ABSTRACT
Loss-of-function mutations in the genes encoding PINK1 and Parkin (also known as PARK2) are the most common causes of recessive Parkinson's disease. Both together mediate the selective degradation of mitochondrial proteins and whole organelles via the proteasome and the autophagy-lysosome pathway (mitophagy). The mitochondrial kinase PINK1 activates and recruits the E3 ubiquitin ligase Parkin to de-energized mitochondria. However, the cognate E2 co-enzymes of Parkin in this ubiquitin-dependent pathway have not been investigated. Here, we discovered a total of four E2s that either positively or negatively regulate the activation, translocation and enzymatic functions of Parkin during mitochondrial quality control. UBE2D family members and UBE2L3 redundantly charged the RING-HECT hybrid ligase Parkin with ubiquitin, resulting in its initial activation and translocation to mitochondria. UBE2N, however, primarily operated through a different mechanism in order to mediate the proper clustering of mitochondria, a prerequisite for degradation. Strikingly, in contrast to UBE2D, UBE2L3 and UBE2N, depletion of UBE2R1 resulted in enhanced Parkin translocation and clustering upon mitochondrial uncoupling. Our study uncovered redundant, cooperative or antagonistic functions of distinct E2 enzymes in the regulation of Parkin and mitophagy that might suggest a putative role in Parkinson's disease pathogenesis.
KEY WORDS: Parkin, PINK1, Mitochondria, Ubiquitin, E2 enzymes, Proteasome, Autophagy, Mitophagy
INTRODUCTION
Parkinson's disease is the most common neurodegenerative movement disorder. Symptoms arise from the selective loss of dopamine-producing neurons in the substantia nigra. The molecular mechanisms for this distinctive neuronal degeneration are poorly understood. Although most Parkinson's disease cases are sporadic, rare familial forms allow insights into potential pathogenic mechanisms, such as failure of protein degradation pathways and mitochondrial dysfunctions (Corti et al., 2011). To date, three recessive parkinsonism genes, PINK1 (Valente et al., 2004), PARKIN (also known as PARK2; Kitada et al., 1998) and FBXO7 (Di Fonzo et al., 2009), have been linked into a single molecular pathway for mitochondrial quality control (Geisler et al., 2010; Matsuda et al., 2010; Narendra et al., 2010b; Vives-Bauza et al., 2010; Burchell et al., 2013). PINK1 encodes a mitochondrial Ser/Thr kinase that is cleaved in healthy mitochondria (Jin et al., 2010; Meissner et al., 2011; Greene et al., 2012) and rapidly degraded (Yamano and Youle, 2013). The gene product of PARKIN is a cytosolic E3 ligase that attaches the small modifier protein ubiquitin to substrate proteins. FBXO7 encodes a putative substrate recognition component of a multi-protein E3 ubiquitin ligase complex but also has ubiquitin-independent functions (Nelson et al., 2013). Strikingly, PINK1, Parkin and FBXO7 physically associate and functionally cooperate to identify, label and target damaged mitochondria for selective degradation. Mutations in either gene disrupt this protective pathway; however, they affect distinct steps of a sequential process. Upon mitochondrial dysfunction, PINK1 protein is stabilized on de-energized organelles. PINK1 accumulation on damaged mitochondria and its kinase activity are prerequisites for the translocation of Parkin from the cytosol. Once localized to mitochondria, Parkin ubiquitylates numerous mitochondrial substrate proteins to facilitate the degradation of individual proteins by the 26S proteasome or of whole organelles by the autophagy-lysosomal system (Chan et al., 2011; Sarraf et al., 2013). Upon ubiquitin modification of mitochondria, adaptor proteins, such as VCP/p97 (Kim et al., 2013), HDAC6 (Lee et al., 2010) or p62/SQSTM1 (Geisler et al., 2010), are co-recruited to decode respective ubiquitin tags and facilitate the removal of substrates. In either case, the E3 ubiquitin ligase activities of Parkin are crucially involved.
Parkin is known as a broadly neuroprotective, multipurpose E3 ligase that is tightly controlled and modifies numerous unrelated substrate proteins (Walden and Martinez-Torres, 2012). Moreover, Parkin has been shown to catalyze the formation of various ubiquitin modifications ranging from (multi-) mono-ubiquitin to poly-ubiquitin chains with distinct characteristics (Sandebring and Cedazo-Mínguez, 2012). Ubiquitin itself contains seven internal lysine residues that all can be used to generate ubiquitin chains of unique topologies and biological functions (Komander and Rape, 2012). In addition, ubiquitin can form linear chains by intermolecular linkage between its C- and N-termini. Parkin has long been regarded as a RING-type E3 ubiquitin ligase that utilizes E2 ubiquitin conjugating enzymes to mediate the direct transfer of ubiquitin from the E2 to a substrate protein. Thereby, E2 enzymes bound to the RING finger domain of an E3 ligase denominate the ubiquitin chain linkages formed. However, recent data has challenged the ubiquitin transfer mechanism for Parkin and other members of the RING-between-RING (RBR) family (Wenzel et al., 2011). Similar to HECT-type E3 ubiquitin ligases, Parkin has been shown to accept ubiquitin from an E2 enzyme in a thioester intermediate on its recently discovered active site C431 before transfer onto a lysine residue of a substrate protein. In this case, the E3 ligase itself dictates the linkage type of the growing poly-ubiquitin chain, largely independent of the E2 enzymes (Sheng et al., 2012). In fact, K48-, K63- and K27-linked ubiquitin chains appear to be successively formed during mitochondrial quality control and might facilitate certain aspects along the course (Geisler et al., 2010; Chan et al., 2011; Birsa et al., 2014). The crystal structures of Parkin (Riley et al., 2013; Spratt et al., 2013; Trempe et al., 2013; Wauer and Komander, 2013), and other RBR-type E3 ubiquitin ligases (Duda et al., 2013), have been recently resolved and show an auto-inhibited, ‘closed’ conformation, consistent with their generally very low enzymatic activity. Recent studies suggest that the activation of Parkin through ‘ubiquitin charging’ is coupled to its enzymatic activity(ies) and its mitochondrial translocation (Iguchi et al., 2013; Lazarou et al., 2013; Zheng and Hunter, 2013). Accordingly, Parkin must receive a ubiquitin moiety from an E2 enzyme and pass this onto a substrate, which might include itself, in order to localize to mitochondria. Besides a recent study that suggests UBE2A is crucially involved (Haddad et al., 2013), the roles of E2 co-enzymes for Parkin activation, translocation to and enzymatic actions on mitochondria have not been investigated.
In this study, we aimed to identify E2 co-enzymes that regulate the activation and mitochondrial translocation of Parkin upon uncoupling of the mitochondrial membrane potential. In total, we have previously analyzed 11 out of 35 human, active E2 enzymes (van Wijk and Timmers, 2010). Strikingly, we identified four E2 cofactors that redundantly, cooperatively or antagonistically regulate the activation and mitochondrial translocation of Parkin. We demonstrate that members of the UBE2D family and UBE2L3 are able to ‘charge’ Parkin with ubiquitin and are essential for its initial activation. UBE2N, however, modifies the translocation of Parkin and, in particular, the subsequent peri-nuclear clustering of ubiquitin-labeled mitochondria, which is thought to facilitate their autophagic removal, through a different mechanism. In contrast to these positive regulatory mechanisms, we also identified UBE2R1 as a negatively acting E2 enzyme. We found that depletion of UBE2R1 enhances the translocation of Parkin to, and clustering of, mitochondria. These findings highlight the crucial involvement of distinct E2 enzymes that differentially regulate PINK1- and Parkin-mediated mitochondrial quality control. Thus, this study establishes the foundation for a crucial evaluation of E2 enzymes in the pathogenesis of Parkinson's disease and a determination of the further molecular details that underlie the activation and E3 ligase functions of Parkin.
RESULTS
Identification of E2 enzymes that regulate activation and mitochondrial translocation of Parkin
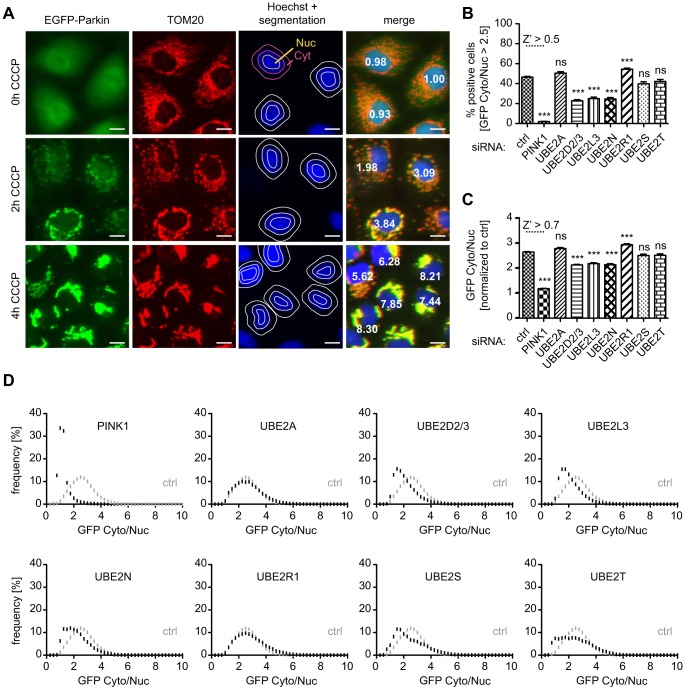
To identify E2 enzymes that might charge Parkin with ubiquitin or might otherwise regulate its activation and mitochondrial translocation, we screened select cofactors by using small interfering RNA (siRNA)-mediated knockdown. From a total of 35 human active E2 enzymes (van Wijk and Timmers, 2010), we selected 11 cofactors based on their previous association with Parkin, their well-defined roles in the formation of distinct ubiquitin linkage types or as controls for specificity [i.e. UBE2A, UBE2C, UBE2D1, UBE2D2, UBE2D3, UBE2D4, UBE2L3, UBE2N, UBE2R1 (also known as CDC34), UBE2S and UBE2T]. Similar to recent studies (Hasson et al., 2013; Lefebvre et al., 2013; McCoy et al., 2014), we have established a High Content Imaging (HCI) assay that allows unbiased monitoring of PINK1-dependent Parkin recruitment from the cytosol onto chemically de-energized mitochondria. Using HeLa cells that stably expressed an enhanced green fluorescent protein (EGFP)-fusion, the distribution of Parkin within cells was quantified as a ratio of the intensity of cytoplasmic and nuclear GFP (Cyto∶Nuc) (Fig. 1A) upon treatment with the uncoupler carbonylcyanide m-chlorophenylhydrazone (CCCP). To calculate the percentage of Parkin-translocation-positive cells, we used a cut-off at 2.5. With this threshold, approximately 50% of the control-silenced cells showed Parkin translocation after 2 h of treatment with CCCP, which allowed us to determine positive and negative modifiers alike (Fig. 1B).
Fig. 1.
High Content Imaging of Parkin translocation upon knockdown of E2 enzymes. (A) HeLa cells that stably expressed EGFP–Parkin were treated with 10 µM CCCP for 2 h, 4 h or left untreated. Images have been acquired using automated microscopy and show mitochondria (TOM20) in red, EGFP–Parkin in green and nuclei (Hoechst) in blue. Parkin translocation was measured as the ratio of cytoplasmic (Cyt) and nuclear (Nuc) GFP signal (given in white for specific cells). The cytoplasmic ring and inner nuclear regions are schematically shown, together with the Hoechst staining. The intensity of GFP in a ring around the nucleus (indicated in pink) was divided by the mean intensity of the nuclear GFP signal (border indicated in yellow). The resulting GFP Cyto∶Nuc ratio is given for each cell in white together with the merge. Scale bars: 10 µm. (B–D) HeLa-EGFP-Parkin cells were treated with siRNAs against UBE2A, UBE2D2/3, UBE2L3, UBE2N, UBE2R1, UBE2S or UBE2T and treated with CCCP for 2 h. (B) The percentages of Parkin-translocation-positive cells with a GFP Cyto∶Nuc ratio >2.5 are shown (n≧24, one-way ANOVA, Bonferroni corrected for multiple testing, P<0.0001, F = 366.6). (C) The averages of the GFP Cyto∶Nuc ratio across all experiments are shown. Knockdown of UBE2D2/3, UBE2L3 or UBE2N significantly inhibited the translocation of Parkin to mitochondria, whereas knockdown of UBE2R1 significantly increased the recruitment of Parkin after 2 h of treatment with CCCP (n≧24, one-way ANOVA, Bonferroni corrected for multiple testing, P<0.0001, F = 460.3). (D) The distribution of the GFP Cyto∶Nuc ratio of all analyzed cells as frequency in percent is shown. Per siRNA, at least 24 wells were measured. Control and PINK1-silenced samples were run on all plates for quality control of siRNA silencing and mitophagy induction upon treatment with CCCP. Both types of analysis received high Z′ scores (>0.5) per plate, indicative of an excellent assay. ***P<0.0005; ns, not significant.
Combined knockdown of the redundant UBE2D family members UBE2D2 and UBE2D3 with a single siRNA that targeted both genes (abbreviated hereafter as UBE2D2/3) significantly reduced the percentage of Parkin-translocation-positive cells. Similarly, knockdown of UBE2L3 or UBE2N each decreased the percentage by about 50% after 2 h of treatment with CCCP (Fig. 1B). Of note, transfection with an siRNA against UBE2R1 resulted in significantly more cells with Parkin translocation compared with control-silenced cells, indicative of an accelerated recruitment to mitochondria. Analyses of the average GFP Cyto∶Nuc ratio (Fig. 1C) and distribution frequency (Fig. 1D) among the individual cells corroborated the inhibitory effects of siRNAs against UBE2D2/3, UBE2L3 and UBE2N, as well as the accelerated Parkin translocation upon UBE2R1 knockdown. Consistent across all measurements, knockdown of UBE2A, UBE2S or UBE2T showed no significant difference compared with control-silenced cells.
Knockdown efficiency for all E2 enzymes was confirmed by using western blot analysis and/or quantitative reverse transcriptase (qRT-) PCR. Most siRNAs significantly reduced transcript and/or protein levels throughout all experiments by 80–90% (supplementary material Fig. S1A–C). Combined knockdown of UBE2D2/3 (supplemental material Fig. S1D) resulted in complete reduction of UBE2D signal, as judged by use of an antibody that recognized all four UBE2D proteins (supplemental material Fig. S1E). Consistently, additional knockdown of UBE2D1 and/or UBE2D4 did not further decrease UBE2D protein levels and Parkin translocation (data not shown).
Taken together, screening of E2 co-enzymes by a highly sensitive HCI assay allowed us to identify positive (UBE2D, UBE2L3 and UBE2N) and negative (UBE2R1) regulators of Parkin translocation upon mitochondrial uncoupling.
E2 enzymes regulate Parkin translocation and mitochondrial clustering differently
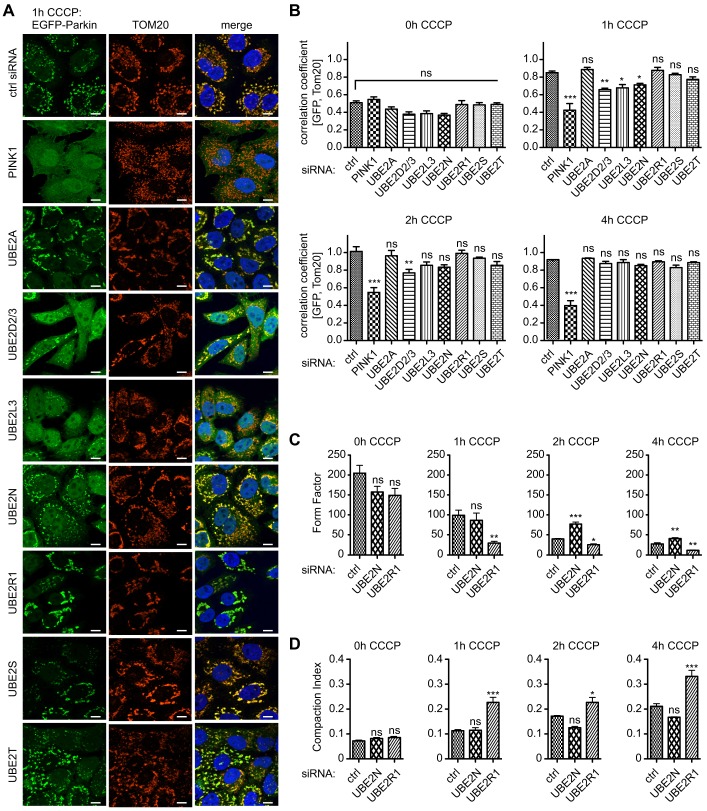
To verify findings from automated HCI microscopy and to yield a higher resolution of events, we employed conventional imaging. For this, HeLa-EGFP-Parkin cells were stained for mitochondria (against TOM20) and analyzed with high magnification. Knockdown of each of the E2 enzymes analyzed did not overly affect mitochondrial morphology or Parkin intracellular localization under non-stress conditions (supplementary material Fig. S2A, 0 h CCCP). Manual inspection confirmed reduced Parkin translocation upon silencing of UBE2D2/3, UBE2L3 or UBE2N, at least at early timepoints of CCCP treatment (Fig. 2A; supplementary material Fig. S2B,C). Consistently, calculations of the Pearson's correlation coefficient between GFP (Parkin) and mitochondrial (TOM20) signal revealed a significant decrease of colocalization after 1 h of treatment with CCCP (Fig. 2B). Although UBE2D2/3 siRNA effects remained significant until 2 h of treatment with CCCP, trends for UBE2L3 and UBE2N silencing persisted beyond this timepoint. Knockdown of UBE2A, UBE2S or UBE2T did not affect Parkin translocation or mitochondrial clustering at any of the analyzed timepoints.
Fig. 2.
Parkin translocation to damaged mitochondria is altered upon knockdown of E2 enzymes. HeLa cells that stably expressed EGFP–Parkin were silenced with siRNAs against E2 co-enzymes and treated with 10 µM CCCP as indicated. (A) Manual inspection of Parkin translocation was used to validate the effects observed by using automated HCI. Shown are representative high magnification (63×) images: EGFP–Parkin in green, mitochondria (TOM20) in red and nuclei (Hoechst) in blue after 1 h of treatment with CCCP (for corresponding images after 0, 2 or 4 h CCCP; see supplementary information Fig. S2). As a positive control, cells were transfected with siRNA against PINK1. Scale bars: 10 µm. (B) The Pearson's correlation coefficient was calculated as a measure of the colocalization of Parkin with mitochondria (n≧4, one-way ANOVA, Tukey's post-hoc; 0 h CCCP: P = 0.0005, F = 3.821; 1 h CCCP: P<0.0001, F = 24.36; 2 h CCCP: P<0.0001, F = 9.443; 4 h CCCP: P<0.0001, F = 41.94). Knockdown of PINK1 strongly inhibited Parkin translocation at all timepoints analyzed, whereas silencing of UBE2D2/3 or UBE2L3 significantly reduced Parkin colocalization with mitochondria only until 2 h of treatment with CCCP. (C,D) TOM20 images were binarized using ImageJ to measure mitochondrial area (Am) and the length of the outline or perimeter (Pm). (C) The form factor was calculated as [(Pm2)/(4πAm)] (n≧4, one-way ANOVA, Tukey's post-hoc; 0 h CCCP: P = 0.0858, F = 3.172; 1 h CCCP: P = 0.0053, F = 9.936; 2 h CCCP: P<0.0001, F = 69.42; 4 h CCCP: P<0.0001, F = 40.74). (D) The compaction index was calculated as [2π×((Am/π)−2))/Pm] (n≧4, one-way ANOVA, Tukey's post-hoc; 0 h CCCP: P = 0.0909, F = 3.078; 1 h CCCP: P = 0.0003, F = 23.18; 2 h CCCP: P = 0.0007, F = 17.88; 4 h CCCP: P<0.0001, F = 38.80). Compared with controls (ctrl), cells that had been treated with siRNA against UBE2R1 showed decreased elongation and branching but increased compaction. Knockdown of UBE2N resulted in the opposite trend, being significant for the mitochondrial form factor at later timepoints. ***P<0.0005; **P<0.005; *P<0.05; ns, not significant.
Upon translocation of Parkin to de-energized mitochondria, organelles are swiftly clustered around perinuclear regions and eventually sequestered into a few bigger ‘mito-aggresomes’. However, we noted a particularly altered mitochondrial clustering in UBE2N-silenced cells. Mitochondria (and Parkin) stayed more dispersed throughout the cell periphery over a longer time in UBE2N-silenced cells. This might result in a less pronounced increase in EGFP–Parkin intensity in a cytoplasmic ring around the nucleus that was analyzed in HCI. Nevertheless, Pearson's correlation also showed reduced colocalization of Parkin with mitochondria upon transfection with UBE2N siRNA, at least at 1 h CCCP (Fig. 2B). Therefore, silencing of UBE2N might affect both Parkin translocation and mitochondrial clustering.
Knockdown of UBE2R1 resulted in significantly accelerated Parkin translocation in HCI and an appreciably enhanced mitochondrial clustering. However, Pearson's correlation did not reveal an enhanced colocalization of Parkin with mitochondria (Fig. 2B). A maximal correlation had already been reached after 2 h of treatment with CCCP in controls. To analyze the effects of UBE2N and UBE2R1 knockdown in greater detail, we calculated the form factor (Fig. 2C) and compaction index (Fig. 2D) as read-outs of mitochondrial morphology and compaction. The form factor is a measure of the elongation and the branching of a mitochondrial network (Koopman et al., 2005), whereas the compaction index measures the circularity of the mitochondrial pool and therefore their clustering (Narendra et al., 2010a). Compared with controls, knockdown of UBE2R1 resulted in decreased connectivity and strongly increased compaction of damaged mitochondria (Fig. 2C,D). Silencing of UBE2N resulted in the opposite trend for both parameters with a significantly increased form factor at later timepoints. These data are consistent with a decrease of mitochondrial clustering upon loss of UBE2N and an increase upon loss of UBE2R1. In summary, the secondary analysis confirmed that loss of UBE2D, UBE2L3 or UBE2N delayed, whereas reduction of UBE2R1 accelerated, the recruitment of Parkin and concomitant mitochondrial clustering.
Knockdown of E2 enzymes delays ubiquitylation and degradation of mitochondrial substrates
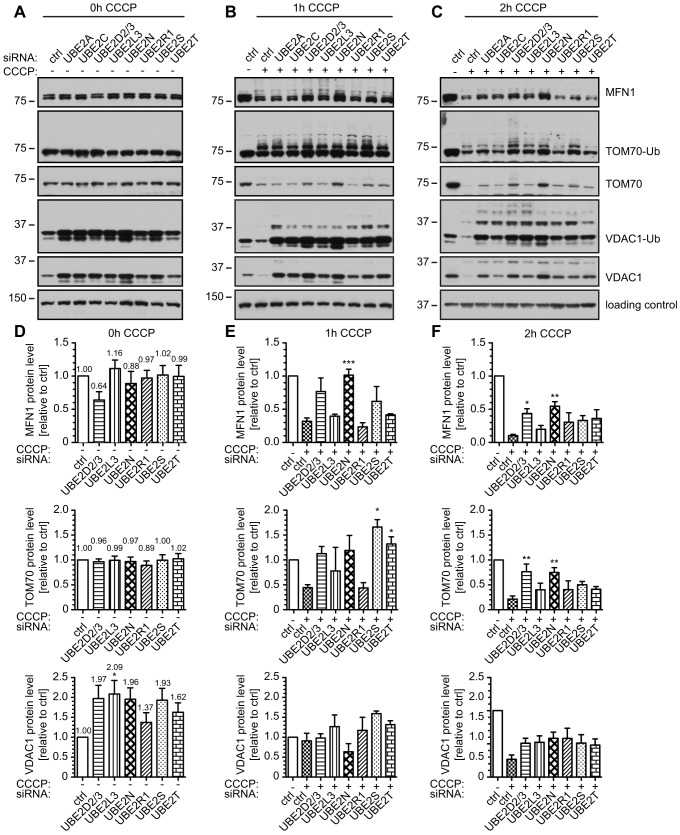
To biochemically validate the effects of altered Parkin translocation to mitochondria, we performed timeline experiments to monitor ubiquitin modification and degradation of mitochondrial Parkin substrates by using western blot profiling at 0, 1, 2, 4 and 6 h upon treatment with CCCP of HeLa cells that expressed untagged Parkin (HeLa-Parkin) (supplementary material Fig. S3A). For this we analyzed different exposures of western blots to capture ubiquitylated species of substrate proteins and, at the same time, to monitor their degradation. If at all, knockdown of UBE2D2 and UBE2D3 only minimally affected the ubiquitylation and degradation of the immediate mitophagy substrates MFN1 and MFN2 compared with cells that had been transfected with control siRNA. By contrast, the subsequent mitophagy substrates TOM70, TOM20 and VDAC1 were clearly less ubiquitylated and degraded, or at the least, modification of these proteins was delayed. All of these outer mitochondrial membrane (OMM) proteins have been described previously as Parkin substrates for proteasomal degradation (Chan et al., 2011; Yoshii et al., 2011; Sarraf et al., 2013). Inner mitochondrial membrane (IMM) proteins, such as the adenine nucleotide transporter (ANT1), as well as the matrix enzymes superoxide dismutase 2 (SOD2) or Cyclophilin D (CypD), probable substrates of actual autophagic clearance, were also (yet) unaffected in control cells at the immediate timepoints (1, 2, 4 and 6 h of treatment with CCCP). We decided to analyze the protein levels of MFN1, TOM70 and VDAC1 as model substrates at different timepoints in more detail. Representative images are shown in Fig. 3A–C. Knockdown of UBE2D2/3, UBE2L3 or UBE2N resulted in delayed substrate ubiquitylation and degradation (as revealed by different exposures). Knockdown of UBE2R1, which accelerates translocation of Parkin and clustering of mitochondria, did not reveal an overly enhanced ubiquitylation and/or degradation rate at the analyzed timepoints. In particular, for VDAC1, we noted a consistent increase of steady-state protein levels at 0 h of treatment with CCCP upon knockdown of several UBE2 enzymes (Fig. 3A). Quantification by densitometry revealed an up to twofold increase depending on the UBE2 enzyme silenced (Fig. 3D). Interestingly, this effect seemed dependent upon the presence of functional Parkin, because it was not observed in HeLa cells that did not express detectable levels of Parkin (supplementary material Fig. S3B). To appropriately quantify the levels of model substrate proteins upon treatment with CCCP, we introduced a ‘correction factor’ based on their steady-state levels (0 h CCCP) (Fig. 3D). Although the observed effects were not significant for all timepoints and substrates analyzed, we noted a trend for increased levels of MFN1 and TOM70 upon silencing of UBE2D2/3, UBE2L3 or UBE2N (Fig. 3E,F). For VDAC1, means were not significant after applying the correction factor. Western blot profiling of substrates also revealed unexpected effects of UBE2S knockdown (and, to a minor extent, knockdown of UBE2T) that did not affect Parkin translocation. Whether these effects are specific remains to be determined. An unambiguous difference between distinct substrates upon knockdown of a particular E2 was not observed. Because our analysis is limited (given that there are hundreds of mitochondrial Parkin substrates), we cannot exclude that a certain E2 enzyme might be solely responsible for the ubiquitylation of a particular substrate.
Fig. 3.
Parkin substrate profiling and quantification upon treatment with siRNAs against E2 proteins. HeLa cells that stably expressed untagged Parkin were treated either with control siRNA or siRNA against E2 cofactors, as indicated. Cells were left untreated or treated with 10 µM CCCP for (A) 0 h, (B) 1 h or (C) 4 h. Western blotting was used to monitor the levels of Parkin substrates. The suffix -Ub denotes the ubiquitylated form of the indicated protein. Labeling of all blots is as C. Vinculin or GAPDH were used as loading controls. Densitometry was used to quantify proteins after (D) 0 h, (E) 1 h and (F) 2 h of treatment with CCCP. In D, ‘correction factors’ calculated for the model substrates (MFN1, TOM70 and VDAC1) and for each E2 siRNA at steady state (0 h CCCP) are shown above the respective bars. Results of further quantification at 1 h and 2 h of treatment with CCCP were adjusted according to protein levels of untreated samples. Compared with control-silenced cells, knockdown of UBE2D2/3, UBE2L3 or UBE2N increased the protein levels of MFN1 and TOM70. At least five independent experiments were analyzed, which had been controlled for knockdown efficiency. n≧5, one-way ANOVA, Tukey's post-hoc; 0 h CCCP (MFN1): P = 0.1920, F = 1.528; 0 h CCCP (TOM70): P = 0.9220, F = 0.3228; 0 h CCCP (VDAC1): P = 0.0212, F = 2.791; 1 h CCCP (MFN1): P<0.0001, F = 13.21; 1 h CCCP (TOM70): P = 0.0002, F = 7.712; 1 h CCCP (VDAC1): P = 0.2742, F = 1.384; 2 h CCCP (MFN1): P<0.0001, F = 21.59; 2 h CCCP (TOM70): P<0.0001, F = 9.338; 2 h CCCP (VDAC1): P<0.0001, F = 5.672. ***P<0.0005; **P<0.005; *P<0.05; ns, not significant.
To investigate a possible effect of treatment with CCCP on E2 protein levels, we performed a timecourse experiment. We analyzed protein levels in HeLa cells with or without overexpression of Parkin. However, we did not observe any alteration in protein levels of the E1 ubiquitin activating enzyme or any of the analyzed E2 ubiquitin conjugating enzymes (supplementary material Fig. S3C).
Knockdown of E2 enzymes delays recruitment of the adaptor protein p62/SQSTM1
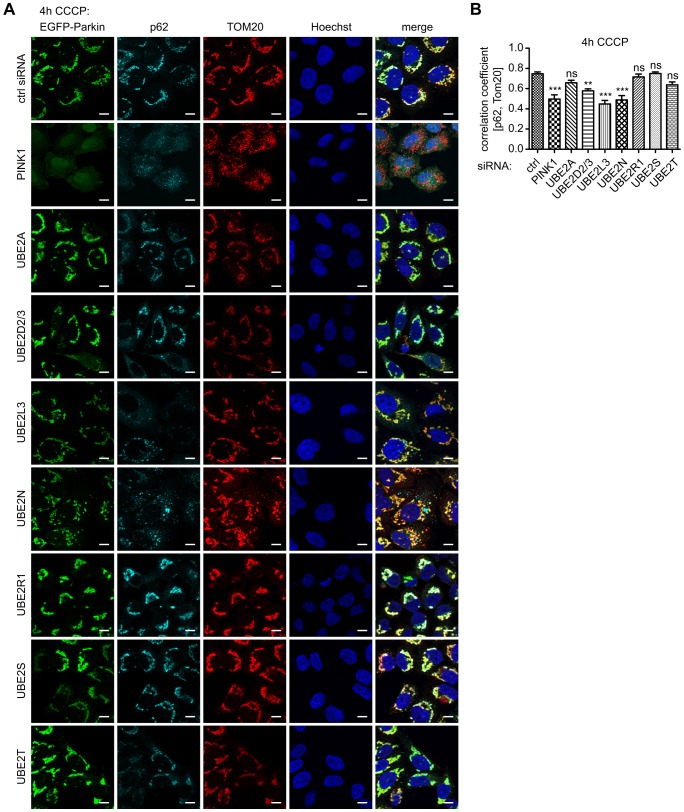
During Parkin-mediated mitophagy, the ubiquitin adaptor protein p62/SQSTM1 is co-recruited to damaged mitochondria to facilitate their clustering and autophagic degradation. p62 is known to bind K63-linked ubiquitin chains and to recruit LC3 and thereby autophagosomal membranes, bridging both cellular degradation systems. To follow up on this subsequent step, we knocked down E2 genes and performed triple labeling of Parkin, mitochondria and p62 to monitor its recruitment and to analyze potential morphological differences among E2 enzymes that regulate activation of Parkin and/or translocation to mitochondria. Although inhibition of Parkin translocation after 4 h of CCCP was not as prominent as that observed at earlier timepoints (for 1 h CCCP, see Fig. 2) and upon PINK1 silencing, UBE2D2/3, UBE2L3 or UBE2N siRNA strongly diminished p62 recruitment to mitochondria (Fig. 4). Consistently, the p62 signal remained diffuse without major colocalization to Parkin and mitochondria. Similar to PINK1 silencing, knockdown of UBE2D2/3, UBE2L3 or UBE2N significantly reduced colocalization of p62 with mitochondria, as measured by using the Pearson's coefficient (Fig. 4B). By contrast, silencing of UBE2A, UBE2S or UBE2T showed no difference to control conditions, suggesting a normal progression of mitophagy. Consistent with an accelerated Parkin translocation, knockdown of UBE2R1 revealed strong p62 accumulation on mitochondria (Fig. 4A). However, colocalization as measured by Pearson's correlation was not significantly enhanced compared with control cells (Fig. 4B).
Fig. 4.
Knockdown of UBE2D, UBE2L3 or UBE2N reduces the recruitment of p62/SQSTM1 to damaged mitochondria. HeLa-EGFP-Parkin cells were treated either with control siRNA or siRNA against E2 cofactors, as indicated. Cells were left untreated or treated with 10 µM CCCP for 4 h, fixed and stained for p62 (shown in cyan) and mitochondria (TOM20, red). (A) Representative images are shown. Scale bars: 10 µm. (B) Pearson's correlation of p62 and TOM20 was analyzed (n≧10, one-way ANOVA, Tukey's post-hoc, P<0.0001, F = 16.80). In contrast with controls and siRNA against UBE2A, UBE2R1, UBE2S or UBE2T, knockdown of UBE2D2/3, UBE2L3 or UBE2N showed significantly less p62 signal on damaged mitochondria. ***P<0.0005; **P<0.005; ns, not significant.
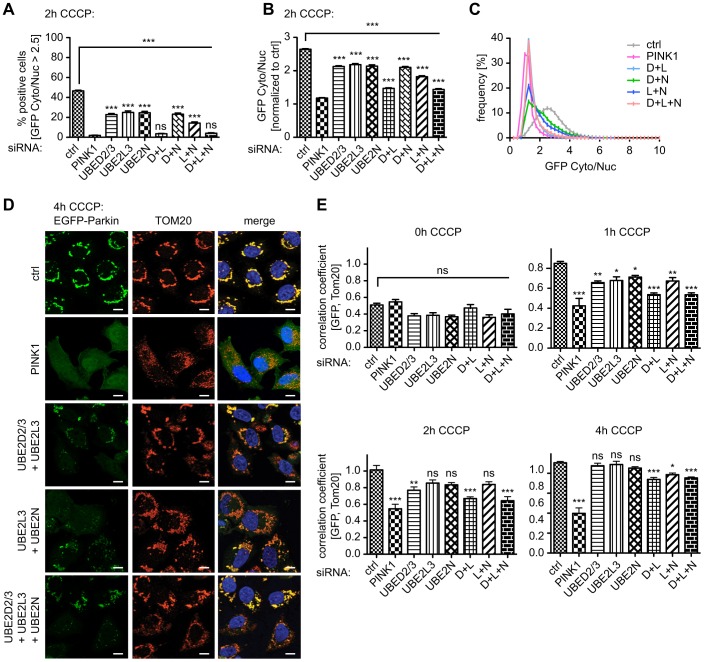
Combined knockdown of UBE2D2/3, UBE2L3 and/or UBE2N strongly impairs functions of Parkin
It has been shown that UBE2D family members and UBE2L3 are able to charge HECT E3 ligases. Although UBE2L3 is reactive to cysteine residues only, UBE2D isoforms are able to discharge ubiquitin also onto lysine residues of substrate proteins (Wenzel et al., 2011). As shown earlier, individual knockdown of UBE2D2/3 and UBE2L3 reduced Parkin translocation by ∼50% of control levels, as measured by HCI, similar to UBE2N siRNA. However, given the different morphological phenotypes with respect to Parkin and mitochondrial clustering, we investigated potential redundant and cooperative effects among these E2 enzymes. We knocked down UBE2D2/3, UBE2L3 and UBE2N in double and triple combinations in HeLa-EGFP-Parkin cells and performed HCI. Simultaneous knockdown of UBE2D2/3 and UBE2L3 further reduced Parkin translocation to ∼4% of all cells, similar to the strong effect observed upon PINK1 knockdown (Fig. 5A). This is consistent with the idea that UBE2D family members and UBE2L3 redundantly function to ‘ubiquitin charge’ Parkin and thereby are involved in its initial activation. This also supports the finding that the enzymatic function(s) and mitochondrial translocation of Parkin are coupled (Iguchi et al., 2013; Lazarou et al., 2013; Zheng and Hunter, 2013).
Fig. 5.
Combined knockdown of UBE2D and UBE2L3 blocks translocation of Parkin upon treatment with CCCP. HeLa-EGFP-Parkin cells were transfected with siRNA as indicated and treated with CCCP for 2 h. Automated HCI was performed and (A) the percentage of Parkin-translocation-positive cells was calculated as the GFP Cyto∶Nuc ratio >2.5 (n≧24, one-way ANOVA, Bonferroni corrected for multiple testing, P<0.0001, F = 366.6). (B) The average of the GFP Cyto∶Nuc ratio across all experiments is shown. Significance compared with controls is indicated on top of the graph, values compared with PINK1-silenced cells are given above the respective bars. For comparison, values for single knockdown are given (n≧24, one-way ANOVA, Bonferroni corrected for multiple testing, P<0.0001, F = 460.3). Simultaneous knockdown of UBE2D2/3 and UBE2L3 (D+L) or UBE2D2/3 with UBE2L3 and UBE2N (D+L+N) almost completely blocked Parkin translocation, similar to silencing of PINK1. Combination of UBE2L3 and UBE2N knockdown (L+N) showed an intermediate phenotype. (C) The distribution of the GFP Cyto∶Nuc ratio for all analyzed cells. (D) Manual inspection of Parkin translocation was used to validate the effects observed by using HCI. Representative high-magnification (63×) images are shown: EGFP–Parkin in green, mitochondria (TOM20) in red and nuclei (Hoechst) in blue, as well as Pearson's correlation between TOM20 and GFP (E). For comparison, values of single siRNA treatments are shown. Compared with single E2 knockdown, double and triple siRNA combinations showed an increased inhibitory effect (n = 10, one-way ANOVA, Tukey's post-hoc; 0 h CCCP: P = 0.0005, F = 3.821; 1 h CCCP: P<0.0001, F = 24.36; 2 h CCCP: P<0.0001, F = 9.443; 4 h CCCP: P<0.0001, F = 41.94). As a positive control, cells were transfected with siRNA against PINK1. Scale bars: 10 µm. ***P<0.0005; **P<0.005; *P<0.05; ns, not significant.
Combination of UBE2L3 with UBE2N siRNA reduced the number of Parkin-translocation-positive cells and the average of the GFP Cyto∶Nuc ratio (Fig. 5A,B) compared with single silencing of these UBE2s; however, this was not significant when compared with single siRNA transfections. Combined knockdown of UBE2D2/3 together with UBE2N did not show any synergistic or additive effect. Compared to the almost abolished translocation upon knockdown of UBE2D2/3 and UBE2L3, triple siRNA transfection of UBE2D2/3, UBE2L3 and UBE2N did not cause any further effects with an almost complete overlap of their respective GFP Cyto∶Nuc ratio distributions (Fig. 5C).
We used qRT-PCR to monitor the silencing efficiency upon combined knockdown of E2s. Double knockdown had a similar silencing efficiency to individual knockdown, but triple knockdown appeared less efficient (supplementary material Fig. S1F). Therefore, a complete loss of all three E2 enzymes might entirely abolish Parkin translocation, similar to PINK1 knockdown.
In line with HCI, higher resolution immunofluorescence and correlation of Parkin and mitochondria confirmed the additive effect of UBE2D2/3 and UBE2L3 siRNAs that resulted in a greatly reduced Parkin translocation and colocalization at all timepoints, even after 4 h of treatment with CCCP (Fig. 5D,E; for other timepoints see supplementary material Fig. S4), compared with either siRNA alone (see Fig. 2A; supplementary material Fig. S2). As seen in the translocation assay, simultaneous knockdown of UBE2N together with UBE2L3 resulted in an intermediate phenotype. However, the morphological distinctions of UBE2N knockdown, such as the particular mitochondrial clustering, appeared less prominent in combination with knockdown of UBE2D2/3 or UBE2L3 (compare Fig. 5D; supplementary material Fig. S2C). This might result from a less overall clustering because of additional delays in Parkin translocation compared with those upon silencing of UBE2N alone.
UBE2D2/3, UBE2L3 and UBE2N cooperate with Parkin through distinct regulatory mechanisms
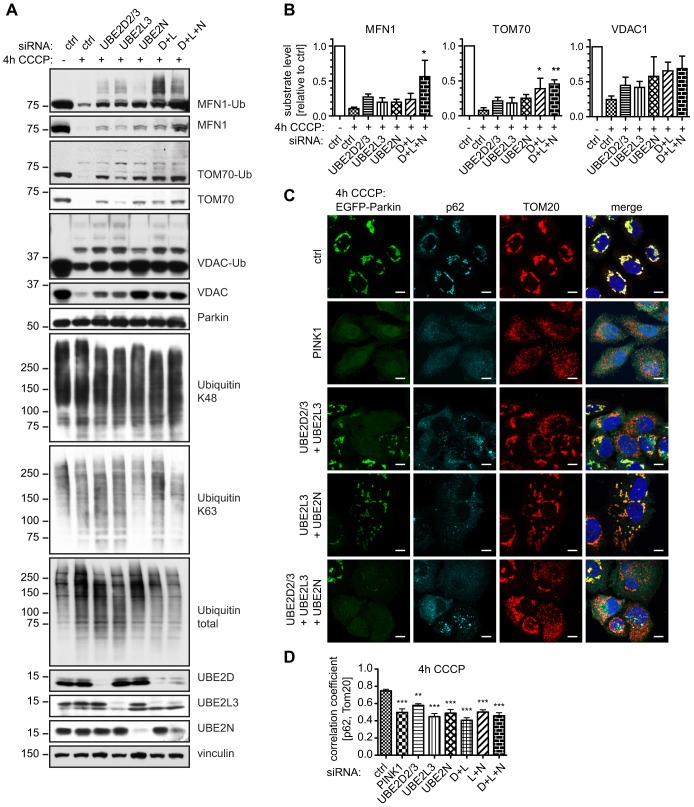
To further elucidate the underlying mechanisms, we performed western blot profiling of substrates upon treatment with combined E2 siRNAs. Levels of OMM protein substrates were greatly stabilized upon combined knockdown of UBE2D2/3 and UBE2L3 compared with those upon knockdown using single siRNAs (Fig. 6A). Furthermore, triple siRNA combinations clearly showed appreciable levels of ubiquitylated species and of unmodified substrate proteins, indicating a delay in both modification and degradation at 4 h CCCP. At this timepoint, substrate proteins have been mostly degraded in control cells, whereas treatment with a single siRNA resulted in intermediate amounts of ubiquitylated and yet unmodified proteins. In line with this, densitometry analysis of double and triple silencing showed significantly increased amounts of unmodified substrate compared with that upon treatment with a single siRNA (Fig. 6B). As expected, combined knockdown of UBE2D2/3 and UBE2L3 more appreciably reduced total cellular ubiquitin levels upon treatment with CCCP (compared with single knockdown) without overly affecting K48 ubiquitin levels, but with a possible shift towards K63 linkage. Consistent with its unique role in the formation of this particular linkage type, knockdown of UBE2N strongly reduced the levels K63-linked poly-ubiquitin chains. Similar to PINK1 knockdown, treatment with an siRNA against a single E2 showed a maximal reduction in the Pearson's correlation between p62 and mitochondria (TOM20) at 4 h of treatment with CCCP. Thus, the colocalization could not be further decreased by double or triple combinatorial knockdown of E2 enzymes (Fig. 6C,D).
Fig. 6.
Combined knockdown of UBE2D and UBE2L3 blocks substrate degradation and p62 recruitment upon treatment CCCP. (A,B) HeLa-Parkin cells were transfected with control, single or a combination of UBE2 siRNAs and left untreated or treated with 10 µM CCCP, as indicated. Western blots were prepared and probed with antibodies against the Parkin substrates MFN1, TOM70 and VDAC1 and those indicated. The suffix -Ub denotes the ubiquitylated form of the indicated protein. (A) Representative western blots are shown. (B) Densitometry was used to quantify the proteins (n≧5, one-way ANOVA, Tukey's post-hoc; 4 h CCCP (MFN1): P<0.0001, F = 13.67; 4 h CCCP (TOM70): P<0.0001, F = 31.70; 4 h CCCP (VDAC1): P = 0.0347, F = 2.952). Combined silencing of UBE2D2/3 with UBE2L3 (D+L) or UBE2D2/3, UBE2L3 and UBE2N (D+L+N) enhanced the effects of the respective single siRNAs. (C) HeLa-EGFP-Parkin cells were stained for mitochondria (TOM20, red) and p62 (shown in cyan). Merged images include nuclear staining (Hoechst, blue) and are shown to the right. Scale bars: 10 µm. (D) Pearson's correlation between p62 and the TOM20 signal was calculated. For comparison, single siRNA values are shown. Combined silencing abrogated colocalization of p62 to a similar extent to that upon silencing of UBE2D2/3, UBE2L3, UBE2N or PINK1 (n≧10, one-way ANOVA, Tukey's post-hoc, P<0.0001, F = 16.80). ***P<0.0005; **P<0.005; *P<0.05.
In summary, double knockdown of UBE2D2/3 and UBE2L3 (with or without additional silencing of UBE2N) significantly reduced Parkin translocation and colocalization with mitochondria, as well as ubiquitylation and/or degradation of substrates and p62 recruitment.
UBE2D2/3, UBE2L3 and UBE2N physically and functionally interact with Parkin
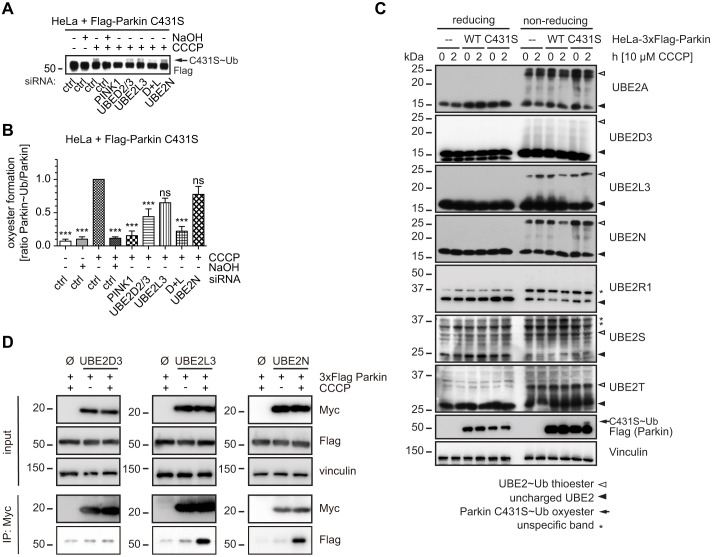
Given the probably redundant functions of UBE2D2/3 and UBE2L3 in the initial activation of Parkin, as well as the potentially cooperative role of UBE2N, we employed an active site mutant of Parkin, C431S, to measure its ‘ubiquitin charging’. Parkin has been suggested to transfer ubiquitin by a novel HECT-RING hybrid mechanism (Wenzel et al., 2011). According to this hypothesis, Parkin accepts a ubiquitin moiety from a ‘charged’ E2 enzyme in a thioester bond on its catalytic center cysteine residue before further transfer onto a lysine residue of a substrate protein, including itself. Thioester intermediates are transient and instable and have not been detected for Parkin, yet they have for other E3 ligases of the RBR family. However, a C431S Parkin mutant can receive ubiquitin similar to wild type, although as an oxyester that traps ubiquitin and disables further transfer. To resolve the obviously differential roles of the E2 co-enzymes of Parkin, we expressed the Parkin C431S mutation and knocked down UBE2D2/3, UBE2L3 and UBE2N, alone and in combination. Treatment with CCCP resulted in a band shift of Parkin signal (Fig. 7A). This was confirmed as an oxyester bonded Parkin C431S–ubiquitin that was sensitive to NaOH treatment. Importantly, PINK1 knockdown completely blocked the ubiquitin charging and thus activation of Parkin. Although knockdown of UBE2D2/3 or UBE2L3 both reduced levels of Parkin C431S–ubiquitin to some degree, combined knockdown of UBE2D2/3 and UBE2L3 strongly diminished ubiquitin loading of Parkin. However, knockdown of UBE2N had no discernable effect on the charging of Parkin with ubiquitin. Indeed, quantification from several experiments confirmed the highly significant reduction in Parkin C431S–ubiquitin levels upon treatment with NaOH, PINK1 siRNA or combined knockdown of UBE2D2/3 and UBE2L3 (Fig. 7B). In contrast to single knockdown of UBE2D2/3, gene silencing of UBE2L3 alone, despite an obvious observed trend, did not reach statistical significance, which is probably related to a limited knockdown efficiency.
Fig. 7.
UBE2D, UBE2L3 and UBE2N are utilized by Parkin during mitophagy. (A) HeLa cells were transfected with Flag–Parkin C431S upon knockdown of UBE2 enzymes. Cells were treated with CCCP for 4 h and harvested. Western blotting was used to monitor the formation of a Parkin-C431S–ubiquitin oxyester (indicated by an arrow), which appears as a band shift and is sensitive to NaOH treatment. Densitometric analysis of western blots is shown in B. Knockdown of UBE2D2/3 significantly reduced the formation of the Parkin C431S–ubiquitin oxyester. Consistent with the stronger effects of two siRNAs, combined knockdown of UBE2D2/3 and UBE2L3 reduced the formation, similar to siRNA against PINK1. Silencing of UBE2N had no effect on oxyester formation (n≧6, one-way ANOVA, Tukey's post-hoc, P<0.0001, F = 9.911). (C) Parental HeLa cells and HeLa cells that stably overexpressed 3×Flag-tagged Parkin wild type (WT) or C431S were left untreated or were treated with 10 µM CCCP for 2 h. Cells were lysed in low pH MES buffer and subjected to SDS-PAGE under reducing and non-reducing conditions. The resulting western blots were probed with antibodies against endogenous UBE2 enzymes to monitor unmodified UBE2 enzymes (closed arrowheads) and 8-kDa shifted ‘charged’ ubiquitin-bound UBE2 enzymes (open arrowheads). HeLa 3×Flag–Parkin WT cells showed a decrease of ubiquitin ‘charged’ species for UBE2D3, UBE2L3 and UBE2N. By contrast, parental HeLa cells and HeLa cells that expressed non-functional C431S Parkin did not show altered thioester levels upon treatment with CCCP. No obvious changes in this ubiquitin-discharge were found for the other E2 enzymes analyzed. (D) Co-immunoprecipitation of Parkin with UBE2 enzymes after treatment with CCCP. HeLa cells that stably expressed 3×FLAG–Parkin wild type were transfected with Myc-tagged E2 enzymes (Myc–UBE2D3, Myc–UBE2L3, Myc–UBE2N) or an empty vector control (Ø). Cells were left untreated or treated with CCCP for 3 h followed by immunoprecipitation for Myc. Upon CCCP treatment, Parkin specifically and strongly co-immunoprecipitated with Myc–UBE2L3 and Myc–UBE2N. ***P<0.0005; ns, not significant.
To further prove the differential roles of E2 enzymes, we performed immunoprecipitation of Parkin coupled to an in vitro ubiquitylation assay. Using stable HEK293E cells that expressed wild-type 3×Flag–Parkin or its inactive C431S mutation, we affinity purified Parkin by immunoprecipitation of Flag from cells that had been treated with CCCP for 1 h or left untreated. Affinity beads were incubated with a complete mix of ATP, recombinant ubiquitin and E1 enzyme, as well as different E2 enzymes. Incubation of wild-type Parkin with the E2 enzymes UBE2D2, UBE2L3 or UBE2N facilitated ubiquitylation, whereas C431S Parkin was unable to generate these ubiquitin modifications (supplementary material Fig. S3D). Consistently, treatment with CCCP slightly increased the formation of ubiquitin modifications catalyzed by wild-type Parkin [as judged by horseradish-peroxidase (HRP)-conjugated Streptavidin]. It is noteworthy that only UBE2L3 was able to catalyze obvious auto-ubiquitylation of wild-type Parkin, but not of the C431S mutation (as judged by using an antibody against Flag). No discernable differences in the auto-ubiquitylation of Parkin could be observed between samples that had been left untreated or had been treated with CCCP.
To show that E2 enzymes are utilized by Parkin in cells, we performed thioester analyses of ubiquitin that was bound to E2 enzymes. Using non-reducing gel conditions, ubiquitin-loaded forms of UBE2 enzymes were visible as 8-kDa shifted bands (Fig. 7C). Upon treatment with CCCP, we found less ubiquitin-charged species of UBE2D3, UBE2L3 or UBE2N when functional Parkin was present. This was neither the case for parental HeLa cells without exogenous Parkin or for HeLa cells that expressed the Parkin C431S mutant. We could not observe specific discharging of the other E2 enzymes analyzed.
In addition, we performed co-immunoprecipitations to unequivocally demonstrate physical interactions of Parkin and select E2 enzymes. Transiently transfected Myc-tagged UBE2 enzymes did not show an interaction with Parkin under basal conditions (Fig. 7D). However, upon treatment with CCCP, Parkin was specifically and robustly co-purified with UBE2L3 or UBE2N. It is noteworthy that we were unable to confirm an interaction of Parkin with UBE2D3 3 h after treatment with CCCP. At this point, it is unclear whether select E2 enzymes are utilized by Parkin for distinct purposes at different steps of the sequential mitophagy process.
Taken together, our results reinforce the idea that UBE2D and UBE2L3 play redundant roles in charging, and thus initial activation, of Parkin. In line with this, UBE2N functions through a different mechanism, probably later in the process after the initial ubiquitin loading of Parkin.
DISCUSSION
In summary, we have characterized the roles of specific E2 enzymes as cofactors for Parkin activation and translocation to de-energized mitochondria, as well as subsequent substrate ubiquitylations and p62 recruitment. Our HCI data indicate that UBE2D, UBE2L3 and UBE2N are positive regulators of the mitochondrial translocation of Parkin. We have validated these results using conventional microscopy and western blot profiling. Knockdown of UBE2D family members, UBE2L3 and UBE2N each delay Parkin translocation, substrate ubiquitylation and degradation, as well as the recruitment of p62 to damaged mitochondria. Although knockdown of PINK1 completely abrogated the mitochondrial translocation of Parkin and is highly significant at any timepoint analyzed (data not shown), depletion of single E2 enzymes showed the most robust phenotype until 2 h after treatment with CCCP. However, at later timepoints, the knockdown effects of individual E2 enzymes became less obvious, which might be due to their incomplete reduction (80–90%). Compared with more stable proteins, the well-documented instability of PINK1 certainly facilitates siRNA-mediated knockdown. Given the transient and dynamic association of E2–E3 pairs, it is conceivable that even greatly reduced amounts of ubiquitin conjugating enzymes might be compensated, at least over time, by few remaining molecules and the partial redundancy among closely related E2s.
The similarity observed between individual knockdown of UBE2D or UBE2L3 is consistent with the idea that Parkin enzymatic actions and mitochondrial translocation are coupled and that both enzymes are involved in its initial ‘charging’ (Iguchi et al., 2013; Lazarou et al., 2013; Zheng and Hunter, 2013). In line with this, double-knockdown experiments showed additive effects of combined silencing of UBE2D with UBE2L3. In fact, simultaneous depletion delayed Parkin translocation, similar to the effects of an siRNA against PINK1. The human E2 enzymes UBE2D1, UBE2D2, UBE2D3 and UBE2D4 are closely related and homologous to the stress-induced yeast E2 enzymes Ubc4 and Ubc5 that can generate chains of various linkages (Seufert and Jentsch, 1990). In addition, UBE2D members have been shown to transfer ubiquitin onto both lysine and cysteine residues (Ye and Rape, 2009; Wenzel et al., 2011). UBE2L3, which has already been identified in an earlier report to interact with Parkin (Shimura et al., 2000), specifically discharges ubiquitin onto the active sites of HECT E3 ligases (Wenzel et al., 2011). The redundant functions of UBE2D and UBE2L3 are further corroborated by analysis of Parkin C431S–ubiquitin oxyester formation. Although UBE2D2/3 knockdown alone significantly reduced the amount of ubiquitin-charged Parkin, combined knockdown together with UBE2L3 greatly enhanced this effect, which was comparable to the effects of silencing PINK1. This is consistent with a previous report that utilized recombinant enzymes for Parkin oxyester formation in vitro (Lazarou et al., 2013).
In contrast to UBE2D and UBE2L3, UBE2N, which is known to mediate K63-linked ubiquitylation (Komander and Rape, 2012), had no effect on ubiquitin charging of Parkin. Knockdown of UBE2N inhibited, primarily, mitochondrial clustering and reduced total K63 poly-ubiquitin levels upon treatment with CCCP. The observed mitochondrial morphology and distribution upon knockdown of UBE2N is indeed phenocopied by overexpression of a K63R ubiquitin variant or by knockdown of the cognate ubiquitin adaptor p62 (Geisler et al., 2010; Narendra et al., 2010a; Okatsu et al., 2010). All of these conditions result in reduced formation or recognition of this particular ubiquitin linkage and corroborates an important role for K63-linked ubiquitin chains in the formation of mito-aggresomes preceding autophagic clearance. Combined knockdown of UBE2N with UBE2D or UBE2L3 showed no strong additive effect, suggesting that UBE2N is not redundant to UBE2D and UBE2L3, but rather functions at a different stage. This is further corroborated by colocalization measurements between Parkin and mitochondria that are independent of organelle morphology and distribution. Interestingly, it was recently suggested that Parkin binds K63-linked chains, which are sufficient to recruit Parkin to mitochondria when cytoplasmic PINK1 is overexpressed (Zheng and Hunter, 2013). Therefore, reducing K63-linked chains by knockdown of UBE2N might delay Parkin translocation to mitochondria in addition to interference with mitochondrial clustering. In summary, this indicates that Parkin activation and translocation, as well as mitochondrial clustering and substrate ubiquitylation are separate, yet linked events. Although UBE2D, UBE2L3 and UBE2N catalyzed the formation of ubiquitin modifications together with wild-type Parkin, but not its inactive mutant in in vitro assays, only UBE2L3 facilitated obvious auto-ubiquitylation of Parkin. Moreover, UBE2L3 and UBE2N strongly and specifically interacted with Parkin from cells after treatment with CCCP. Whether our inability to co-immunoprecipitate UBE2D3 with Parkin after 3 h of treatment with CCCP reflects a time-dependent change in the utilization of E2 enzymes is unclear. Nevertheless, it would be interesting to measure Parkin–E2 interaction upon mitochondrial depolarization with greater temporal resolution. This would also be in line with a previous study that suggests a higher binding affinity of Parkin to UBE2N upon phosphorylation by PINK1 (Sha et al., 2010). Although an interaction of UBE2D3 could not be demonstrated in co-immunoprecipitation experiments, E2-discharging experiments show that, in cells, functional Parkin indeed utilizes all three E2 enzymes.
Interestingly, we discovered UBE2R1/CDC34 as a negative regulator of Parkin-mediated mitophagy. RNA interference (RNAi)-mediated silencing of UBE2R1 enhanced Parkin translocation and increased mitochondrial clustering. However, western-blot-based substrate profiling did not unequivocally show a significant difference to that using a control siRNA. In yeast, cdc34 is the dedicated E2 enzyme for Skp–Cullin–Fbox (SCF)-type multiprotein E3 ubiquitin ligases and promotes the synthesis of K48-linked ubiquitin chains (Lydeard et al., 2013). Cdc34 is known to be important for cell cycle progression. Indeed, in our HCI assay, we had, consistently, approximately 35% less cells in wells upon UBE2R1 knockdown compared with those of control. It is noteworthy that FBXO7, the third protein associated with Parkinson's disease that is implicated in mitophagy (Burchell et al., 2013), can function as the substrate recruitment adaptor in a SCF complex. This is of particular interest given that Parkin itself has been described in an earlier study to function in a SCF complex (Staropoli et al., 2003). In addition, a SCF complex has been shown to generate stabilizing ubiquitin modifications of MFN proteins that result in mitochondrial fusion (Anton et al., 2013). These activating modifications however are counterbalanced by destabilizing ubiquitylation events that are mediated through HUWE1 (Leboucher et al., 2012) or Parkin (Gegg et al., 2010; Poole et al., 2010; Glauser et al., 2011) upon stress. Therefore, a reduction of UBE2R1 levels might shift the balance towards degradation of MFN proteins resulting in mitochondrial fragmentation, which in turn could cause activation and translocation of Parkin (Narendra et al., 2008). Although this would be consistent with a slightly reduced mitochondrial form factor upon UBE2R1 silencing, it does not rule out other direct or indirect effects.
Silencing of other E2 enzymes had no effect on the mitochondrial translocation of Parkin or p62 recruitment. This includes the K11-linkage specific UBE2S, which has an important role for cell cycle regulation (Williamson et al., 2009), and UBE2T, which is involved in DNA damage repair (Machida et al., 2006). However, we noted increased levels of VDAC1 in particular upon knockdown of several E2 enzymes already under steady-state conditions (the absence of CCCP). This effect was only visible in the presence of high levels of exogenous Parkin. Although we cannot rule out Parkin-independent effects, an upregulation of endogenous VDAC1 might serve to counteract its constant basal turnover in the presence of Parkin. Upon silencing of UBE2S and UBE2T, we also noted significantly enhanced levels of TOM70. Whether these effects indeed are specific remains to be determined. However, the assay relies on detection by antibodies that do not recognize higher molecular weight species to the same extent as the respective unmodified proteins. We thus restricted our analysis to the unmodified species of substrate proteins only. However, this does not take into account higher molecular weight (i.e. ubiquitylated) species of the respective proteins and might thus underestimate the remaining levels of substrate proteins.
Surprisingly, knockdown of UBE2A did not affect Parkin translocation, mitochondrial clustering or p62 recruitment in HeLa cells. However, we noted a slight discharge of UBE2A upon treatment with CCCP, yet this appeared to be rather independent of the presence of functional Parkin. Loss of UBE2A was recently reported to inhibit Parkin translocation upon treatment with CCCP in knockout mouse embryonic fibroblasts (Haddad et al., 2013), but the underlying mechanisms remained unclear. Loss of UBE2A causes mental retardation, but no parkinsonian syndromes (de Leeuw et al., 2010). The apparent discrepancy between both studies might result from different cellular models (murine versus human cells), different timepoints of treatment with CCCP or from non-complete knockdown versus knockout. In human HeLa cells, the closely related UBE2B/RAD6B might also be able to compensate for the knockdown of UBE2A. In either case this underlines the importance of cell type specificity and regulation of Parkin mitophagy depending on the cellular context of the E2 enzymes.
Interestingly, during the preparation of this manuscript, a novel study identified positive and negative regulators of Parkin translocation by a genome-wide HCI siRNA approach (Hasson et al., 2013). Consistent with our data, UBE2D3 and UBE2L3 were identified as positive regulators of Parkin translocation in an approach similar to the present study. In addition to the co-enzymes studied here, the endoplasmic reticulum (ER)-anchored UBE2J2/NCUBE2, which has been implicated in ER-associated degradation (ERAD) pathway, and the orphan E2 UBE2O/E2-230K were also identified as Parkin regulators, however the mechanisms remain elusive. Nevertheless, neither UBE2A nor UBE2R1 have been recovered from this screen, but knockdown efficiencies were not monitored. It is thus possible that additional E2 enzymes play important roles during Parkin-mediated mitophagy. In fact, our findings show that specific E2 enzymes operate at distinct steps during this process and probably with different purposes.
In summary, our study uncovered four E2 enzymes that regulate the activation of Parkin and its translocation to and enzymatic activities at mitochondria. Moreover, we have identified distinct molecular mechanisms, including redundant, cooperative or antagonistic functions, among E2 enzymes. Together with two recent reports (Haddad et al., 2013; Hasson et al., 2013), our study underscores the importance of distinct E2 enzymes as direct or indirect regulators of Parkin and mitophagy. The involvement of several E2 enzymes in Parkin-mediated mitophagy is also consistent with the sequential appearance of distinct ubiquitin linkage types (K48, K63 and K27) on select substrates along the course (Geisler et al., 2010; Chan et al., 2011; Birsa et al., 2014). The very recent and striking discovery of PINK1-dependent phosphorylation of ubiquitin itself (Kazlauskaite et al., 2014), which acts as a cofactor for the activation of Parkin (together with the PINK1-dependent phosphorylation of the UBL domain of Parkin), emphasizes the importance of the involved modifications, as well as their complexity. It will thus be interesting to further dissect the roles of phosphorylated ubiquitin and to identify the E2 enzymes that are involved in its conjugation.
In light of the newly identified RING-HECT-hybrid mechanism for Parkin-mediated ubiquitin transfer (Wenzel et al., 2011) and its recently resolved structure (Riley et al., 2013; Spratt et al., 2013; Trempe et al., 2013; Wauer and Komander, 2013), knowledge of the cognate E2s will certainly help to better understand the activation of Parkin and its enzymatic functions, as well as the biological roles of the ubiquitin modifications generated. Although E2s were believed to be ubiquitin-charged at steady-state (Jin et al., 2007), surprising differences in the ratio of charged and uncharged E2 enzymes (UBE2D2 versus UBE2N) in particular cells have recently been suggested (Wiener et al., 2013). Moreover, cellular stress increases ubiquitin charging of some E2s (Takada et al., 2001), whereas others are redox-regulated (Jahngen-Hodge et al., 1997) or sensitive to oxidative stress (Doris et al., 2012). Thus, similar to Parkin (Dawson and Dawson, 2014), E2 enzymes might be inactivated in sporadic disease by modification of their active center. Given that E2 enzymes are key mediators of ubiquitin chain assembly (Ye and Rape, 2009) that can be selectively targeted by small molecules (Ceccarelli et al., 2011), in future it will be important to further expand our understanding of their functions as essential Parkin cofactors and modifiers, as well as to elucidate their relevance and contribution to disease pathogenesis.
MATERIALS AND METHODS
Cloning and mutagenesis
Wild-type Flag–Parkin has been described previously (Geisler et al., 2010). Parkin C431S was cloned using site-directed mutagenesis into pcDNA3-Flag. The pCMV-Myc-E2 constructs were cloned using BglII and NotI. Overexpression constructs were sequence verified using BigDye Terminator v.3.1 and an ABI 3100 Genetic Analyzer (Applied Biosystems). Primer sequences can be obtained on request.
Cell culture and RNA silencing
Human epithelial cancer cells (HeLa) were obtained from the American Type Culture Collection, and human embryonic kidney (HEK293E) cells were obtained from Invitrogen. Parental HEK293E and HeLa cells or clonal cells that stably expressed untagged Parkin (HeLa-Parkin), EGFP-Myc Parkin (HeLa-GFP-Parkin) or 3×Flag–Parkin wild type or C431S were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (PAA) at 37°C under humidified 5% CO2/air. DNA transfections were performed using Lipofectamine 2000 (Invitrogen) or XtremeGene9 (Roche) according to the manufacturers' instructions. siRNA transfections were performed on two consecutive days using HiPerfect with 5 nmol control, PINK1- or UBE2-specific siRNAs (all from Qiagen): PINK1 5′-GACGCTGTTCCTCGTTATGAA-3′; UBE2A 5′-GCGUGUUUCUGCAAUAGUAUU-3′; UBE2C 5′-AGCCCUUGUAUAAUUAAAUAAA-3′; UBE2D1 5′- UCCACUGGCAAGCCACUAUUA-3′; UBE2D2/3 5′-AACAGUAAUGGCAGCAUUUGU-3′; UBE2D4 5′-CCGAAUGACAGUCCUUACCAA-3′; UBE2L3 5′-UGAAGAGUUUACAAAGAAA-3′; UBE2N 5′-AACCAGAUGAUCCAUUAGCAA-3′; UBE2R1 5′-AACAAGUUCCCCAUCGACUAC-3′; UBE2S 5′-AAGGCACUGGGACCUGGAUUU-3′; UBE2T 5′-AUCCUCAGAAUUUAAAUAUAA-3′. As a control, AllStars negative control siRNA (Qiagen) was used. For combined knockdown with two or three siRNAs, the amount of each siRNA was halved (2.5 nmol).
High Content Imaging
For HCI HeLa-GFP-Parkin cells were seeded with 4000 cells/100 µl per well in 96-well imaging plates (BD Biosciences). At 24 h and 48 h after plating, cells were transfected with siRNA. Transfection mixes were prepared for at least 12 wells per siRNA and contained, per well, 8.33 µl Opti-MEM (Invitrogen), 1.6 nmol siRNA (Qiagen) and 1 µl HiPerfect. At 48 h after the second transfection, cells were treated with or without 10 µM CCCP for 2 h. Cells were washed once in PBS and fixed for 20 min in 4% paraformaldehyde. Nuclei were stained with Hoechst 33342 (1∶5000, Invitrogen) for 10 min, and cells were then washed twice in PBS. Plates were imaged on a BD Pathway 855 system by using a 20× objective and a 3×3 montage (no gaps) with laser autofocus every second frame. Raw images were processed using the built-in AttoVision V1.6 software. Regions of interest (ROIs) were defined as nucleus (Nuc) and cytoplasm (Cyto) using the built-in ‘RING - 2 outputs’ segmentation for the Hoechst channel after applying a shading algorithm. As a measure of Parkin translocation, the ratio of the GFP intensity Cyto∶Nuc signal was calculated. Translocation-positive cells were defined to have a GFP Cyto∶Nuc ratio greater than 2.5. Per siRNA, we analyzed at least four independent experiments with six wells per condition. PINK1 and negative control siRNA were present on all plates for quality control. Resulting values were normalized to the control siRNA average value of all plates.
SDS-PAGE and western blot analysis
Cells were harvested and lysed in RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS) plus Complete proteinase inhibitor, 10 mM N-ethylmaleimide, 0.2 mM sodium orthovanadate, 20 mM sodium fluoride. Protein concentration was determined by use of bicinchoninic acid (Pierce Biotechnology). Proteins were subjected to SDS-PAGE using 14% or 8–16% Tris-Glycine or 4–12% Bis-Tris NuPAGE gels (Invitrogen) and transferred onto polyvinylidene fluoride or nitrocellulose membranes (both Millipore). Membranes were incubated with primary antibodies overnight at 4°C followed by HRP-conjugated secondary antibodies (1∶15,000; Jackson ImmunoResearch Laboratories). Bands were visualized by using ImmobilonWestern Chemiluminescent HRP Substrate (Millipore) on Blue Devil Lite X-ray films (Genesee Scientific). For densitometric analysis, ImageJ software version 1.46R was used. Total ubiquitin blots were microwaved 10 min in ddH2O before incubation with primary antibody for antigen retrieval.
For thioester recovery, cells were harvested and lysed in MES buffer {50 mM MES [2-(N-morpholino) ethanesulfonic acid] pH 4, 150 mM NaCl, 0.2% NP-40} containing Complete proteinase inhibitor. Samples were loaded in either reducing or non-reducing Laemmli loading buffer.
Immunostaining
Cells were plated onto glass coverslips coated with poly-d-lysine (Sigma), fixed with 4% (w/v) paraformaldehyde and permeabilized with 1% Triton-X-100. Cells were incubated with primary antibodies and then with secondary antibodies against mouse IgGs conjugated to Alexa Fluor 568 or against rabbit IgGs conjugated to Alexa Fluor 568 or 647 (Molecular Probes) diluted 1∶2000. Nuclei were stained with Hoechst 33342 diluted 1∶5000. Coverslips were mounted onto microscope slides using fluorescent mounting medium (Dako). Confocal fluorescent images were taken with an AxioObserver microscope equipped with an ApoTome Imaging System (Zeiss). For analysis of Pearson's correlation, raw images were converted into 8-bit tif files and analyzed using ImageJ and the Manders' Coefficients Plugin (Manders et al., 1993). For analysis of the form factor [(Pm2)/(4πAm)] (Koopman et al., 2005) and the compaction index [2π×((Am/π)−2))/Pm] (Narendra et al., 2010a), images were binarized and analyzed using the built-in analyze particle function of ImageJ to measure mitochondrial area (Am) and length of the outline or perimeter (Pm).
Antibodies
The following antibodies were used for western blot analysis (WB) or immunofluorescence (IF): mouse against ANT1 (WB, 1∶1000, Abcam ab110322), mouse against CypD (WB, 1∶5000, Abcam ab110324), mouse against Flag (WB, 1∶20,000–1∶100,000; Sigma F3165), mouse against Flag-HRP (WB, 1∶50,000–250,000; Sigma A8592), mouse against GAPDH (WB, 1∶1 50,000; Meridian Life Science H86504M), mouse against Mitofusin 1 (WB, 1∶5000; Abcam ab57602), mouse against Mitofusin 2 (WB, 1∶5000; Abcam ab56889), mouse against Myc antibody (WB, 1∶5000; Roche 11667149001), mouse against p62 (WB, 1∶5000; IF, 1∶500; BD Bioscience 610832), rabbit against p62 (WB, 1∶10,000: ProteinTech Group 18420-1-AP), rabbit against Parkin (WB, 1∶5000, Abcam ab15954), rabbit against PINK1 (WB, 1∶500, Novus Biologicals BC100-494), rabbit against SOD2 (WB, 1∶100,000, Abcam ab13533), rabbit against TOM20 (IF, 1∶2000; ProteinTech Group 11802-1-AP), mouse against TOM20 (WB, 1∶500; Santa Cruz sc-17764), rabbit against TOM70 (WB, 1∶5000; ProteinTech Group 14528-1-AP), mouse against ubiquitin (WB, 1∶1000; Millipore MAB1510), rabbit against ubiquitin K48-specific (WB, 1∶2500; Millipore 05-1307), rabbit against ubiquitin K63-specific (WB, 1∶2500; Millipore 05-1308), mouse against VDAC1 (WB, 1∶5000; Abcam ab14734), mouse against vinculin (WB, 1∶100,000; Sigma V9131), rabbit against UBE2A (WB, 1∶5000; ProteinTech Group 11080-1-AP), goat against UBE2C (WB, 1∶2000; Santa Cruz sc-100611), UBE2D (WB, 1∶5000; Santa Cruz sc-15000), rabbit against UBE2D3 (WB, 1∶5000; Cell Signaling Technology no. 4330), rabbit against UBE2L3 (WB, 1∶2000; ProteinTech Group 14415-1-AP), goat against UBE2L3 (WB, 1∶5000; Santa Cruz sc-47549), rabbit against UBE2N (WB, 1∶5000; Cell Signaling Technology no. 6999), mouse against UBE2N (Ubc13) (WB, 1∶1000; Invitrogen 37-1100), rabbit against UBE2R1 (WB, 1∶1000; ProteinTech Group 10964-2-AP), goat against UBE2R2 (WB, 1∶5000; Santa Cruz sc-167440), goat against UBE2S (WB, 1∶2000; Santa Cruz sc-131354), goat against UBE2T (WB, 1∶2000; Santa Cruz sc-103913).
qRT-PCR
Cellular RNA was extracted using the RNeasy Mini Kit (Qiagen) following the manufacturer's instruction. For qRT-PCR, 1000 ng of total RNA was reverse transcribed using Transcriptor High Fidelity cDNA Synthesis Kit (Roche) and anchored oligo-dT primer. 1 in 10 dilutions were used in triplicate with 0.2 µM primer and 5 µl iTaq Universal SYBR Green Supermix (Bio-Rad) in a 10 µl reaction, and qPCR analyses were executed in a 384-well block on a LightCycler 480 system (Roche). Absolute transcript levels for UBE2A, UBE2C, UBE2D1, UBE2D2, UBE2D3, UBE2D4, UBE2L3, UBE2N, UBE2R1, UBE2S, UBE2T and PBGD (also known as HMBS) were obtained by second derivative method. Relative transcript levels were calculated as UBE2∶PBGD ratio and normalized to the relative expression level of the control-transfected sample. Primer sequences are available upon request.
In vitro ubiquitylation assay
HEK293E cells that stably expressed 3×Flag–Parkin wild type or the C431S mutant were incubated with or without 10 µM CCCP for 1 h before lysis. Cells were washed once in cold 1×HBS (20 mM HEPES pH 7.4, 150 mM NaCl) and lysed in immunoprecipitation buffer (50 mM HEPES pH 7.5, 10 mM KCl, 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.2% NP-40) plus Complete proteinase inhibitors. Protein (750 µg) was immunoprecipitated for 4 h using 20 µl of anti-Flag EZview beads (Sigma). Beads were washed twice in cold immunoprecipitation buffer and once in 1× ubiquitylation buffer (20 mM HEPES pH 7.4, 50 mM NaCl, 5 mM MgCl2) before the supernatant was completely removed. Recombinant proteins were purchased from Boston Biochem. Ubiquitylation reactions contained 100 nM E1 enzyme (GST-Ube1, no. E306), 1 µM E2 enzyme (no. E2-622, no. E2-640 or no. E2-664), 2.5 mM ATP, 2 mM DTT, 5 µg untagged ubiquitin (no. U-100H), 1 µg N-terminally biotinylated ubiquitin (no. UB-560) in 1×ubiquitylation buffer in a 20 µl reaction. Ubiquitylation reactions were carried out for 1.5 h at 37°C. Laemmli buffer was added and samples boiled for 10 min at 95°C. Samples were run on a gel, blotted and probed with HRP-coupled Streptavidin (1∶100,000–1∶250,000; Jackson Immunoresearch 016-030-084) and an antibody against Flag.
Oxyester analysis
HeLa cells were transfected on two consecutive days with siRNA. At 24 h after the second siRNA transfection, cells were transfected with Flag–Parkin C431S using Lipofectamine 2000 according to the manufacturer's protocol, and the medium was replaced 4 h later. The next day, cells were treated with or without 10 µM CCCP for 4 h. Cells were harvested in preheated (95°C) SDS lysis buffer (50 mM Tris pH 7.6, 150 mM NaCl, 1% SDS). Lysates were homogenized by ten strokes through a 20 G needle. To verify the band shift by oxyester formation, aliquots of lysates were treated with or without NaOH (100 mM final concentration) for 1 h at 37°C before loading them onto 4–12% Bis-Tris gels.
Co-immunoprecipitation
HeLa cells that stably expressed 3×Flag–Parkin wild type were transfected with pCMV-Myc empty vector, or Myc-tagged UBE2D3, UBE2L3 or UBE2N using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions for 2 days. Cells were left untreated or treated with 10 µM CCCP for 3 h and lysed in co-immunoprecipitation buffer (50 mM HEPES pH 7.5, 10 mM KCl, 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1.5 mM MgCl2, 10% Glycerol and 0.2% NP-40) plus Complete proteinase inhibitors. Protein lysate (700 µg) for each condition was immunoprecipitated using 20 µl of anti-Myc EZview beads (Sigma E6654) at 4°C for 3 h. Beads were washed three times in cold co-immunoprecipitation buffer before the supernatant was completely removed. Beads were eluted in 50 µl 2×Laemmli buffer and boiled at 95°C. Immunoprecipitates and input cell lysates were analyzed by western blotting with mouse antibodies against Flag–HRP (1∶10,000; Sigma A8592) and or Myc (1∶5000; Roche 11667149001).
Statistical analysis
Statistical analysis was performed using one-way ANOVA for all experiments. Error bars indicate the s.e.m. Significance levels are as indicated: ***P<0.0005; **P<0.005; *P<0.05; ns, not significant.
Supplementary Material
Acknowledgments
We are grateful to Curtis Younkin (Mayo Clinic, Florida) for help with HCI data extraction and Michael Heckman (Mayo Clinic, Florida) for statistical consultation. We thank Philipp Kahle and Sven Geisler (University of Tübingen, Germany) for generously providing Myc-tagged UBE2 constructs.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
W.S. conceived the study. F.C.F. and W.S. designed the experiments. F.C.F., E.L.M.-L. and M.A. performed experiments. F.C.F., E.L.M.-L. and W.S. analyzed the data. F.C.F. and W.S. wrote the manuscript.
Funding
This work was supported by grants from the National Institutes of Health - National Institute of Neurological Disorders and Stroke [grant number R01NS085070]; the Michael J. Fox Foundation; Mayo Clinic Foundation and the Center for Individualized Medicine (CIM); the Marriott Family Foundation; and a Gerstner Family Career Development Award to W.S. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.147520/-/DC1
References
- Anton F., Dittmar G., Langer T., Escobar-Henriques M. (2013). Two deubiquitylases act on mitofusin and regulate mitochondrial fusion along independent pathways. Mol. Cell 49, 487–498 10.1016/j.molcel.2012.12.003 [DOI] [PubMed] [Google Scholar]
- Birsa N., Norkett R., Wauer T., Mevissen T. E., Wu H. C., Foltynie T., Bhatia K., Hirst W. D., Komander D., Plun-Favreau H. et al. (2014). K27 ubiquitination of the mitochondrial transport protein Miro is dependent on serine 65 of the Parkin ubiquitin ligase. J. Biol. Chem 10.1074/jbc.M114.563031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchell V. S., Nelson D. E., Sanchez-Martinez A., Delgado-Camprubi M., Ivatt R. M., Pogson J. H., Randle S. J., Wray S., Lewis P. A., Houlden H. et al. (2013). The Parkinson's disease-linked proteins Fbxo7 and Parkin interact to mediate mitophagy. Nat. Neurosci. 16, 1257–1265 10.1038/nn.3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli D. F., Tang X., Pelletier B., Orlicky S., Xie W., Plantevin V., Neculai D., Chou Y. C., Ogunjimi A., Al-Hakim A. et al. (2011). An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell 145, 1075–1087 10.1016/j.cell.2011.05.039 [DOI] [PubMed] [Google Scholar]
- Chan N. C., Salazar A. M., Pham A. H., Sweredoski M. J., Kolawa N. J., Graham R. L., Hess S., Chan D. C. (2011). Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum. Mol. Genet. 20, 1726–1737 10.1093/hmg/ddr048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti O., Lesage S., Brice A. (2011). What genetics tells us about the causes and mechanisms of Parkinson's disease. Physiol. Rev. 91, 1161–1218 10.1152/physrev.00022.2010 [DOI] [PubMed] [Google Scholar]
- Dawson T. M., Dawson V. L. (2014). Parkin plays a role in sporadic Parkinson's Disease. Neurodegener. Dis. 13, 69–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw N., Bulk S., Green A., Jaeckle-Santos L., Baker L. A., Zinn A. R., Kleefstra T., van der Smagt J. J., Vianne Morgante A. M., de Vries B. B. et al. (2010). UBE2A deficiency syndrome: Mild to severe intellectual disability accompanied by seizures, absent speech, urogenital, and skin anomalies in male patients. Am. J. Med. Genet. A. 152A, 3084–3090 10.1002/ajmg.a.33743 [DOI] [PubMed] [Google Scholar]
- Di Fonzo A., Dekker M. C., Montagna P., Baruzzi A., Yonova E. H., Correia Guedes L., Szczerbinska A., Zhao T., Dubbel-Hulsman L. O., Wouters C. H. et al. (2009). FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology 72, 240–245 10.1212/01.wnl.0000338144.10967.2b [DOI] [PubMed] [Google Scholar]
- Doris K. S., Rumsby E. L., Morgan B. A. (2012). Oxidative stress responses involve oxidation of a conserved ubiquitin pathway enzyme. Mol. Cell. Biol. 32, 4472–4481 10.1128/MCB.00559-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda D. M., Olszewski J. L., Schuermann J. P., Kurinov I., Miller D. J., Nourse A., Alpi A. F., Schulman B. A. (2013). Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism. Structure 21, 1030–1041 10.1016/j.str.2013.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegg M. E., Cooper J. M., Chau K. Y., Rojo M., Schapira A. H., Taanman J. W. (2010). Mitofusin 1 and mitofusin 2 are ubiquitinated in a PINK1/parkin-dependent manner upon induction of mitophagy. Hum. Mol. Genet. 19, 4861–4870 10.1093/hmg/ddq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010). PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 10.1038/ncb2012 [DOI] [PubMed] [Google Scholar]
- Glauser L., Sonnay S., Stafa K., Moore D. J. (2011). Parkin promotes the ubiquitination and degradation of the mitochondrial fusion factor mitofusin 1. J. Neurochem. 118, 636–645 10.1111/j.1471-4159.2011.07318.x [DOI] [PubMed] [Google Scholar]
- Greene A. W., Grenier K., Aguileta M. A., Muise S., Farazifard R., Haque M. E., McBride H. M., Park D. S., Fon E. A. (2012). Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 13, 378–385 10.1038/embor.2012.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad D. M., Vilain S., Vos M., Esposito G., Matta S., Kalscheuer V. M., Craessaerts K., Leyssen M., Nascimento R. M., Vianna-Morgante A. M. et al. (2013). Mutations in the intellectual disability gene Ube2a cause neuronal dysfunction and impair Parkin-dependent mitophagy. Mol. Cell 50, 831–843 [DOI] [PubMed] [Google Scholar]
- Hasson S. A., Kane L. A., Yamano K., Huang C. H., Sliter D. A., Buehler E., Wang C., Heman-Ackah S. M., Hessa T., Guha R. et al. (2013). High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature 504, 291–295 10.1038/nature12748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi M., Kujuro Y., Okatsu K., Koyano F., Kosako H., Kimura M., Suzuki N., Uchiyama S., Tanaka K., Matsuda N. (2013). Parkin catalyzed ubiquitin-ester transfer is triggered by PINK1-dependent phosphorylation. J. Biol. Chem. 288, 22019–22032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahngen-Hodge J., Obin M. S., Gong X., Shang F., Nowell T. R., Jr, Gong J., Abasi H., Blumberg J., Taylor A. (1997). Regulation of ubiquitin-conjugating enzymes by glutathione following oxidative stress. J. Biol. Chem. 272, 28218–28226 10.1074/jbc.272.45.28218 [DOI] [PubMed] [Google Scholar]
- Jin J., Li X., Gygi S. P., Harper J. W. (2007). Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature 447, 1135–1138 10.1038/nature05902 [DOI] [PubMed] [Google Scholar]
- Jin S. M., Lazarou M., Wang C., Kane L. A., Narendra D. P., Youle R. J. (2010). Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 191, 933–942 10.1083/jcb.201008084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A., Kondapalli C., Gourlay R., Campbell D. G., Ritorto M. S., Hofmann K., Alessi D. R., Knebel A., Trost M., Muqit M. M. (2014). Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Serine65. Biochem. J. 460, 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N. C., Tresse E., Kolaitis R. M., Molliex A., Thomas R. E., Alami N. H., Wang B., Joshi A., Smith R. B., Ritson G. P. et al. (2013). VCP is essential for mitochondrial quality control by PINK1/Parkin and this function is impaired by VCP mutations. Neuron 78, 65–80 10.1016/j.neuron.2013.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. (1998). Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 10.1038/33416 [DOI] [PubMed] [Google Scholar]
- Komander D., Rape M. (2012). The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- Koopman W. J., Visch H. J., Verkaart S., van den Heuvel L. W., Smeitink J. A., Willems P. H. (2005). Mitochondrial network complexity and pathological decrease in complex I activity are tightly correlated in isolated human complex I deficiency. Am. J. Physiol. 289, C881–C890 10.1152/ajpcell.00104.2005 [DOI] [PubMed] [Google Scholar]
- Lazarou M., Narendra D. P., Jin S. M., Tekle E., Banerjee S., Youle R. J. (2013). PINK1 drives Parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J. Cell Biol. 200, 163–172 10.1083/jcb.201210111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leboucher G. P., Tsai Y. C., Yang M., Shaw K. C., Zhou M., Veenstra T. D., Glickman M. H., Weissman A. M. (2012). Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol. Cell 47, 547–557 10.1016/j.molcel.2012.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Nagano Y., Taylor J. P., Lim K. L., Yao T. P. (2010). Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy. J. Cell Biol. 189, 671–679 10.1083/jcb.201001039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V., Du Q., Baird S., Ng A. C., Nascimento M., Campanella M., McBride H. M., Screaton R. A. (2013). Genome-wide RNAi screen identifies ATPase inhibitory factor 1 (ATPIF1) as essential for PARK2 recruitment and mitophagy. Autophagy 9, 1770–1779 10.4161/auto.25413 [DOI] [PubMed] [Google Scholar]
- Lydeard J. R., Schulman B. A., Harper J. W. (2013). Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 14, 1050–1061 10.1038/embor.2013.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida Y. J., Machida Y., Chen Y., Gurtan A. M., Kupfer G. M., D'Andrea A. D., Dutta A. (2006). UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol. Cell 23, 589–596 10.1016/j.molcel.2006.06.024 [DOI] [PubMed] [Google Scholar]
- Manders E. M. M., Verbeek F. J., Aten J. A. (1993). Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 169, 375–382 10.1111/j.1365-2818.1993.tb03313.x [DOI] [PubMed] [Google Scholar]
- Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C. A., Sou Y. S., Saiki S., Kawajiri S., Sato F. et al. (2010). PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221 10.1083/jcb.200910140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy M. K., Kaganovich A., Rudenko I. N., Ding J., Cookson M. R. (2014). Hexokinase activity is required for recruitment of parkin to depolarized mitochondria. Hum. Mol. Genet. 23, 145–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner C., Lorenz H., Weihofen A., Selkoe D. J., Lemberg M. K. (2011). The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J. Neurochem. 117, 856–867 10.1111/j.1471-4159.2011.07253.x [DOI] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008). Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Kane L. A., Hauser D. N., Fearnley I. M., Youle R. J. (2010a). p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 6, 1090–1106 10.4161/auto.6.8.13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D. P., Jin S. M., Tanaka A., Suen D. F., Gautier C. A., Shen J., Cookson M. R., Youle R. J. (2010b). PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. E., Randle S. J., Laman H. (2013). Beyond ubiquitination: the atypical functions of Fbxo7 and other F-box proteins. Open Biology 3, 130131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K., Saisho K., Shimanuki M., Nakada K., Shitara H., Sou Y. S., Kimura M., Sato S., Hattori N., Komatsu M. et al. (2010). p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells 15, 887–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole A. C., Thomas R. E., Yu S., Vincow E. S., Pallanck L. (2010). The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS ONE 5, e10054 10.1371/journal.pone.0010054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley B. E., Lougheed J. C., Callaway K., Velasquez M., Brecht E., Nguyen L., Shaler T., Walker D., Yang Y., Regnstrom K. et al. (2013). Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat. Commun. 4, 1982 10.1038/ncomms2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandebring A., Cedazo-Mínguez A. (2012). Parkin- an E3 ubiquitin ligase with multiple substrates. J. Alzheimers Dis. Parkinsonism S10, 002 10.4172/2161-0460.S10-002 [DOI] [Google Scholar]
- Sarraf S. A., Raman M., Guarani-Pereira V., Sowa M. E., Huttlin E. L., Gygi S. P., Harper J. W. (2013). Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, 372–376 10.1038/nature12043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert W., Jentsch S. (1990). Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 9, 543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha D., Chin L. S., Li L. (2010). Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-kappaB signaling. Hum. Mol. Genet. 19, 352–363 10.1093/hmg/ddp501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng Y., Hong J. H., Doherty R., Srikumar T., Shloush J., Avvakumov G. V., Walker J. R., Xue S., Neculai D., Wan J. W. et al. (2012). A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen. Mol. Cell. Proteomics 11, 329–341 10.1074/mcp.O111.013706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H., Hattori N., Kubo S., Mizuno Y., Asakawa S., Minoshima S., Shimizu N., Iwai K., Chiba T., Tanaka K. et al. (2000). Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25, 302–305 10.1038/77060 [DOI] [PubMed] [Google Scholar]
- Spratt D. E., Martinez-Torres R. J., Noh Y. J., Mercier P., Manczyk N., Barber K. R., Aguirre J. D., Burchell L., Purkiss A., Walden H. et al. (2013). A molecular explanation for the recessive nature of parkin-linked Parkinson's disease. Nat. Commun. 4, 1983 10.1038/ncomms2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staropoli J. F., McDermott C., Martinat C., Schulman B., Demireva E., Abeliovich A. (2003). Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron 37, 735–749 10.1016/S0896-6273(03)00084-9 [DOI] [PubMed] [Google Scholar]
- Takada K., Hirakawa T., Yokosawa H., Okawa Y., Taguchi H., Ohkawa K. (2001). Isolation of ubiquitin-E2 (ubiquitin-conjugating enzyme) complexes from erythroleukaemia cells using immunoaffinity techniques. Biochem. J. 356, 199–206 10.1042/0264-6021:3560199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe J. F., Sauvé V., Grenier K., Seirafi M., Tang M. Y., Ménade M., Al-Abdul-Wahid S., Krett J., Wong K., Kozlov G. et al. (2013). Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 340, 1451–1455 10.1126/science.1237908 [DOI] [PubMed] [Google Scholar]
- Valente E. M., Abou-Sleiman P. M., Caputo V., Muqit M. M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A. R., Healy D. G. et al. (2004). Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304, 1158–1160 10.1126/science.1096284 [DOI] [PubMed] [Google Scholar]
- van Wijk S. J., Timmers H. T. (2010). The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 24, 981–993 10.1096/fj.09-136259 [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R. L., Kim J., May J., Tocilescu M. A., Liu W., Ko H. S. et al. (2010). PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl. Acad. Sci. USA 107, 378–383 10.1073/pnas.0911187107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden H., Martinez-Torres R. J. (2012). Regulation of Parkin E3 ubiquitin ligase activity. Cell. Mol. Life Sci. 69, 3053–3067 10.1007/s00018-012-0978-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauer T., Komander D. (2013). Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 32, 2099–2112 10.1038/emboj.2013.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel D. M., Lissounov A., Brzovic P. S., Klevit R. E. (2011). UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108 10.1038/nature09966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener R., DiBello A. T., Lombardi P. M., Guzzo C. M., Zhang X., Matunis M. J., Wolberger C. (2013). E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat. Struct. Mol. Biol. 20, 1033–1039 10.1038/nsmb.2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A., Wickliffe K. E., Mellone B. G., Song L., Karpen G. H., Rape M. (2009). Identification of a physiological E2 module for the human anaphase-promoting complex. Proc. Natl. Acad. Sci. USA 106, 18213–18218 10.1073/pnas.0907887106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K., Youle R. J. (2013). PINK1 is degraded through the N-end rule pathway. Autophagy 9, 1758–1769 10.4161/auto.24633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Rape M. (2009). Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10, 755–764 10.1038/nrm2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii S. R., Kishi C., Ishihara N., Mizushima N. (2011). Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J. Biol. Chem. 286, 19630–19640 10.1074/jbc.M110.209338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Hunter T. (2013). Parkin mitochondrial translocation is achieved through a novel catalytic activity coupled mechanism. Cell Res. 23, 886–897 10.1038/cr.2013.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.