Abstract
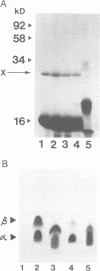
From in vitro translation studies we have previously demonstrated the existence of an apparent efficient UAG (amber) suppressor tRNA in the dimorphic fungus Candida albicans (Santos et al., 1990). Using an in vitro assay for termination codon readthrough the tRNA responsible was purified to homogeneity from C.albicans cells. The determined sequence of the purified tRNA predicts a 5'-CAG-3' anticodon that should decode the leucine codon CUG and not the UAG termination codon as originally hypothesized. However, the tRNA(CAG) sequence shows greater nucleotide homology with seryl-tRNAs from the closely related yeast Saccharomyces cerevisiae than with leucyl-tRNAs from the same species. In vitro tRNA-charging studies demonstrated that the purified tRNA(CAG) is charged with Ser. The gene encoding the tRNA was cloned from C.albicans by a PCR-based strategy and DNA sequence analysis confirmed both the structure of the tRNA(CAG) and the absence of any introns in the tRNA gene. The copy number of the tRNA(CAG) gene (1-2 genes per haploid genome) is in agreement with the relatively low abundance (< 0.5% total tRNA) of this tRNA. In vitro translation studies revealed that the purified tRNA(CAG) could induce apparent translational bypass of all three termination codons. However, peptide mapping of in vitro translation products demonstrated that the tRNA(CAG) induces translational misreading in the amino-terminal region of two RNA templates employed, namely the rabbit alpha- and beta-globin mRNAs. These results suggest that the C.albicans tRNA(CAG) is not an 'omnipotent' suppressor tRNA but rather may mediate a novel non-standard translational event in vitro during the translation of the CUG codon. The possible nature of this non-standard translation event is discussed in the context of both the unusual structural features of the tRNA(CAG) and its in vitro behaviour.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Weiss R. B., Gesteland R. F. Ribosome gymnastics--degree of difficulty 9.5, style 10.0. Cell. 1990 Aug 10;62(3):413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins J. F., Weiss R. B., Thompson S., Gesteland R. F. Towards a genetic dissection of the basis of triplet decoding, and its natural subversion: programmed reading frame shifts and hops. Annu Rev Genet. 1991;25:201–228. doi: 10.1146/annurev.ge.25.120191.001221. [DOI] [PubMed] [Google Scholar]
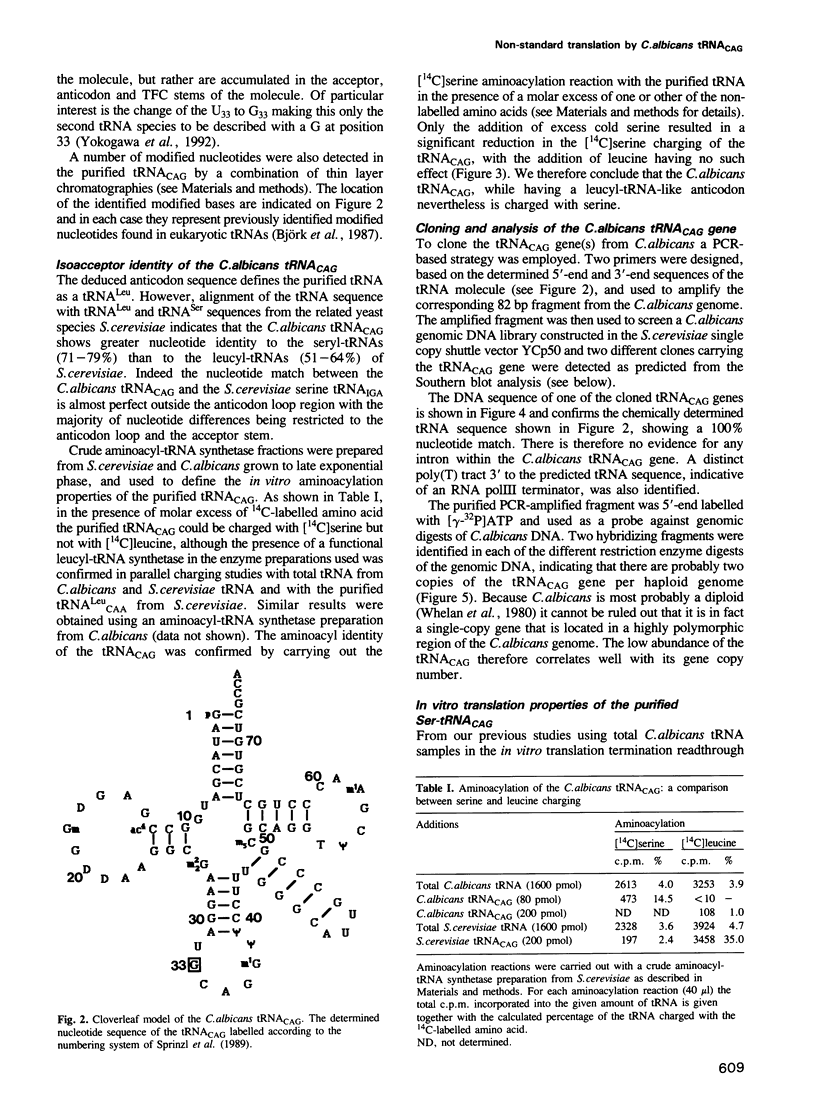
- Björk G. R., Ericson J. U., Gustafsson C. E., Hagervall T. G., Jönsson Y. H., Wikström P. M. Transfer RNA modification. Annu Rev Biochem. 1987;56:263–287. doi: 10.1146/annurev.bi.56.070187.001403. [DOI] [PubMed] [Google Scholar]
- Brown A. J., Bertram G., Feldmann P. J., Peggie M. W., Swoboda R. K. Codon utilisation in the pathogenic yeast, Candida albicans. Nucleic Acids Res. 1991 Aug 11;19(15):4298–4298. doi: 10.1093/nar/19.15.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böck A., Forchhammer K., Heider J., Leinfelder W., Sawers G., Veprek B., Zinoni F. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991 Mar;5(3):515–520. doi: 10.1111/j.1365-2958.1991.tb00722.x. [DOI] [PubMed] [Google Scholar]
- Colthurst D. R., Schauder B. S., Hayes M. V., Tuite M. F. Elongation factor 3 (EF-3) from Candida albicans shows both structural and functional similarity to EF-3 from Saccharomyces cerevisiae. Mol Microbiol. 1992 Apr;6(8):1025–1033. doi: 10.1111/j.1365-2958.1992.tb02168.x. [DOI] [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Diamond A., Dudock B., Hatfield D. Structure and properties of a bovine liver UGA suppressor serine tRNA with a tryptophan anticodon. Cell. 1981 Aug;25(2):497–506. doi: 10.1016/0092-8674(81)90068-4. [DOI] [PubMed] [Google Scholar]
- Fox T. D. Natural variation in the genetic code. Annu Rev Genet. 1987;21:67–91. doi: 10.1146/annurev.ge.21.120187.000435. [DOI] [PubMed] [Google Scholar]
- Gillam I., Blew D., Warrington R. C., von Tigerstrom M., Tener G. M. A general procedure for the isolation of specific transfer ribonucleic acids. Biochemistry. 1968 Oct;7(10):3459–3468. doi: 10.1021/bi00850a022. [DOI] [PubMed] [Google Scholar]
- Gillam I., Millward S., Blew D., von Tigerstrom M., Wimmer E., Tener G. M. The separation of soluble ribonucleic acids on benzoylated diethylaminoethylcellulose. Biochemistry. 1967 Oct;6(10):3043–3056. doi: 10.1021/bi00862a011. [DOI] [PubMed] [Google Scholar]
- Gorman J. A., Chan W., Gorman J. W. Repeated use of GAL1 for gene disruption in Candida albicans. Genetics. 1991 Sep;129(1):19–24. doi: 10.1093/genetics/129.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D. L., Smith D. W., Lee B. J., Worland P. J., Oroszlan S. Structure and function of suppressor tRNAs in higher eukaryotes. Crit Rev Biochem Mol Biol. 1990;25(2):71–96. doi: 10.3109/10409239009090606. [DOI] [PubMed] [Google Scholar]
- Hatfield D., Diamond A., Dudock B. Opal suppressor serine tRNAs from bovine liver form phosphoseryl-tRNA. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6215–6219. doi: 10.1073/pnas.79.20.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hizi A., Henderson L. E., Copeland T. D., Sowder R. C., Hixson C. V., Oroszlan S. Characterization of mouse mammary tumor virus gag-pro gene products and the ribosomal frameshift site by protein sequencing. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7041–7045. doi: 10.1073/pnas.84.20.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes W. M., Hurd R. E., Reid B. R., Rimerman R. A., Hatfield G. W. Separation of transfer ribonucleic acid by sepharose chromatography using reverse salt gradients. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1068–1071. doi: 10.1073/pnas.72.3.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 1982 Jul 15;158(4):573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- Jacks T., Power M. D., Masiarz F. R., Luciw P. A., Barr P. J., Varmus H. E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988 Jan 21;331(6153):280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- Jacks T., Townsley K., Varmus H. E., Majors J. Two efficient ribosomal frameshifting events are required for synthesis of mouse mammary tumor virus gag-related polyproteins. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4298–4302. doi: 10.1073/pnas.84.12.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Varmus H. E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985 Dec 13;230(4731):1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- Jakubowski H., Goldman E. Editing of errors in selection of amino acids for protein synthesis. Microbiol Rev. 1992 Sep;56(3):412–429. doi: 10.1128/mr.56.3.412-429.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y., Honda H., Taniguchi-Morimura J., Iwasaki S. The codon CUG is read as serine in an asporogenic yeast Candida cylindracea. Nature. 1989 Sep 14;341(6238):164–166. doi: 10.1038/341164a0. [DOI] [PubMed] [Google Scholar]
- Keith G., Pixa G., Fix C., Dirheimer G. Primary structure of three tRNAs from brewer's yeast: tRNAPro2, tRNAHis1 and tRNAHis2. Biochimie. 1983 Nov-Dec;65(11-12):661–672. doi: 10.1016/s0300-9084(84)80030-9. [DOI] [PubMed] [Google Scholar]
- Leinfelder W., Zehelein E., Mandrand-Berthelot M. A., Böck A. Gene for a novel tRNA species that accepts L-serine and cotranslationally inserts selenocysteine. Nature. 1988 Feb 25;331(6158):723–725. doi: 10.1038/331723a0. [DOI] [PubMed] [Google Scholar]
- Mizutani T., Tachibana Y. Possible incorporation of phosphoserine into globin readthrough protein via bovine opal suppressor phosphoseryl-tRNA. FEBS Lett. 1986 Oct 20;207(1):162–166. doi: 10.1016/0014-5793(86)80032-1. [DOI] [PubMed] [Google Scholar]
- Morrison M. R., Brinkley S. A., Gorski J., Lingrel J. B. The separation and identification of alpha- and beta-globin messenger ribonucleic acids. J Biol Chem. 1974 Aug 25;249(16):5290–5295. [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Rovera G., Magarian C., Borun T. W. Resolution of hemoglobin subunits by electrophoresis in acid urea polyacrylamide gels containing Triton X-100. Anal Biochem. 1978 Apr;85(2):506–518. doi: 10.1016/0003-2697(78)90248-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M., Colthurst D. R., Wills N., McLaughlin C. S., Tuite M. F. Efficient translation of the UAG termination codon in Candida species. Curr Genet. 1990 Jun;17(6):487–491. doi: 10.1007/BF00313076. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. The use of nuclease P1 in sequence analysis of end group labeled RNA. Nucleic Acids Res. 1977 Dec;4(12):4091–4108. doi: 10.1093/nar/4.12.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberklang M., Prochiantz A., Haenni A. L., Rajbhandary U. L. Studies on the sequence of the 3'-terminal region of turnip-yellow-mosaic-virus RNA. Eur J Biochem. 1977 Feb;72(3):465–478. doi: 10.1111/j.1432-1033.1977.tb11270.x. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Hartmann T., Weber J., Blank J., Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17 (Suppl):r1–172. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite M. F., Bower P. A., McLaughlin C. S. A novel suppressor tRNA from the dimorphic fungus Candida albicans. Biochim Biophys Acta. 1986 Feb 24;866(1):26–31. doi: 10.1016/0167-4781(86)90096-5. [DOI] [PubMed] [Google Scholar]
- Tuite M. F., Plesset J. mRNA-dependent yeast cell-free translation systems: theory and practice. Yeast. 1986 Mar;2(1):35–52. doi: 10.1002/yea.320020103. [DOI] [PubMed] [Google Scholar]
- Valle R. P., Morch M. D. Stop making sense: or Regulation at the level of termination in eukaryotic protein synthesis. FEBS Lett. 1988 Aug 1;235(1-2):1–15. doi: 10.1016/0014-5793(88)81225-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan W. L., Partridge R. M., Magee P. T. Heterozygosity and segregation in Candida albicans. Mol Gen Genet. 1980;180(1):107–113. doi: 10.1007/BF00267358. [DOI] [PubMed] [Google Scholar]
- Wilson W., Braddock M., Adams S. E., Rathjen P. D., Kingsman S. M., Kingsman A. J. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell. 1988 Dec 23;55(6):1159–1169. doi: 10.1016/0092-8674(88)90260-7. [DOI] [PubMed] [Google Scholar]
- Yokogawa T., Suzuki T., Ueda T., Mori M., Ohama T., Kuchino Y., Yoshinari S., Motoki I., Nishikawa K., Osawa S. Serine tRNA complementary to the nonuniversal serine codon CUG in Candida cylindracea: evolutionary implications. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7408–7411. doi: 10.1073/pnas.89.16.7408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H. J., Donahue T. F. The suil suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNA(iMet) recognition of the start codon. Mol Cell Biol. 1992 Jan;12(1):248–260. doi: 10.1128/mcb.12.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Adlouni C., Desgrès J., Dirheimer G., Keith G. Sequence of a new tRNA(Leu)(U*AA) from brewer's yeast. Biochimie. 1991 Nov;73(11):1355–1360. doi: 10.1016/0300-9084(91)90165-w. [DOI] [PubMed] [Google Scholar]