Pro/Hyp)(Hyp/Pro)(Thr/Val), Thr(Pro/Hyp)(Hyp/Pro)Val, and Tyr(Pro/Hyp)(Hyp/Pro)Thr (Hijazi et al., 2012). The monosaccharide linked to Hyp is an hexose which is most probably a Gal based on the monosaccharide analysis of the purified protein (53.2% Gal, 39.5% Ara, 2.2% Xyl, 1.9% Fuc, 1.8% Glc, 1.3% Man, 0.3% GlcUA). It should be noted that this global analysis includes O-glycans linked to the AGP domain of AtAGP31 and N-glycans linked to its PAC domain. The O-glycan linked to the H/PRP domain of AtAGP31 is not recognized by the β-D-glucosyl Yariv reagent, but it interacts with the Peanut Agglutinin (PNA), a lectin having a high affinity for Gal residues (Hijazi et al., 2012). It was called Gal/Ara-rich motif (Hijazi et al., 2012). Nicotiana alata NaPRP4 shares the same type of H/PRP domain and a PAC domain with AtAGP31 (Sommer-Knudsen et al., 1996). The predominant monosaccharide of this O-glycoprotein is Gal (83%) wheareas Ara, GlcNac, Man, Xyl are in minor amounts (7, 4, 4, 1% respectively). The linkage analysis has shown the presence of terminal Araf (6%), terminal Galp (48%), 1,3-Galp (4%), 1,6-Galp (14%), 1,3,6 Galp (25%), 1,2-Manp (1%) and Xylp (1%). Altogether, H/PRPs with X(Pro/Hyp)n ≥ 2X motifs are O-glycosylated with Gal-Ara-rich glycans which seems to be slightly different from the previously described type I, II and III AGs. Further characterization, especially by NMR will be required to fully describe these structures.
Interactions of H/PRPs with polysaccharides
H/PRPs are assumed to be cross-linked in cell walls, but direct evidence is still lacking (Bradley et al., 1992; Brisson et al., 1994; Frueauf et al., 2000). Nothing is known about the possible roles of O-glycosylations. AtAGP31was recently proposed to be involved in non-covalent interactions networks (Hijazi et al., 2014). Consistently and unlike HRGPs which are covalently insolubilized in cell walls, AtAGP31 is easily extracted from cell walls of etiolated hypocotyls (Hijazi et al., 2012). It should be noted that NaPRP4 is not insolubilized in cell walls as well (Sommer-Knudsen et al., 1996). AtAGP31 was shown to interact in vitro with RGI type I AG branches through its PAC domain and with methyl-esterified polygalacturonic acid, probably through its His-stretch. Protein/protein interactions were also assumed for AtAGP31, with (i) self-recognition between its PAC domain and its H/PRP domain O-glycans, and (ii) interaction with cell wall lectins. It was proposed that the AtAGP31 multi-domain organization results in complex supra-molecular scaffolds with different cell wall components, thus contributing to the strengthening of cell walls of quickly growing organs like etiolated hypocotyls. Such non-covalent networks have not been described before for HRGPs. A similar behavior may exist for proteins sharing features with AtAGP31 (Hijazi et al., 2014). However, as mentioned above, except NaPRP4 whose glycosylation has been characterized (Sommer-Knudsen et al., 1996), these proteins were not described at the molecular level and their interactions with cell wall polysaccharides were not investigated. TTS-1 and TTS-2 (Transmitting Tissue-Specific) from N. tabacum, and DcAGP1 from D. carota were shown to display an ellipsoidal shape and to self-assemble into higher-order structures using microscopy techniques (Baldwin et al., 2000, 2001; Wu et al., 2001). Interestingly, the deglycosylation of TTS disrupts its ability to aggregate, suggesting a regulation of self-association by its level of O-glycosylation (Wu et al., 1995). Self-assembly in a head-to-tail fashion through interactions between the O-glycans of H/PRP domain and the PAC domain can be proposed for proteins like TTS and DcAGP1, similarly to AtAGP31.
Concluding remarks and future developments
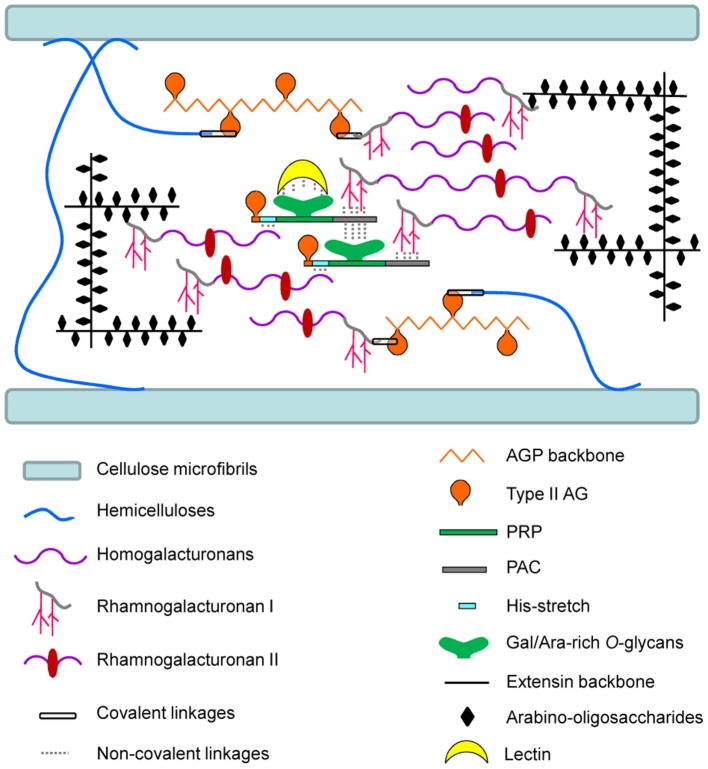
In this review, we have focused on some structural features of HRGP O-glycans and we have highlighted their possible interactions in muro through covalent glycosidic linkages or non-covalent interactions. As proposed in the model shown in Figure 2, HRGPs could serve as cross-linkers in cell walls, connecting non-cellulosic polysaccharides, thus forming a continuous network. Large covalent complexes connecting AGP, hemicelluloses and pectins, as proposed in APAP1, are represented (Tan et al., 2013). However, the relevance of such covalent complexes in cell walls need to be confirmed. EXTs appear to form covalent linkages with pectins as reported (Qi et al., 1995; Nuñez et al., 2009). The precise moieties involved in these linkages have not been identified so far. Finally, chimeric HRGPs with H/PRP and PAC domains like AtAGP31 may form non-covalent networks with a set of cell wall components, including polysaccharides and lectins (Hijazi et al., 2014). It can be speculated that these protein/polysaccharide networks contribute to the cell wall architecture, by reinforcing the polysaccharide scaffold and by controlling its porosity. A recent high-resolution solid-state NMR study elucidating the 3D-architecture of the polysaccharides and proteins in muro revealed that the structural proteins in the primary cell wall are separated from the polysaccharides by more than one nanometer (Wang et al., 2012). This corroborates the assumption that O-glycans acts as spacers between HRGP backbones and cell wall polysaccharides.
Figure 2.
Schematic representation of interactions between HRGPs and cell wall polysaccharides in muro. This model proposes an overview of the interactions assumed or demonstrated between HRGPs and polysaccharides according to most relevant publications in this field. For clarity, the model does not represent the whole complexity of the polysaccharide networks. AGPs are represented with covalent linkages with pectins and hemicelluloses, as proposed by Tan et al. (2013) for the so-called APAP1 complex. EXTs are drawn attached covalently with pectins as proposed by Qi et al. (1995). Finally, non-covalent networks between chimeric HRGPs and polysaccharides are represented according to Hijazi et al. (2014) for AtAGP31. Lectins assumed to bind to Gal/Ara-rich O-glycans of AtAGP31 are also integrated into the model.
These new features render even more complex the cell wall architecture. Plant cell walls contain a variety of complex macromolecules, possibly interconnected, resulting from a sophisticated metabolism. A tremendous set of carbohydrate active enzymes is required to achieve (i) polysaccharide synthesis and assembly, (ii) protein glycosylation, and (iii) possible polysaccharide/protein linkages. Non-cellulosic polymer synthesis occurs in the Golgi (Mohnen, 2008; Brown et al., 2011), and HRGP synthesis starts in the ER and continues in the Golgi (Basu et al., 2013; Knoch et al., 2014). An important issue is now to determine in which sub-cellular compartment covalent HRGP/polysaccharide complexes are formed and by which mechanism. Is there a code for establishing these links or are they occurring randomly? Which enzymes are involved? Answering these questions constitutes a real challenge toward a better understanding of cell wall biosynthesis and architecture. Further studies will also be necessary to elucidate the molecular basis of HRGP functions in cell walls and their involvement in physiological processes like cell plate formation or root hair cell expansion (Cannon et al., 2008; Velasquez et al., 2011).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
May Hijazi, Cécile Albenne, and Elisabeth Jamet are thankful to Université Paul Sabatier (Toulouse, France), CNRS for supporting their research work and to the Lebanon Ecological Association (grant to May Hijazi). Part of this work has been done at LRSV which belongs to the Laboratoire d'Excellence (LabEx) entitled TULIP (ANR-10-LABX-41; ANR-11-IDEX-0002-02). This work was also supported by PICT FONCyT 2011-0054, Argentina (JME) and Mizutani Foundation Glycoscience Grant 2013, Japan (JME). The authors wish to apologize for the papers not quoted in this review, due to lack of space.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00395/abstract
References
- Albenne C., Canut H., Jamet E. (2013). Plant cell wall proteomics: the leadership of Arabidopsis thaliana. Front. Plant Sci. 4:111 10.3389/fpls.2013.00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif M. H., Trivedi P. K., Misra P., Nath P. (2009). Prolyl-4-hydroxylase (AtP4H1) mediates and mimics low oxygen response in Arabidopsis thaliana. Funct. Integr. Genomics 9, 525–535 10.1007/s10142-009-0118-y [DOI] [PubMed] [Google Scholar]
- Baldwin T. C., Domingo C., Schindler T., Seetharaman G., Stacey N., Roberts K. (2001). DcAGP1, a secreted arabinogalactan protein, is related to a family of basic proline-rich proteins. Plant Mol. Biol. 45, 421–435 10.1023/A:1010637426934 [DOI] [PubMed] [Google Scholar]
- Baldwin T. C., Hengel A., Roberts K. (2000). The C-terminal PAC domain of a secreted arabinogalactan protein from carrot defines a family of basic proline-rich proteins, in Cell and Developmental Biology of Arabinogalactan Proteins, eds Nothnagel E. A., Bacic A., Clarke A. E. (New York, NY: Kluwer Academic Publishers; ), 43–50 10.1007/978-1-4615-4207-0_4 [DOI] [Google Scholar]
- Basu D., Liang Y., Liu X., Himmeldirk K., Faik A., Kieliszewski M., et al. (2013). Functional identification of a hydroxyproline-O-galactosyltransferase specific for arabinogalactan protein biosynthesis in Arabidopsis. J. Biol. Chem. 288, 10132–10143 10.1074/jbc.M112.432609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M., Solorzano R. M., Hernandez M., Cuellar-Ortiz S., Garcia-Gomez B., Marquez J., et al. (2007). Proline-rich cell wall proteins accumulate in growing regions and phloem tissue in response to water deficit in common bean seedlings. Planta 225, 1121–1133 10.1007/s00425-006-0423-9 [DOI] [PubMed] [Google Scholar]
- Baumberger N., Ringli C., Keller B. (2001). The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev. 15, 1128–1139 10.1101/gad.200201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N., Steiner M., Ryser U., Keller B., Ringli C. (2003). Synergistic interaction of the two paralogous Arabidopsis genes LRX1 and LRX2 in cell wall formation during root hair development. Plant J. 35, 71–81 10.1046/j.1365-313X.2003.01784.x [DOI] [PubMed] [Google Scholar]
- Bernhardt C., Tierney M. L. (2000). Expression of AtPRP3, a proline-rich structural cell wall protein from arabidopsis, is regulated by cell-type-specific developmental pathways involved in root hair formation. Plant Physiol. 122, 705–714 10.1104/pp.122.3.705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D. J., Kjellbom P., Lamb C. J. (1992). Elicitor-induced and wound induced oxidative cross-linking of a proline rich plant cell wall protein: a novel, rapid defense response. Cell 70, 21–30 10.1016/0092-8674(92)90530-P [DOI] [PubMed] [Google Scholar]
- Brady J. D., Sadler I. H., Fry S. C. (1996). Di-isodityrosine, a novel tetrametric derivative of tyrosine in plant cell wall proteins: a new potential cross-link. Biochem. J. 315, 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson L. F., Tenhaken R., Lamb C. (1994). Function of oxidative cross linking of cell wall structural proteins in plant disease resistance. Plant Cell 6, 1703–1712 10.1105/tpc.6.12.1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D., Wightman R., Zhang Z., Gomez L., Atanassov I., Bukowski J. P., et al. (2011). Arabidopsis genes IRREGULAR XYLEM (IRX15) and IRX15L encode DUF579-containing proteins that are essential for normal xylan deposition in the secondary cell wall. Plant J. 66, 401–403 10.1111/j.1365-313X.2011.04501.x [DOI] [PubMed] [Google Scholar]
- Bucher M., Brunner S., Zimmermann P., Zardi G. I., Amrhein N., Willmitzer L., et al. (2002). The expression of an extensin-like protein correlates with cellular tip growth in tomato. Plant Physiol. 128, 911–923 10.1104/pp.010998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon M. C., Terneus K., Hall Q., Tan L., Wang Y., Wegenhart B. L., et al. (2008). Self-assembly of the plant cell wall requires an extensin scaffold. Proc. Natl. Acad. Sci. U.S.A. 105, 2226–2231 10.1073/pnas.0711980105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita N. C., Gibeaut D. M. (1993). Structural models of primary cell walls in flowering plants, consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3, 1–30 10.1111/j.1365-313X.1993.tb00007.x [DOI] [PubMed] [Google Scholar]
- Cavalier D. M., Lerouxel O., Neumetzler L., Yamauchi K., Reinecke A., Freshour G., et al. (2008). Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20, 1519–1537 10.1105/tpc.108.059873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861 10.1038/nrm1746 [DOI] [PubMed] [Google Scholar]
- Dick-Perez M., Zhang Y., Hayes J., Salazar A., Zabotina O. A., Hong M. (2011). Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry 50, 989–1000 10.1021/bi101795q [DOI] [PubMed] [Google Scholar]
- Diet A., Link B., Seifert G. J., Schellenberg B., Wagner U., Pauly M., et al. (2006). The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-L-rhamnose synthase. Plant Cell 18, 1630–1641 10.1105/tpc.105.038653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edens W. A., Sharling L., Cheng G., Shapira R., Kinkade J. M., Lee T., et al. (2001). Tyrosine cross-linking of extracellular matrix is catalyzed by Duox, a multidomain oxidase/peroxidase with homology to the phagocyte oxidase subunit gp91phox. J. Cell Biol. 154, 879–891 10.1083/jcb.200103132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelund J., Obel N., Ulvskov P., Geshi N., Pauly M., Bacic A., et al. (2007). Molecular characterization of two Arabidopsis thaliana glycosyltransferase mutants, rra1 and rra2, which have a reduced residual arabinose content in a polymer tightly associated with the cellulosic wall residue. Plant Mol. Biol. 64, 439–451 10.1007/s11103-007-9162-y [DOI] [PubMed] [Google Scholar]
- Ellis M., Egelund J., Schultz C. J., Bacic A. (2010). Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol. 153, 403–419 10.1104/pp.110.156000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertl H., Hallmann A., Wenzl S., Sumper M. (1992). A novel extensin that may organize extracellular matrix biogenesis in Volvox carteri. EMBO J. 11, 2055–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez J. M., Kieliszewski M. J., Khitrov N., Somerville C. (2006). Characterization of synthetic hydroxyproline-rich proteoglycans with arabinogalactan protein and extensin motifs in Arabidopsis. Plant Physiol. 142, 458–470 10.1104/pp.106.084244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P. J., Woessner J. P., Waffenschmidt S., Kilz S., Drees J., Goodenough U. W. (2001). Glycosylated polyproline II rods with kinks as a structural motif in plant hydroxyproline-rich glycoproteins. Biochemistry 40, 2978–2987 10.1021/bi0023605 [DOI] [PubMed] [Google Scholar]
- Frueauf J. B., Dolata M., Leykam J. F., Lloyd E., Gonzales M., Vandenbosch K., et al. (2000). Peptides isolated from cell walls of Medicago truncatula nodules and uninfected root. Phytochemistry 55, 429–438 10.1016/S0031-9422(00)00336-8 [DOI] [PubMed] [Google Scholar]
- Fry S. C. (2004). Primary cell wall metabolism: tracking the careers of wall polymers in living plant cells. New Phytol. 161, 641–675 10.1111/j.1469-8137.2004.00980.x [DOI] [PubMed] [Google Scholar]
- Gille S., Hansel U., Ziemann M., Pauly M. (2009). Identification of plant cell wall mutants by means of a forward chemical genetic approach using hydrolases. Proc. Natl. Acad. Sci. U.S.A. 106, 14699–14704 10.1073/pnas.0905434106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J. S., Tsai A. Y., Xue H., Voiniciuc C., Sola K., Seifert G., et al. (2014). SALT-OVERLY SENSITIVE5 mediates Arabidopsis seed coat mucilage adherence and organization through pectins. Plant Physiol. 165, 991–1004 10.1104/pp.114.239400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzardi P., Genot G., Jamet E. (2004). The Nicotiana sylvestris extensin gene, Ext 1.2A, is expressed in the root transition zone and upon wounding. Biochim. Biophys. Acta 1680, 83–92 10.1016/j.bbaexp.2004.08.012 [DOI] [PubMed] [Google Scholar]
- Hall Q., Cannon M. C. (2002). The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell 14, 1161–1172 10.1105/tpc.010477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieta R., Myllyharju J. (2002). Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana. Effective hydroxylation of proline-rich, collagen-like, and hypoxia-inducible transcription factor alpha-like peptides. J. Biol. Chem. 277, 23965–23971 10.1074/jbc.M201865200 [DOI] [PubMed] [Google Scholar]
- Hijazi M., Durand J., Pichereaux C., Pont F., Jamet E., Albenne C. (2012). Characterization of the arabinogalactan protein 31 (AGP31) of Arabidopsis thaliana: new advances on the Hyp-O-glycosylation of the Pro-rich domain. J. Biol. Chem. 287, 9623–9632 10.1074/jbc.M111.247874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijazi M., Roujol D., Nguyen-Kim H., Del Rocio Cisneros Castillo L., Saland E., Jamet E., et al. (2014). Arabinogalactan protein 31 (AGP31), a putative network-forming protein in Arabidopsis thaliana cell walls? Ann Bot. [Epub ahead of print]. 10.1093/aob/mcu038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsinger C., Salva I., Marbach J., Durr A., Fleck J., Jamet E. (1999). The tobacco extensin gene Ext 1.4 is expressed in cells submitted to mechanical constraints and in cells proliferating under hormone control. J. Exp. Bot. 50, 343–355 [Google Scholar]
- Immerzeel P., Eppinka M. M., De Vriesb S. C., Scholsa H. A., Voragen A. G. J. (2006). Carrot arabinogalactan proteins are interlinked with pectins. Physiol. Plant. 128, 18–28 10.1111/j.1399-3054.2006.00712.x [DOI] [Google Scholar]
- Jackson P. A., Galinha C. I., Pereira C. S., Fortunato A., Soares N. C., Amancio S. B., et al. (2001). Rapid deposition of extensin during the elicitation of grapevine callus cultures is specifically catalyzed by a 40-kilodalton peroxidase. Plant Physiol. 127, 1065–1076 10.1104/pp.010192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauh G. Y., Lord E. M. (1996). Localization of pectins and arabinogalactan-proteins in lily (Lilium longittorum L.) pollen tube and style, and their possible roles in pollination. Planta 199, 251–261 10.1007/BF00196566 [DOI] [Google Scholar]
- Keegstra K., Talmadge K. W., Bauer W. D., Albersheim P. (1973). The structure of plant cell walls: III. A model of the walls of suspension-cultured sycamore cells based on the interconnections of the macromolecular components. Plant Physiol. 51, 188–197 10.1104/pp.51.1.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskiaho K., Hieta R., Sormunen R., Myllyharju J. (2007). Chlamydomonas reinhardtii has multiple prolyl 4-hydroxylases, one of which is essential for proper cell wall assembly. Plant Cell 19, 256–269 10.1105/tpc.106.042739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieliszewski M. J. (2001). The latest hype on Hyp-O-glycosylation codes. Phytochemistry 57, 319–323 10.1016/S0031-9422(01)00029-2 [DOI] [PubMed] [Google Scholar]
- Kieliszewski M. J., Lamport D. T., Tan L., Cannon M. C. (2011). Hydroxyproline-rich glycoproteins: form and function, in Plant Polysaccharides: Biosynthesis and Bioengineering, ed. Ulvskov P. (Oxford: Wiley-Blackwell; ), 321–342 [Google Scholar]
- Kieliszewski M. J., Leykam J. F., Lamport D. T. (1989). Trypsin cleaves lysylproline in a hydroxyproline-rich glycoprotein from Zea mays. Pept. Res. 2, 246–248 [PubMed] [Google Scholar]
- Kitazawa K., Tryfona T., Yoshimi Y., Hayashi Y., Kawauchi S., Antonov L., et al. (2013). β-galactosyl Yariv reagent binds to the β-1,3-galactan of arabinogalactan proteins. Plant Physiol. 161, 1117–1126 10.1104/pp.112.211722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoch E., Dilokpimol A., Geshi N. (2014). Arabinogalactan proteins: focus on carbohydrate active enzymes. Front. Plant Sci. 5:198 10.3389/fpls.2014.00198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski M. K., Hieta R., Bollner C., Kivirikko K. I., Myllyharju J., Wierenga R. K. (2007). The active site of an algal prolyl 4-hydroxylase has a large structural plasticity. J. Biol. Chem. 282, 37112–37123 10.1074/jbc.M706554200 [DOI] [PubMed] [Google Scholar]
- Koski M. K., Hieta R., Hirsila M., Ronka A., Myllyharju J., Wierenga R. K. (2009). The crystal structure of an algal prolyl 4-hydroxylase complexed with a proline-rich peptide reveals a novel buried tripeptide binding motif. J. Biol. Chem. 284, 25290–25301 10.1074/jbc.M109.014050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan J. S., Morvan H. (1995). Characterization of extracellular beta-(1,4)-xylan backbone O-substituted by arabinogalactans type-II in a plant-cell suspension. Carbohydr. Polym. 26, 99–107 10.1016/0144-8617(94)00098-E [DOI] [Google Scholar]
- Lamport D. T. (1973). Galactosylserine in extensin. Biochem. J. 133, 125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport D. T. (2001). Life behind cell walls: paradigm lost, paradigm regained. Cell. Mol. Life Sci. 58, 1363–1385 10.1007/PL00000782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport D. T., Kieliszewski M. J., Chen Y., Cannon M. C. (2011). Role of the extensin superfamily in primary cell wall architecture. Plant Physiol. 156, 11–19 10.1104/pp.110.169011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport D. T., Kieliszewski M. J., Showalter A. M. (2006). Salt stress upregulates periplasmic arabinogalactan proteins: using salt stress to analyse AGP function. New Phytol. 169, 479–492 10.1111/j.1469-8137.2005.01591.x [DOI] [PubMed] [Google Scholar]
- Lamport D. T., Northcote D. H. (1960). Hydroxyproline in primary cell walls of higher plants. Nature 188, 665–666 10.1038/188665b013757790 [DOI] [Google Scholar]
- Lamport D. T., Várnai P. (2013). Periplasmic arabinogalactan glycoproteins act as a calcium capacitor that regulates plant growth and development. New Phytol. 197, 58–64 10.1111/nph.12005 [DOI] [PubMed] [Google Scholar]
- Léonard R., Petersen B. O., Himly M., Kaar W., Wopfner N., Kolarich D., et al. (2005). Two novel types of O-glycans on the mugwort pollen allergen Art v 1 and their role in antibody binding. J. Biol. Chem. 280, 7932–7940 10.1074/jbc.M410407200 [DOI] [PubMed] [Google Scholar]
- Léonard R., Wopfner N., Pabst M., Stadlmann J., Petersen B. O., Duus J. O., et al. (2010). A new allergen from ragweed (Ambrosia artemisiifolia) with homology to Art v 1 from mugwort. J. Biol. Chem. 285, 27192–27200 10.1074/jbc.M110.127118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. Q., Faleri C., Geitmann A., Zhang H. Q., Cresti M. (1995). Immunogold localization of arabinogalactan proteins, unesterified and esterified pectins in pollen grains and pollen tubes of Nicotiana tabacum L. Protoplasma 189, 26–36 10.1007/BF01280289 [DOI] [Google Scholar]
- Liu C., Mehdy M. C. (2007). A nonclassical arabinogalactan protein gene highly expressed in vascular tissues, AGP31, is transcriptionally repressed by methyl jasmonic acid in Arabidopsis. Plant Physiol. 145, 863–874 10.1104/pp.107.102657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna C., Al-Assaf S., Phillips G. O., Funami T. (2006). The protein component in pectin - is it a AGP? Foods Food Ingred. J. Jpn. 211, 264–271 [Google Scholar]
- Merkouropoulos G., Shirsat A. H. (2003). The unusual Arabidopsis extensin gene atExt1 is expressed throughout plant development and is induced by a variety of biotic and abiotic stresses. Planta 217, 356–366 10.1007/s00425-003-1002-y [DOI] [PubMed] [Google Scholar]
- Micheli F. (2001). Pectin methylesterases: cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 6, 414–419 10.1016/S1360-1385(01)02045-3 [DOI] [PubMed] [Google Scholar]
- Mohnen D. (2008). Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 11, 266–277 10.1016/j.pbi.2008.03.006 [DOI] [PubMed] [Google Scholar]
- Mollet J. C., Kim S., Jauh G. Y., Lord E. M. (2002). Arabinogalactan proteins, pollen tube growth, and the reversible effects of Yariv phenylglycoside. Protoplasma 219, 89–98 10.1007/s007090200009 [DOI] [PubMed] [Google Scholar]
- Nguema-Ona E., Vicré-Gibouin M., Cannesan M. A., Driouich A. (2013). Arabinogalactan proteins in root-microbe interactions. Trends Plant Sci. 18, 440–449 10.1016/j.tplants.2013.03.006 [DOI] [PubMed] [Google Scholar]
- Nuñez A., Fishman M. L., Fortis L. L., Cooke P. H., Hotchkiss A. T. J. (2009). Identification of extensin protein associated with sugar beet pectin. J. Agric. Food Chem. 57, 10951–10958 10.1021/jf902162t [DOI] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M., Matsushita W., Matsubayashi Y. (2013). Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nat. Chem. Biol. 9, 726–730 10.1038/nchembio.1351 [DOI] [PubMed] [Google Scholar]
- Park S., Szumlanski A. L., Gu F., Guo F., Nielsen E. (2011). A role for CSLD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat. Cell Biol. 13, 973–980 10.1038/ncb2294 [DOI] [PubMed] [Google Scholar]
- Park Y. B., Cosgrove D. J. (2012). A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol. 158, 1933–1943 10.1104/pp.111.192880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J., Altmann F., Graf M., Stadlmann J., Reski R., Decker E. L. (2013). A gene responsible for prolyl-hydroxylation of moss-produced recombinant human erythropoietin. Sci. Rep. 3:319 10.1038/srep03019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerin P., Pellerin P., Vidal S., Williams P., Brillouet J. M. (1995). Characterization of five type II arabinogalactan-protein fractions from red wine of increasing uronic acid content. Carbohydr. Res. 277, 135–143 10.1016/0008-6215(95)00206-9 [DOI] [PubMed] [Google Scholar]
- Pena M. J., Kong Y., York W. S., O'Neill M. A. (2012). A galacturonic acid-containing xyloglucan is involved in Arabidopsis root hair tip growth. Plant Cell 24, 4511–4524 10.1105/tpc.112.103390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price N. J., Pinheiro C., Soares C. M., Ashford D. A., Ricardo C. P., Jackson P. A. (2003). A biochemical and molecular characterization of LEP1, an extensin peroxidase from lupin. J. Biol. Chem. 278, 41389–41399 10.1074/jbc.M304519200 [DOI] [PubMed] [Google Scholar]
- Qi X. Y., Behrens B. X., West P. R., Mort A. J. (1995). Solubilization and partial characterization of extensin fragments from cell walls of cotton suspension-cultures, evidence for a covalent cross-link between extensin and pectin. Plant Physiol. 108, 1691–1701 10.1104/pp.108.4.1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees D. A., Wight N. J. (1969). Molecular cohesion in plant cell walls. Methylation analysis of pectic polysaccharides from the cotyledons of white mustard. Biochem. J. 115, 431–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringli C. (2010). The hydroxyproline-rich glycoprotein domain of the Arabidopsis LRX1 requires Tyr for function but not for insolubilization in the cell wall. Plant J. 63, 662–669 10.1111/j.1365-313X.2010.04270.x [DOI] [PubMed] [Google Scholar]
- Rose J. K. C., Lee S.-J. (2010). Straying off the highway: trafficking of secreted plant proteins and complexity in the plant cell wall proteome. Plant Physiol. Biochem. 153, 433–436 10.1104/pp.110.154872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein A. L., Marquez J., Suarez-Cervera M., Bedinger P. A. (1995). Extensin-like glycoproteins in the maize pollen tube wall. Plant Cell 7, 2211–2225 10.1105/tpc.7.12.2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito F., Suyama A., Oka T., Yoko-O T., Matsuoka K., Jigami Y., et al. (2014). Identification of novel peptidyl serine O-galactosyltransferase gene family in plants. J. Biol. Chem. 289, 20405–20420 10.1074/jbc.M114.553933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvà I., Jamet E. (2001). Expression of the tobacco Ext1.4 extensin gene upon mechanical constraints and localization of regulatory regions. Plant Biol. 3, 1–10 10.1055/s-2001-11746 [DOI] [Google Scholar]
- Scheller H. V., Ulvskov P. (2010). Hemicelluloses. Annu. Rev. Plant Biol. 61, 263–289 10.1146/annurev-arplant-042809-112315 [DOI] [PubMed] [Google Scholar]
- Schnabelrauch L. S., Kieliszewski M. J., Upham B. L., Alizedeh H., Lamport D. T. (1996). Isolation of pI 4.6 extensin peroxidase from tomato cell suspension cultures and identification of Val-Tyr-Lys as putative intermolecular cross-link site. Plant J. 9, 477–489 10.1046/j.1365-313X.1996.09040477.x [DOI] [PubMed] [Google Scholar]
- Seifert G. J., Roberts K. (2007). The biology of arabinogalactan proteins. Annu. Rev. Plant Biol. 58, 137–161 10.1146/annurev.arplant.58.032806.103801 [DOI] [PubMed] [Google Scholar]
- Shirsat A. H., Wieczorek D., Kozbial P. (1996). A gene for Brassica napus extensin is differentially expressed on wounding. Plant Mol. Biol. 30, 1291–1300 10.1007/BF00019559 [DOI] [PubMed] [Google Scholar]
- Showalter A. M., Keppler B., Lichtenberg J., Gu D., Welch L. R. (2010). A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 153, 485–513 10.1104/pp.110.156554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E., Barbar E., Leykam J. F., Kieliszewski M. J. (2001). Contiguous hydroxyproline residues direct hydroxyproline arabinosylation in Nicotiana tabacum. J. Biol. Chem. 276, 11272–11278 10.1074/jbc.M011323200 [DOI] [PubMed] [Google Scholar]
- Sommer-Knudsen J., Clarke A. E., Bacic A. (1996). A galactose-rich, cell-wall glycoprotein from styles of Nicotiana alata. Plant J. 9, 71–83 10.1046/j.1365-313X.1996.09010071.x [DOI] [PubMed] [Google Scholar]
- Stratford S., Barne W., Hohorst D. L., Sagert J. G., Cotter R., Golubiewski A., et al. (2001). A leucine-rich repeat region is conserved in pollen extensin-like (Pex) proteins in monocots and dicots. Plant Mol. Biol. 46, 43–56 10.1023/A:1010659425399 [DOI] [PubMed] [Google Scholar]
- Tan L., Eberhard S., Pattathil S., Warder C., Glushka J., Yuan C., et al. (2013). An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25, 270–287 10.1105/tpc.112.107334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Qiu F., Lamport D. T., Kieliszewski M. J. (2004). Structure of a hydroxyproline (Hyp)-arabinogalactan polysaccharide from repetitive Ala-Hyp expressed in transgenic Nicotiana tabacum. J. Biol. Chem. 279, 13156–13165 10.1074/jbc.M311864200 [DOI] [PubMed] [Google Scholar]
- Tan L., Showalter A. M., Egelund J., Hernandez-Sanchez A., Doblin M. S., Bacic A. (2012). Arabinogalactan-proteins and the research challenges for these enigmatic plant cell surface proteoglycans. Front. Plant Sci. 3:140 10.3389/fpls.2012.00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Varnai P., Lamport D. T., Yuan C., Xu J., Qiu F., et al. (2010). Plant O-hydroxyproline arabinogalactans are composed of repeating trigalactosyl subunits with short bifurcated side chains. J. Biol. Chem. 285, 24575–24583 10.1074/jbc.M109.100149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiainen P., Myllyharju J., Koivunen P. (2005). Characterization of a second Arabidopsis thaliana prolyl 4-hydroxylase with distinct substrate specificity. J. Biol. Chem. 280, 1142–1148 10.1074/jbc.M411109200 [DOI] [PubMed] [Google Scholar]
- Tryfona T., Liang H. C., Kotake T., Tsumuraya Y., Stephens E., Dupree P. (2012). Structural characterization of Arabidopsis leaf arabinogalactan polysaccharides. Plant Physiol. 160, 563–666 10.1104/pp.112.202309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin R., Cerclier C., Geneix N., Aguie-Beghin V., Gaillard C., Ralet M. C., et al. (2010). Elaboration of extensin-pectin thin film model of primary plant cell wall. Langmuir 26, 9891–9898 10.1021/la100265d [DOI] [PubMed] [Google Scholar]
- Velasquez S. M., Ricardi M. M., Dorosz J. G., Fernandez P. V., Nadra A. D., Pol-Fachin L., et al. (2011). O-glycosylated cell wall proteins are essential in root hair growth. Science 332, 1401–1403 10.1126/science.1206657 [DOI] [PubMed] [Google Scholar]
- Velasquez S. M., Salgado Salter J., Petersen B. L., Estevez J. M. (2012). Recent advances on the post-translational modifications of EXTs and their roles in plant cell walls. Front. Plant Sci. 3:93 10.3389/fpls.2012.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F., Spano T., Vlad D., Daher F. B., Ouelhadj A., Fragkostefanakis S., et al. (2007). Involvement of Arabidopsis prolyl 4 hydroxylases in hypoxia, anoxia and mechanical wounding. Plant Signal. Behav. 2, 368–369 10.4161/psb.2.5.4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F., Tiainen P., Owen C., Spano T., Daher F. B., Oualid F., et al. (2010). Characterization of two carnation petal prolyl 4 hydroxylases. Physiol. Plant. 140, 199–207 10.1111/j.1399-3054.2010.01390.x [DOI] [PubMed] [Google Scholar]
- Voragen A. G. J., Coenen G. J., Verhoef R. P., Schols H. A. (2009). Pectin, a versatile polysaccharide present in plant cell walls. Struct. Chem. 20, 263–275 10.1007/s11224-009-9442-z [DOI] [Google Scholar]
- Wang T., Zabotina O., Hong M. (2012). Pectin-cellulose interactions in the Arabidopsis primary cell wall from two-dimensional magic-angle-spinning solid-state nuclear magnetic resonance. Biochemistry 51, 9846–9856 10.1021/bi3015532 [DOI] [PubMed] [Google Scholar]
- Willats W. G. T., Knox P., Mikkelsen J. D. (2006). Pectin: new insights into an old polymer are starting to gel. Trends Food Sci. Technol. 17, 97–104 10.1016/j.tifs.2005.10.008 [DOI] [Google Scholar]
- Wu H., De Graaf B., Mariani C., Cheung A. Y. (2001). Hydroxyproline-rich glycoproteins in plant reproductive tissues: structure, functions and regulation. Cell. Mol. Life Sci. 58, 1418–1429 10.1007/PL00000785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Wang H., Cheung A. Y. (1995). A pollen tube growth stimulatory glycoprotein is deglycosylated by pollen tubes and displays a glycosylation gradient in the flower. Cell 82, 395–403 10.1016/0092-8674(95)90428-X [DOI] [PubMed] [Google Scholar]
- Yuasa K., Toyooka K., Fukuda H., Matsuoka K. (2005). Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. Plant J. 41, 81–94 10.1111/j.1365-313X.2004.02279.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.