Abstract
Several types of genetic and epigenetic regulation have been implicated in the development of drug resistance, one significant challenge for cancer therapy. Although changes in the expression of non-coding RNA are also responsible for drug resistance, the specific identities and roles of them remain to be elucidated. Long non-coding RNAs (lncRNAs) are a type of ncRNA (> 200 nt) that influence the regulation of gene expression in various ways. In this study, we aimed to identify differentially expressed lncRNAs in 5-fluorouracil-resistant colon cancer cells. Using two pairs of 5-FU-resistant cells derived from the human colon cancer cell lines SNU-C4 and SNU-C5, we analyzed the expression of 90 lncRNAs by qPCR-based profiling and found that 19 and 23 lncRNAs were differentially expressed in SNU-C4R and SNU-C5R cells, respectively. We confirmed that snaR and BACE1AS were downregulated in resistant cells. To further investigate the effects of snaR on cell growth, cell viability and cell cycle were analyzed after transfection of siRNAs targeting snaR. Down-regulation of snaR decreased cell death after 5-FU treatment, which indicates that snaR loss decreases in vitro sensitivity to 5-FU. Our results provide an important insight into the involvement of lncRNAs in 5-FU resistance in colon cancer cells.
Keywords: 5-Fluorouracil, cell viability, drug resistance, long non-coding RNAs, snaR
INTRODUCTION
Drug resistance is considered a multifactorial phenomenon that results from a variety of factors, including individual variations in patients and genetic and/or epigenetic differences in tumors (Raguz and Yague, 2008; Tan et al., 2010). Although chemotherapy has been widely used for cancer treatment, acquisition of drug resistance is considered a substantial obstacle in effective chemotherapy (Kang et al., 2013). Altered expression or mutation of transporter proteins that increase drug efflux from cancer cells, reduce uptake of drugs, increase repair of DNA damage, decrease sensitivity resulting from induction of apoptosis, and accelerate drug metabolism are responsible for the development of drug resistance (Fojo, 2007; Glasspool et al., 2006; Roberti et al., 2006; Tan et al., 2010). Recently, several studies have shown that non-mutational regulation of gene expression by microRNAs (miRNAs) is also largely involved in the acquisition of drug resistance (Fojo, 2007; Mishra, 2012; Mishra and Bertino, 2009; Xu et al., 2013).
5-Fluorouracil (5-FU), a classical anti-metabolite, is widely used for cancer treatment and results in cytotoxic effects that cause cell death by affecting nucleoside metabolism [International Multicentre Polled Analysis of Colon Cancer Trials (IMPACT) investigators, 1995; Grem, 2000; Zhang et al., 2008]. However, clinical applications of 5-FU have been limited by drug resistance (Peters et al., 2002; Zhang et al., 2008). Although several efforts have been made to elucidate the molecular events causing the 5-FU-resistant phenotype, no convincing findings have been reported with regard to simple gene expression changes (Boyer et al., 2006; Karasawa et al., 2009; Kurokawa et al., 2012; Mariadason et al., 2003; Ooyama et al., 2006). Therefore, multiple and distinct factors may be related to primary or acquired 5-FU resistance.
Genome-wide studies have revealed that different types of non-coding RNAs (ncRNAs) regulate gene expression (Djebali et al., 2012; Taft et al., 2010). Small ncRNAs such as siRNAs, miRNAs, and piRNAs are highly conserved and engage in transcriptional and posttranscriptional gene silencing by interacting with their targets. Long non-coding RNAs (lncRNAs) have a length greater than 200 nt, are poorly conserved, and belong to a novel heterogeneous class of ncRNAs that includes thousands of different species (Mercer et al., 2009; Whitehead et al., 2009; Wilusz et al., 2009). lncRNAs are emerging as new players in gene regulation that affect various stages of gene expression through diverse mechanisms that have not yet been fully elucidated (Batista and Chang, 2013; Rinn and Chang, 2012). Several studies have indicated that the expression of lncRNAs is tightly controlled with regard to cell and tissue distribution, and aberrant expression is involved in development and diseases, including cancers (Esteller, 2011; Fatica and Bozzoni, 2013; Gibb et al., 2011; Wang and Chang, 2011). However, the functional link between lncRNAs and the acquisition of drug resistance is unclear.
In this study, we explored the altered expression of lncRNAs in two different 5-FU-resistant human colon cancer cell lines and further investigated the role of snaR, which is a downregulated lncRNA in 5-FU-resistant cells. Our results provide experimental evidence that differential expression of lncRNAs such as snaR is responsible for altered chemosensitivity to 5-FU and provide a possible link between lncRNAs and 5-FU resistance.
MATERIALS AND METHODS
Cell culture and transfection
The human colon cancer cell lines SNU-C4 and SNU-C5 (Oh et al., 1999; Park et al., 1987), and their individual 5-FU-resistant cell lines, SNU-C4R and SNU-C5R, were obtained from the Korean Cell Line Bank (Korea) (Choi et al., 2011; Shin et al., 2005; 2009). SNU-C4, SNU-C5, and HCT-116 cells were maintained in RPMI medium (Thermo Scientific Inc.) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Resistant cells were maintained in RPMI/10% FBS/1% penicillin/streptomycin and exposed to IC50 value to 5-FU (Sigma) for 72 h. siRNAs (siCTRL, 5′-AAUUCUCCGAACGUGUCACGU-3′; and sisnaR, 5′-CCACAUGGGUCGGAAAAAAUU-3′) were synthesized (Genolution, Korea) and transfected using Lipofectamine 2000 (Life Technologies).
MTT assay
A colorimetric assay using the tetrazolium salt, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), was used to assess cell viability. After 24 h of transfection with siRNA, cells were exposed to 10 μM 5-FU for 72 h. Additionally, 0.5 mg/mL of MTT was added to each well and incubated for 3 h. Plates were centrifuged at 450 × g for 5 min at room temperature, the medium was removed, and then 100 μl of 40 mM acidic isopropanol was added to solubilize the crystals. The absorbance was measured at 570 mm using a microplate reader, Victor 3 (Perkin Elmer, Finland).
RNA analysis and RT-qPCR
Total RNA was prepared from whole cells by using TRIzol reagent (Invitrogen). After reverse transcription (RT) using random hexamers and reverse transcriptase (Toyobo, Japan), the abundance of transcripts was assessed by quantitative PCR (qPCR) analysis by using SYBR green PCR master mix (Kapa Biosystems) and the following gene-specific primer sets: snaR_Fwd: 5′-TGGAGCCATTGTGGCTCCGGCC-3′, snaR_Rev: 5′-CCCATGTGGACCAGGTTGGCCT-3′; PRAS_Fwd: 5′-GCCTCGGGTTGTAGATTTCA-3′, PRAS_Rev: 5′-AGGTCCGGTAATTGGGGTAG-3′; BACE1AS_Fwd: 5′-ATTTCACCCT GTTGGTCAGG-3′, BACE1AS_Rev: 5′-TCAGCAACAGCCA AGATGTC-3′; GAPDH_Fwd: 5′-TGCACCACCAACTGCTTAGC-3′, GAPDH_Rev: 5′-GGCATGGACTGTGGTCATGAG-3′. RT-qPCR analysis was performed using a StepOne Plus™ instrument (Life Technologies). GAPDH mRNA was used as the internal control for normalization. The relative expression of transcripts was analyzed using the 2−ΔΔCt method.
Flow cytometric analysis
The cell cycle was evaluated by fluorescence-activated cell-sorting (FACS) analysis of propidium iodide-stained nuclei as previously described (Fulda et al., 1998). After transfection of siRNA and/or 5-FU treatment, cells were stained with propidium iodide (Sigma), and the cell cycle was analyzed by flow cytometry (FACSCalibur). Cell death was determined by staining Annexin V using Aposcan kit (Biobud, Korea) according to the manufacturers’ protocol.
lncRNA profiling
lncRNA profiling was performed using lncRNA profiler™ qPCR arrays (System Bioscience, Inc., USA) consisting of 90 lncRNAs. Total RNA was isolated from each cell line by using TRIzol reagent (Invitrogen). cDNAs for lncRNA profiling were synthesized by reverse transcription after polyadenylation and annealing of oligo-dTs, and RT-qPCR reaction was performed according to the manufacturer’s protocol.
RESULTS
Chemosensitivity of SNU-C4R and SNU-C5R cells to 5-FU
Two 5-FU-resistant SNU-C4R and SNU-C5R cell lines were previously established from human colon cancer cells, SNU-C4 and SNU-C5, respectively (Choi et al., 2011; Jung et al., 2007; Shin et al., 2005; 2009). We confirmed the relative chemosensitivity of these resistant cell lines against 5-FU using MTT assay. 5-FU treatment for 72 h resulted in a dose-dependent suppression of cell growth (Fig. 1). The IC50 values for 5-FU in SNU-C4R and SNU-C5R cells were 105.0 ± 14.5 μM and 118.7 ± 4.9 μM, respectively, and the corresponding values for their parental cells, SNU-C4 and SNU-C5 cells were 8.3 ± 4.7 μM and 23.2 ± 3.4 μM, respectively.
Fig. 1.
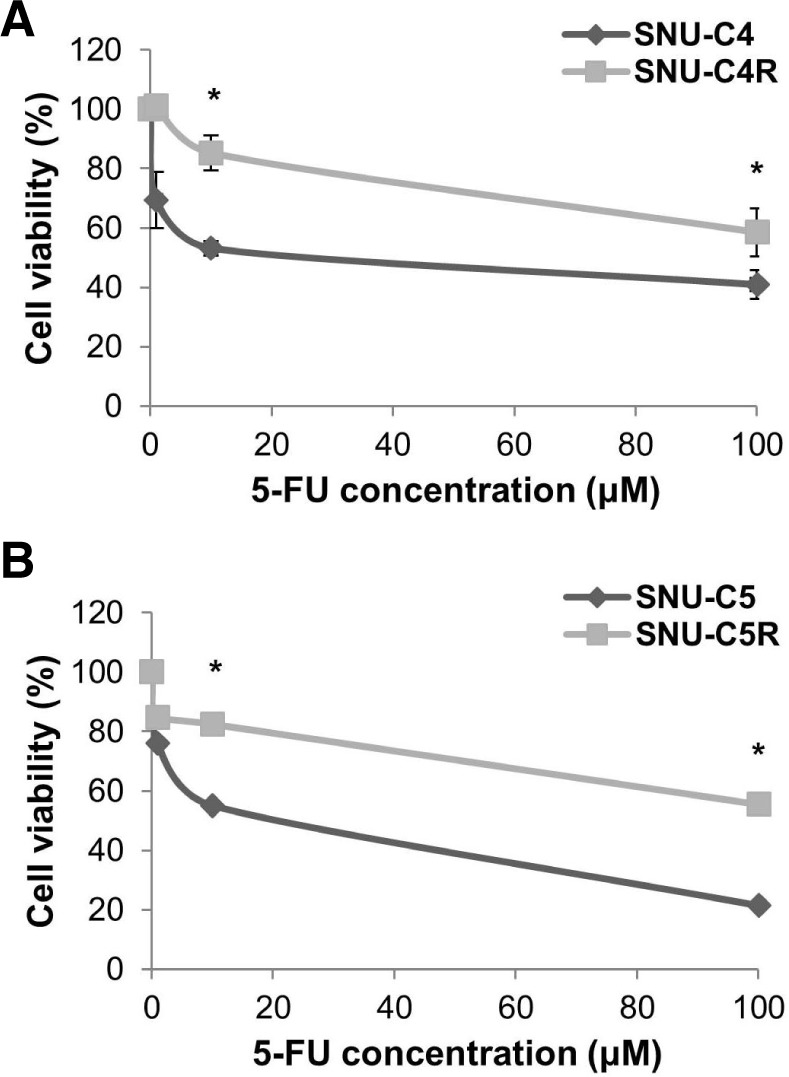
Chemosensitivity of 5-FU-resistant human colon cancer cells, SNU-C4R and SNU-C5R. Human colon cancer cells (SNU-C4 and SNU-C5) and their 5-FU-resistant cells (SNU-C4R and SNU-C5R) were exposed to the indicated concentration of 5-FU. After 72 h, cell viability was assessed by MTT assay. (A) SNU-C4 vs. SNU-C4R, (B) SNU-C5 vs. SNU-C5R. Data represent the mean ± SEM from 3 independent experiments. *p < 0.05.
Differential expression of lncRNAs in 5-FU-resistant cells
To explore the potential role of long non-coding RNAs (lnc-RNAs) in 5-FU resistance, we investigated the differential expression of lncRNAs between 5-FU-resistant cells and their parental cells by performing qPCR-based lncRNA profiling according to the manufacturer’s protocol (System Bioscience, Inc.). We analyzed the relative expression of 90 lncRNAs and observed differential expression of lncRNAs in the 5-FU-resistant cell lines. In all, in SNU-C4R and SNU-C5R cells, 16 and 12 lncRNAs were down-regulated by more than 1.5-fold, whereas 3 and 10 lncRNAs were up-regulated by more than 1.5-fold, respectively (Tables 1 and 2). These results suggest that these lncRNAs may play roles in the development of 5-FU resistance in human colon cancer cells.
Table 1.
Differentially expressed lncRNAs in SNU-C4R cells
| NCBI Reference sequence | lncRNAs | Expression in SNU-C4R/SNU-C4 |
|---|---|---|
| N/A | PRAS | 0.20 |
| NR_002819.2 | Malat1 | 0.34 |
| N/A | NTT | 0.44 |
| NR_003245.1 | HAR1B | 0.48 |
| N/A | DHFR upstream transcripts | 0.49 |
| NR_002795.2 | Hoxa11as | 0.51 |
| NR_002578.2 | GAS5-family | 0.53 |
| NR_004435.1 | snaR | 0.53 |
| NR_004428.1 | EGO B | 0.54 |
| NR_002196.1 | H19 | 0.59 |
| NR_002770.1 | Dio3os | 0.62 |
| N/A | E2F4 antisense | 0.63 |
| NR_047514.1 | Air | 0.66 |
| NR_037803.1 | BACE1AS (family) | 0.66 |
| NR_023917.1 | PTENP1 | 0.64 |
| N/A | TEA ncRNAs | 0.64 |
| NR_001568.1 | BC200 | 2.38 |
| NR_015391.1 | LOC285194 | 2.12 |
| NR_023388.1 | PRINS | 2.10 |
N/A, NCBI reference sequence information is not available.
Table 2.
Differentially expressed lncRNAs in SNU-C5R cells
| NCBI Reference sequence | lncRNAs | Expression in SNU-C5R/SNU-C5 |
|---|---|---|
| NR_003529.3 | ANRIL | 0.42 |
| NR_004435.1 | snaR | 0.47 |
| NR_024582.1 | Jpx | 0.55 |
| NR_002791.2 | Emx2os | 0.60 |
| N/A | lincRNA-VLDLR | 0.61 |
| NR_003141.3 | SNHG4 | 0.61 |
| N/A | HULC | 0.62 |
| NR_002323.1 | TUG1 (family) | 0.63 |
| NR_024281 | RNCR3 | 0.64 |
| NR_015391.1 | LOC285194 | 0.65 |
| NR_002795.2 | Hoxa11as | 0.65 |
| NR_015379.3 | UCA1 | 0.66 |
| NR_037803.1 | BACE1AS (family) | 0.83 |
| N/A | Evf1 and EVF2 | 3.04 |
| NR_004428.1 | EGO B | 2.77 |
| N/A | PRAS | 2.40 |
| N/A | E2F4 antisense | 1.96 |
| N/A | lincRNA-p21 | 1.91 |
| NR_002196.1 | H19 | 1.64 |
| N/A | L1PA16 | 1.58 |
| N/A | IPW | 1.56 |
| NR_023920 | WT1-AS | 1.54 |
| NR_002578.2 | GAS5-family | 1.52 |
N/A, NCBI reference sequence information is not available.
Validation of array data by RT-qPCR
To validate the results from the profiling analysis, we selected three lncRNAs, snaR, BACE1AS, and PRAS, and we detected their expression by RT-qPCR using specific primer sets. SnaR and BACE1AS were significantly down-regulated in both resistant cell lines (SNU-C4R and SNU-C5R), whereas PRAS was down-regulated in SNU-C4R cells but not in SNU-C5R cells (Fig. 2). These results indicate that the expression of the three lncRNAs assessed by RT-qPCR was consistent with the lncRNA profiling analysis shown in Tables 1 and 2. From these results, we hypothesized that differential expression of lncRNAs may have functions in the 5-FU resistance in colon cancer cells.
Fig. 2.
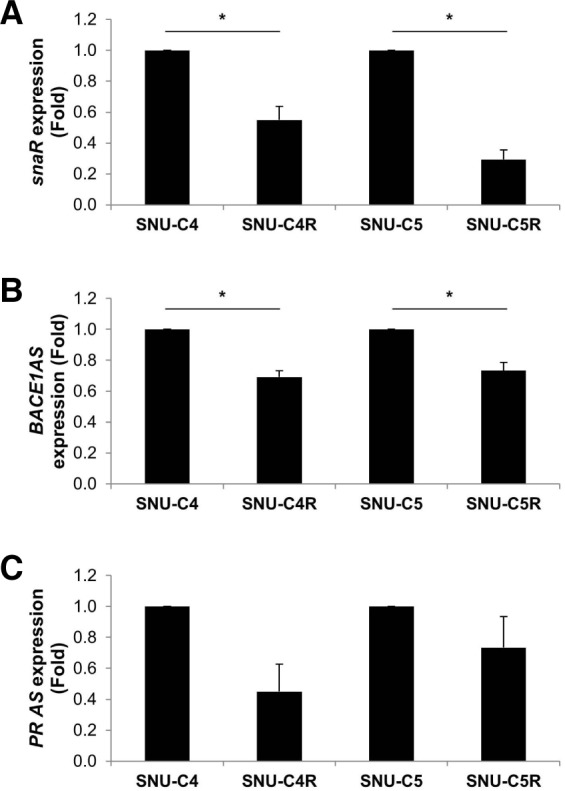
Validation of differentially expressed lncRNAs in 5-FU-resistant cells. Total RNAs were isolated from 5-FU-resistant cell lines (SNU-C4R and SNU-C5R) and their parental cells (SNU-C4 and SNU-C5), and cDNAs were synthesized by reverse transcripttion. Relative expression of (A) snaR, (B) BACE1AS, and (C) PRAS among cell lines was analyzed by RT-qPCR. Data represent the mean ± SEM from 3 independent experiments. *p < 0.05.
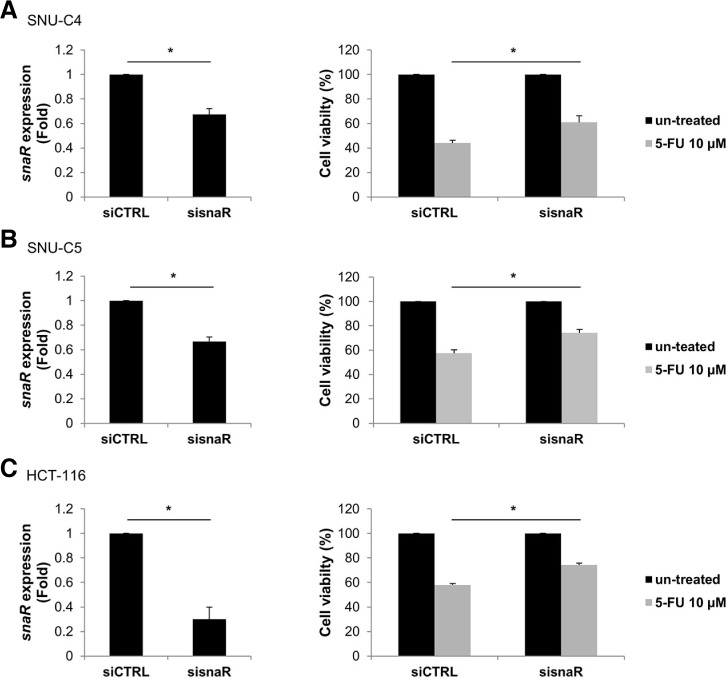
Effect of snaR on cell growth after 5-FU treatment
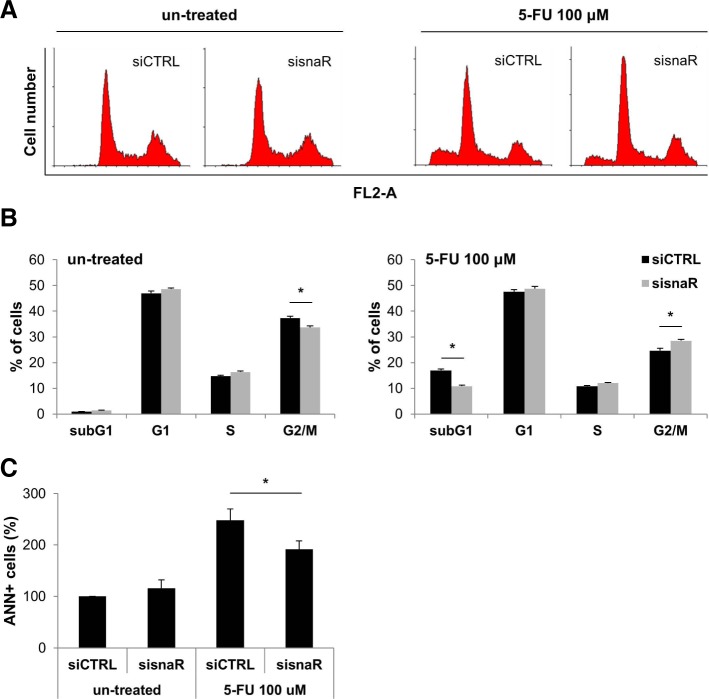
To explore whether the differentially expressed lncRNAs are involved in the regulation of 5-FU responsiveness, we further examined the effect of snaR, which was down-regulated in both 5-FU-resistant cell lines, on cell growth. Because resistant cells showed down-regulation of snaR, we hypothesized that the loss of snaR is responsible for the increase in cell viability after 5-FU treatment. To validate this hypothesis, we investigated the effect of snaR down-regulation on cell viability after 5-FU treatment. Small interfering RNA against snaR was synthesized and transfected into SNU-C4, SNU-C5 and HCT-116 cells, and snaR expression level was then analyzed by RT-qPCR. Transfection of siRNA targeting snaR significantly down-regulated snaR expression in each cell lines (Figs. 3A, 3B, and 3C, left). After transfections of sisnaR or siCTRL, cells were exposed to 5-FU for 72 h, and then cell viability was assessed by MTT assay. Down-regulation of snaR resulted in increased cell viability after 5-FU treatment (Figs. 3A, 3B and 3C, right). To further investigate the effect of snaR on cell cycle regulation, the cell cycle distribution and cell death were analyzed by flow cytometry (Fig. 4). snaR down-regulation did not produce significant changes in cell cycle distribution at the baseline (Figs. 4A and 4B, left); however, it resulted in a significant decrease of sub-G1 phase of the cell cycle after 5-FU treatment (Figs. 4A and 4B, right). Additionally, snaR down-regulation decreased Annexin V-positive (ANN+) apoptotic cells after 5-FU treatment (Fig. 4C). These results indicate that loss of snaR, which was observed in 5-FU-resistant cells, is responsible for the development of 5-FU resistance in colon cancer cells.
Fig. 3.
Down-regulation of snaR decreases in the vitro sensitivity of human colon cancer cells to 5-FU. SNU-C4 and HCT-116 cells were transfected with siRNAs targeting snaR. After 96 h of transfection, relative expression of snaR in (A, left) SNU-C4, (B, left) SNU-C5 or (C, left) HCT-116 cells was measured by RTqPCR. GAPDH mRNA was used as an internal control. After transfection of siRNAs, cells were exposed to 10 μM 5-FU for 72 h and cell viability was assessed by MTT assay (A, right) SNU-C4, (B, right) SNU-C5 or (C, right) HCT-116 cells. Data represent the mean ± SEM from 3 independent experiments. *p < 0.05.
Fig. 4.
Down-regulation of snaR decreases cell death after 5-FU treatment. After transfection of siRNA targeting snaR or control siRNA, HCT-116 cells were exposed to 100 μM 5-FU for 16 h. (A and B) Cells were stained with propidium iodide and the distribution of cell cycles between untreated (left) and 5-FU-treated (right) cells were analyzed by flow cytometry. The cytograms are representative of three independent experiments. (C) For measure of apoptotic cells, the cells were stained with both propidium iodide and Annexin V and analyzed by flow cytometry. The results represent the mean ± SEM from 3 independent experiments. *p < 0.05.
DISCUSSION
Anti-cancer drug resistance remains one of the most significant challenges to successful treatment of cancer (Gottesman, 2002). Although the mechanism of drug resistance has not been fully elucidated, many studies have revealed that genetic alterations of several genes encoding membrane transporters, drug metabolizers, and cell cycle regulators, and those involved in the DNA repair process are responsible for the development of drug resistance (Ganguly et al., 2011). In addition, numerous studies have also indicated the involvement of substantial epigenetic regulation by DNA methylation or miRNAs in drug resistance (Ma et al., 2010; Rukov and Shomron, 2011; Strathdee, 2007; Wang et al., 2010; Zheng et al., 2010).
In addition to miRNAs, lncRNAs also participate in a wide spectrum of biological processes through diverse mechanisms that affect transcription, mRNA splicing, mRNA decay, translation, and protein-protein interaction (Batista and Chang, 2013; Gibb et al., 2011; Wang and Chang, 2011). Recent studies have shown that lncRNAs such as HOTAIR and UCA1 contribute to cisplatin resistance (Fan et al., 2014; Liu et al., 2013). However, lncRNA expression in drug resistance and its physiopathological significance have not yet been fully elucidated.
Here, we hypothesized that treatment of an anti-cancer drug, 5-FU, may be associated with the alteration of lncRNAs profiles. Therefore, we investigated differentially expressed lncRNAs in two 5-FU-resistant human colon cancer cell lines. We found that several lncRNAs such as snaR, BACE1AS, and PRAS were differentially expressed in 5-FU-resistant cells. Although both resistant cells showed chemoresistance to 5-FU, there was little correlation in the expression of lncRNAs, which might be resulted from the different genetic background of their parental cells (Oh et al., 1999; Park et al., 1987). Our results suggested that the differential expression of these lncRNAs is involved in the development of 5-FU resistance in human colon cancer cells. Furthermore, the potential role of snaR in the regulation of 5-FU responsiveness was implicated. SnaR is an lncRNA (~117 nt) transcribed by RNA polymerase III that associates with nuclear factor 90 (NF90; a double-stranded RNA-binding protein implicated in multiple cellular functions) (Parrott and Mathews, 2007). Several snaR transcripts have been found in diverse cell lines and human tissues, and they associate with ribosomes in the cytoplasm (Parrott and Mathews, 2009; Parrott et al., 2011). Although previous reports suggest that snaR transcripts are involved in tissue- and species-specific regulation of cell growth and translation, the detailed functions of snaR have not yet been elucidated.
In this study, we found that snaR was down-regulated in 5-FU-resistant colon cancer cells, and snaR loss increased cell viability after 5-FU treatment, which suggests that snaR has a potential role as a negative regulator of cell growth in response to 5-FU. Further experiments will be necessary to obtain more information regarding the roles of snaR in 5-FU resistance. We believe that our results may be useful as a starting point for further studies to elucidate the molecular functions of lncRNAs in drug resistance and to assess their therapeutic potentials.
Acknowledgments
This work is supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government [Ministry of Education, Science and Technology (MEST)] (2012M3A9 D105451, 2012R1A5A2047939).
REFERENCES
- Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer J, Allen WL, McLean EG, Wilson PM, McCulla A, Moore S, Longley DB, Caldas C, Johnston PG. Pharmacogenomic identification of novel determinants of response to chemotherapy in colon cancer. Cancer Res. 2006;66:2765–2777. doi: 10.1158/0008-5472.CAN-05-2693. [DOI] [PubMed] [Google Scholar]
- Choi CH, Lee TB, Lee YA, Choi S, Kim KJ. Upregulation of cyclooxygenase-2-derived prostaglandin E(2) in colon cancer cells resistant to 5-fluorouracil. J Korean Surg Soc. 2011;81:115–121. doi: 10.4174/jkss.2011.81.2.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y. Long non-coding RNA UCA1 increases chemoresistances of bladder cancer cells by regulating Wnt signaling. FEBS J. 2014;281:1750–1758. doi: 10.1111/febs.12737. [DOI] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2013;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- Fojo T. Multiple paths to a drug resistance phenotype: mutations, translocations, deletions and amplification of coding genes or promoter regions, epigenetic changes and microRNAs. Drug Resist Updat. 2007;10:59–67. doi: 10.1016/j.drup.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Fulda S, Susin SA, Kroemer G, Debatin KM. Molecular ordering of apoptosis induced by anticancer drugs in neuroblastoma cells. Cancer Res. 1998;58:4453–4460. [PubMed] [Google Scholar]
- Ganguly A, Banerjee K, Chakraborty P, Das S, Sarkar A, Hazra A, Banerjee M, Maity A, Chatterjee M, Mondal NB, et al. Overcoming multidrug resistance (MDR) in cancer in virto and in vivo by a quinoline derivative. Biomed Pharmacother. 2011;65:387–394. doi: 10.1016/j.biopha.2011.04.024. [DOI] [PubMed] [Google Scholar]
- Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasspool RM, Teodoridis JM, Brown R. Epigenetics as a mechanism driving polygenic clinical drug resistance. Br J Cancer. 2006;94:1087–1092. doi: 10.1038/sj.bjc.6603024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman MM. Mechanisms of cancer drug resistance. Ann Rev Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- Grem JL. 5-Fluorouracil: forty-plus and still ticking. A review of its preclinical and clinical development. Invest New Drugs. 2000;18:299–313. doi: 10.1023/a:1006416410198. [DOI] [PubMed] [Google Scholar]
- International Multicentre Pooled Analysis of Colon Cancer Trials (IMPACT) investigators Efficacy of adjuvant fluorouracil and folinic acid in colon cancer. Lancet. 1995;345:939–944. [PubMed] [Google Scholar]
- Jung GR, Kim KJ, Choi CH, Lee TB, Han SI, Han HK, Lim SC. Effect of betulinic acid on anticancer drug-resistant colon cancer cells. Basic Clin Pharmacol Toxicol. 2007;101:277–285. doi: 10.1111/j.1742-7843.2007.00115.x. [DOI] [PubMed] [Google Scholar]
- Kang H, Kim C, Lee H, Kim W, Lee EK. Post-transcriptional controls by ribonucleoprotein complexes in the acquisition of drug resistance. Int J Mol Sci. 2013;14:17204–17220. doi: 10.3390/ijms140817204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasawa H, Miura K, Fujibuchi W, Ishida K, Kaneko N, Kinouchi M, Okabe M, Ando T, Murata Y, Sasaki H, et al. Down-regulation of cIAP2 enhances 5-FU sensitivity through the apoptotic pathway in human colon cancer cells. Cancer Sci. 2009;100:903–913. doi: 10.1111/j.1349-7006.2009.01112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K, Tanahashi T, Iima T, Yamamoto Y, Akaike Y, Nishida K, Masuda K, Kuwano Y, Murakami Y, Fukushima M, et al. Role of miR-19b and its target mRNAs in 5-fluorouracil resistance in colon cancer cells. J Gastroenterol. 2012;47:883–895. doi: 10.1007/s00535-012-0547-6. [DOI] [PubMed] [Google Scholar]
- Liu Z, Sun M, Lu K, Liu J, Zhang M, Wu W, De W, Wang Z, Wang R. The long noncoding RNA HOTAIR contributes to cisplatin resistance of human lung adenocarcinoma cells via downregualtion of p21(WAF1/CIP1) expression. PLoS One. 2013;8:e77293. doi: 10.1371/journal.pone.0077293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Dong C, Ji C. MicroRNA and drug resistance. Cancer Gene Ther. 2010;17:523–531. doi: 10.1038/cgt.2010.18. [DOI] [PubMed] [Google Scholar]
- Mariadason JM, Arango D, Shi Q, Wilson AJ, Corner GA, Nicholas C, Aranes MJ, Lesser M, Schwartz EL, Augenlicht LH. Gene expression profiling-based prediction of response of colon carcinoma cells to 5-fluorouracil and camptothecin. Cancer Res. 2003;63:8791–8812. [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Mishra PJ. The miRNA-drug resistance connection: a new era of personalized medicine using noncoding RNA begins. Pharmacogenomics. 2012;13:1321–1324. doi: 10.2217/pgs.12.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PJ, Bertino JR. MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine. Pharmacogenomics. 2009;10:399–416. doi: 10.2217/14622416.10.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JH, Ku JL, Yoon KA, Kwon HJ, Kim WH, Park HS, Yeo KS, Song SY, Chung JK, Park JG. Establishment and characterization of 12 human colorectal-carcinoma cell lines. International journal of cancer. J Int Cancer. 1999;81:902–910. doi: 10.1002/(sici)1097-0215(19990611)81:6<902::aid-ijc11>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Ooyama A, Takechi T, Toda E, Nagase H, Okayama Y, Kitazato K, Sugimoto Y, Oka T, Fukushima M. Gene expression analysis using human cancer xenografts to identify novel predictive marker genes for the efficacy of 5-fluorouracil-based drugs. Cancer Sci. 2006;97:510–522. doi: 10.1111/j.1349-7006.2006.00204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JG, Oie HK, Sugarbaker PH, Henslee JG, Chen TR, Johnson BE, Gazdar A. Characteristics of cell lines established from human colorectal carcinoma. Cancer Res. 1987;47:6710–6718. [PubMed] [Google Scholar]
- Parrott AM, Mathews MB. Novel rapidly evolving hominid RNAs bind nuclear factor 90 and display tissue-restricted distribution. Nucleic Acids Res. 2007;35:6249–6258. doi: 10.1093/nar/gkm668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott AM, Mathews MB. snaR genes: recent descendants of Alu involved in the evolution of chorionic gonadotropins. Cold Spring Harbor Symp Quant Biol. 2009;74:363–373. doi: 10.1101/sqb.2009.74.038. [DOI] [PubMed] [Google Scholar]
- Parrott AM, Tsai M, Batchu P, Ryan K, Ozer HL, Tian B, Mathews MB. The evolution and expression of the snaR family of small non-coding RNAs. Nucleic Acids Res. 2011;39:1485–1500. doi: 10.1093/nar/gkq856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters GJ, Backus HH, Freemantle S, van Triest B, Codacci-Pisanelli G, van der Wilt CL, Smid K, Lunec J, Calvert AH, Marsh S, et al. Induction of thymidylate synthase as a 5-fluorouracil resistance mechanism. Biochim Biophys Acta. 2002;1587:194–205. doi: 10.1016/s0925-4439(02)00082-0. [DOI] [PubMed] [Google Scholar]
- Raguz S, Yague E. Resistance to chemotherapy: new treatments and novel insights into an old problem. Br J Cancer. 2008;99:387–391. doi: 10.1038/sj.bjc.6604510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Ann Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberti A, La Sala D, Cinti C. Multiple genetic and epigenetic interacting mechanisms contribute to clonally selection of drug-resistant tumors: current views and new therapeutic prospective. J Cell Physiol. 2006;207:571–581. doi: 10.1002/jcp.20515. [DOI] [PubMed] [Google Scholar]
- Rukov JL, Shomron N. MicroRNA pharmacogenomics: post-transcriptional regulation of drug response. Trends Mol Med. 2011;17:412–423. doi: 10.1016/j.molmed.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Shin YK, Yoo BC, Chang HJ, Jeon E, Hong SH, Jung MS, Lim SJ, Park JG. Down-regulation of mitochondrial F1F0-ATP synthase in human colon cancer cells with induced 5-fluorouracil resistance. Cancer Res. 2005;65:3162–3170. doi: 10.1158/0008-5472.CAN-04-3300. [DOI] [PubMed] [Google Scholar]
- Shin YK, Yoo BC, Hong YS, Chang HJ, Jung KH, Jeong SY, Park JG. Upregulation of glycolytic enzymes in proteins secreted from human colon cancer cells with 5-fluorouracil resistance. Electrophoresis. 2009;30:2182–2192. doi: 10.1002/elps.200800806. [DOI] [PubMed] [Google Scholar]
- Strathdee G. Epigenetic markers and response to chemotherapy in cancer. Dis Markers. 2007;23:43–49. doi: 10.1155/2007/610815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- Tan DS, Gerlinger M, Teh BT, Swanton C. Anticancer drug resistance: understanding the mechanisms through the use of integrative genomics and functional RNA interference. Eur J Cancer. 2010;46:2166–2177. doi: 10.1016/j.ejca.2010.03.019. [DOI] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li Y, Ahmad A, Azmi AS, Kong D, Banerjee S, Sarkar FH. Targeting miRNAs involved in cancer stem cell and EMT regulation: an emerging concept in overcoming drug resistance. Drug Resist Updat. 2010;13:109–118. doi: 10.1016/j.drup.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead J, Pandey GK, Kanduri C. Regulation of the mammalian epigenome by long noncoding RNAs. Biochim Biophys Acta. 2009;1790:936–947. doi: 10.1016/j.bbagen.2008.10.007. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chen H, Lin Y, Hu Z, Mao Y, Wu J, Xu X, Zhu Y, Li S, Zheng X, et al. MicroRNA-409-3p inhibits migration and invasion of bladder cancer cells via targeting c-Met. Mol Cells. 2013;36:62–68. doi: 10.1007/s10059-013-0044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Yin Y, Xu SJ, Chen WS. 5-Fluorouracil: mechanisms of resistance and reversal strategies. Molecules. 2008;13:1551–1569. doi: 10.3390/molecules13081551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T, Wang J, Chen X, Liu L. Role of micro-RNA in anticancer drug resistance. Int J Cancer. 2010;126:2–10. doi: 10.1002/ijc.24782. [DOI] [PubMed] [Google Scholar]