Abstract
Among the iron-sulfur cluster assembly proteins encoded by gene cluster iscSUA-hscBA-fdx in Escherichia coli, IscA has a unique and strong iron binding activity and can provide iron for iron-sulfur cluster assembly in proteins in vitro. Deletion of IscA and its paralogue SufA results in an E. coli mutant that fails to assemble [4Fe-4S] clusters in proteins under aerobic conditions, suggesting that IscA has a crucial role for iron-sulfur cluster biogenesis. Here we report that among the iron-sulfur cluster assembly proteins, IscA also has a strong and specific binding activity for Cu(I) in vivo and in vitro. The Cu(I) center in IscA is stable and resistant to oxidation under aerobic conditions. Mutation of the conserved cysteine residues that are essential for the iron binding in IscA abolishes the copper binding activity, indicating that copper and iron may share the same binding site in the protein. Additional studies reveal that copper can compete with iron for the metal binding site in IscA and effectively inhibits the IscA-mediated [4Fe-4S] cluster assembly in E. coli cells. The results suggest that copper may not only attack the [4Fe-4S] clusters in dehydratases, but also block the [4Fe-4S] cluster assembly in proteins by targeting IscA in cells.
Keywords: IscA, copper toxicity, iron-sulfur cluster assembly
Introduction
IscA is a key member of the iron-sulfur cluster assembly machinery encoded by the housekeeping gene cluster iscSUA-hscBA-fdx in Escherichia coli (Zheng et al., 1998, Roche et al., 2013), and is highly conserved among aerobic organisms from bacteria to humans (Vinella et al., 2009). Biochemical studies have shown that IscA may act as an alternative scaffold (Krebs et al., 2001, Ollagnier-de-Choudens et al., 2001) or intermediate carrier for iron-sulfur cluster biogenesis (Mapolelo et al., 2012b, Mapolelo et al., 2013, Vinella et al., 2013). However, unlike other scaffold proteins such as IscU (Agar et al., 2000, Unciuleac et al., 2007, Raulfs et al., 2008), E. coli IscA has a unique and strong iron binding activity in vitro (Ding & Clark, 2004, Ding et al., 2005b, Yang et al., 2006, Landry et al., 2013) and in vivo (Wang et al., 2010). Recent studies further showed that iron binding activity of IscA is conserved, as IscA homologues from Azotobacter vinelandii (Mapolelo et al., 2012a), Saccharomyces cerevisiae (Muhlenhoff et al., 2011), and humans (Lu et al., 2010) also have a strong iron binding activity. Furthermore, the iron center in IscA can be mobilized by L-cysteine (Ding et al., 2005a, Landry et al., 2013) and transferred for iron-sulfur cluster assembly in target proteins in vitro (Yang et al., 2006), suggesting that IscA may act as an iron chaperone to deliver iron for iron-sulfur cluster biogenesis. In human cells, depletion of IscA1 results in deficiency of iron-sulfur cluster assembly in mitochondria and cytosol (Song et al., 2009). In A. vinelandii, depletion of IscA homologue leads to a null growth phenotype when cells are cultured under the oxygen-elevated conditions (Johnson et al., 2006). In E. coli, deletion of IscA and its paralogue SufA also produces a null growth phenotype in M9 minimal media under aerobic conditions (Lu et al., 2008, Mettert et al., 2008). Further studies revealed that deletion of IscA and SufA blocks the [4Fe-4S] cluster assembly without significant effect on the [2Fe-2S] cluster assembly in E. coli cells under aerobic growth conditions (Tan et al., 2009), suggesting that IscA/SufA has a crucial role for the [4Fe-4S] cluster assembly in E. coli cells. Consistent with this idea, other research groups have also reported that IscA homologues are essential for the [4Fe-4S] cluster assembly in S. cerevisiae (Muhlenhoff et al., 2011) and human cells (Sheftel et al., 2012).
E. coli IscA is a homodimer with three conserved cysteine residues (Cys-35, Cys-99 and Cys-101) from each monomer forming a “cysteine pocket” between two monomers (Bilder et al., 2004, Cupp-Vickery et al., 2004). The site-directed mutagenesis studies showed that the “cysteine pocket” is essential for the iron binding activity in vitro (Ding et al., 2004) and the physiological function of IscA in E. coli cells (Lu et al., 2008). The “cysteine pocket” in IscA appears to be highly flexible to accommodate a mononuclear iron or an iron-sulfur cluster without significant change of the structure (Wada et al., 2005). The flexibility of the “cysteine pocket” led us to postulate that IscA may also bind other transition metal ions such as copper in its metal binding site. While copper is an essential element for all living cells, excess copper is highly toxic (Rodriguez-Montelongo et al., 1993, Karlsson et al., 2008). Recent studies further suggested that excess copper may disrupt the labile [4Fe-4S] clusters in dehydratases (Macomber & Imlay, 2009) and block iron-sulfur cluster biogenesis in E. coli (Fung et al., 2013, Outten & Munson, 2013) and Bacillus subtilis (Chillappagari et al., 2010). Since iron-sulfur proteins are involved in diverse physiological processes from energy metabolism to DNA repair and replication (Johnson et al., 2005, White & Dillingham, 2012), the copper-mediated inhibition of iron-sulfur cluster biogenesis would have a broad impact on multiple cellular functions. Nevertheless, the molecular mechanism underlying the copper-mediated toxicity on iron-sulfur proteins has not been fully understood. Here, we report that among the iron-sulfur cluster assembly proteins encoded by the gene cluster iscSUA-hscBA-fdx (Zheng et al., 1998), IscA has a strong and specific copper binding activity in E. coli cells and in vitro. Copper and iron appear to share the same binding site in IscA, as mutation of the three conserved cysteine residues in IscA abolishes the iron and copper binding activities. Furthermore, excess copper can compete with iron for the metal binding site in IscA and effectively inhibit the IscA-mediated [4Fe-4S] cluster assembly without significant effect on the [2Fe-2S] cluster assembly in E. coli cells. The results suggest that copper may not only attack the labile [4Fe-4S] clusters in dehydratases as reported previously (Macomber & Imlay, 2009), but also block the [4Fe-4S] cluster assembly in E. coli cells by targeting the iron-sulfur cluster assembly protein IscA.
Results
IscA has a unique and strong copper binding activity among the iron-sulfur cluster assembly proteins
To prevent or alleviate copper toxicity, E. coli has three copper homeostatic systems to maintain low intracellular copper content: CopA, a P-type ATPase that pumps copper ion out of the cytoplasm (Fan & Rosen, 2002); CueO, an oxidase that oxidizes Cu(I) to Cu(II) in the periplasm to prevent adventitious entry into the cytoplasm (Stoyanov et al., 2001); and a copper pump that transports copper ion from the periplasm to the extracellular environment (Munson et al., 2000). Deletion of CopA, CueO, and CusA, a subunit of the copper pump (Munson et al., 2000), results in an E. coli strain that is hypersensitive to copper in growth media (Grass & Rensing, 2001, Macomber & Imlay, 2009).
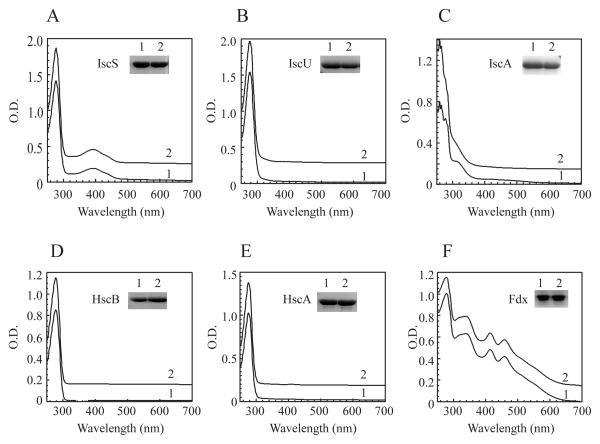
To explore the copper binding activity of iron-sulfur cluster assembly proteins in vivo, we expressed each protein encoded by the gene cluster iscSUA-hscBA-fdx in the constructed E. coli copA/cueO/cusA mutant cells grown in LB media under aerobic conditions. CuSO4 (200 μM) was added to the cell cultures ten min before the expression of recombinant protein was induced. CuSO4 at 200 μM was chosen as it reduced cell growth of the E. coli copA/cueO/cusA mutant in LB by about 20% and did not significantly affect protein synthesis in the cells. Each of the iron-sulfur cluster assembly proteins was produced in the E. coli copA/cueO/cusA mutant cells grown in LB media supplemented with or without 200 μM CuSO4. Purified proteins were then subjected to the UV-visible absorption measurements and metal content analyses. As shown in Figure 1A, addition of CuSO4 (200 μM) to LB media had little or no effect on the UV-visible absorption spectrum of IscS, a cysteine desulfurase that catalyzes desulfurization of L-cysteine and provides sulfide for iron-sulfur cluster assembly in proteins (Smith et al., 2001, Marinoni et al., 2012). Similarly, addition of CuSO4 (200 μM) to LB media did not significantly change the UV-visible absorption spectra of the iron-sulfur cluster assembly scaffold protein IscU (Agar et al., 2000, Unciuleac et al., 2007, Raulfs et al., 2008) (Figure 1B), and heat shock cognate proteins HscB (Figure 1D) and HscA (Figure 1E) (Kim et al., 2012). Ferredoxin (Fdx) [2Fe-2S] cluster (Ta & Vickery, 1992, Chandramouli et al., 2007, Yan et al., 2013, Kim et al., 2013) was also not affected by addition of CuSO4 (200 μM) in LB media (Figure 1F). Only IscA showed a new absorption peak at 258 nm (Figure 1C) when CuSO4 (200 μM) was added to LB media. As the absorption peak at 258 nm has been attributed to the Cu(I) binding site with two cysteine residues as ligands in the Parkinsonism-associated protein DJ-1 (Puno et al., 2013), we propose that IscA may bind Cu(I) via cysteine residues.
Figure 1. IscA has a unique copper binding activity among the iron-sulfur cluster assembly proteins.
Each protein encoded by the gene cluster iscSUA-hscBA-fdx was expressed in the E. coli copA/cueO/cusA mutant cells grown in LB media supplemented with or without 200 μM CuSO4. Proteins were purified from the cells and subjected to UV-visible absorption measurements. A), IscS; B), IscU; C), IscA; D), HscB; E), HscA; F), Ferredoxin. In each panel: spectrum 1, without CuSO4 in LB media; spectrum 2, with CuSO4 in LB media. Insert in each panel is a photograph of SDS-PAGE gel of purified proteins. The results are representatives of three independent protein preparations.
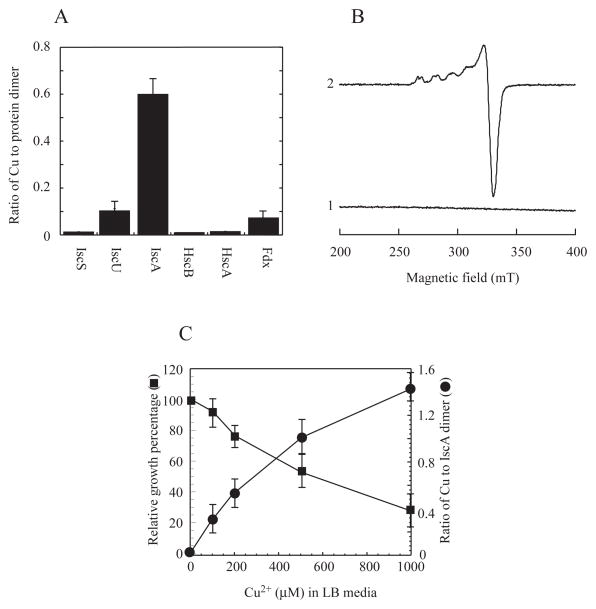
The copper content analyses using neocuproine or ICP-ES (the Inductively Coupled Plasma-Emission Spectrometry) showed that purified IscA contained 0.60±0.09 copper atoms per IscA dimer, while IscS, HscA and HscB had very little or no detectable amounts of copper (Figure 2A). IscU and ferredoxin retained a small but reproducible amount of copper (~ 0.1 copper atoms per dimer). However, unlike IscA, the copper content in IscU and ferredoxin was further decreased after additional purification steps, indicating that copper binding in IscU and ferredoxin was not stable. Thus, among the iron-sulfur cluster assembly proteins encoded by iscSUA-hscBA-fdx, IscA has a unique and strong copper binding activity.
Figure 2. Relative copper binding activity of IscA in the E. coli copA/cueO/cusA mutant cells.
A), the copper content of the iron-sulfur cluster assembly proteins encoded by the gene cluster iscSUA-hscBA-fdx purified from the E. coli copA/cueO/cusA mutant cells grown in LB media supplemented with 200 μM CuSO4. B), the EPR spectra of IscA purified from the E. coli copA/cueO/cusA mutant cells grown in LB media supplemented with 200 μM CuSO4. Purified IscA (50 μM) (spectrum 1) was treated with 2.5% nitric acid (spectrum 2). C), correlation of the copper binding in IscA and the relative cell growth in LB media supplemented with 0, 100, 200, 500, and 1000 μM CuSO4. The copper content in purified IscA was plotted as a function of the CuSO4 concentration in LB media (closed circles). The relative cell growth was defined as the percentage of the cell growth in LB media with CuSO4 over that without CuSO4, and was plotted as a function of the CuSO4 concentration in LB media (closed squares). The 100% cell growth represented the cell density of O.D. at 600 nm of ~ 1.0 after 3 hours at 37°C in LB media with aeration. The results are the mean ± standard deviations from three independent experiments.
As copper-binding proteins often have electron paramagnetic resonance (EPR) signals (Ve et al., 2012), purified IscA was subjected to EPR measurements. As shown in Figure 2B, IscA purified from the E. coli copA/cueO/cusA mutant cells grown in LB media supplemented with CuSO4 (200 μM) had no EPR signal. However, when purified IscA was treated with 2.5% (v/v) nitric acid to oxidize Cu(I) in the protein as described in (Ve et al., 2012), an EPR signal representing a typical Cu(II) center (Smith et al., 2008, Ve et al., 2012) appeared (Figure 2B), demonstrating that purified IscA indeed binds Cu(I) that can be oxidized by the strong oxidant nitric acid. Quantification of the Cu(II) EPR signal showed that purified IscA contained 0.55±0.12 copper atoms per IscA dimer, which is close to that determined using neocuproine (0.60±0.09 copper atoms per IscA dimer).
To further determine the copper binding activity of IscA in vivo, recombinant IscA was expressed in the E. coli copA/cueO/cusA mutant cells grown in LB media supplemented with increased concentrations of CuSO4. Figure 2C shows that as the concentration of CuSO4 in LB media was gradually increased from 0 to 1.0 mM, the copper binding of IscA was progressively increased from 0 to about 1.4 copper atoms per IscA dimer. On the other hand, the cell growth of the E. coli copA/cueO/cusA mutant was gradually decreased to about 30% when the concentration of CuSO4 in LB media was increased from 0 to 1.0 mM (Figure 2C). Because the cell growth of the E. coli copA/cueO/cusA mutant in LB media was severely inhibited by 1.0 mM CuSO4 (Figure 2C), we were unable to obtain the maximum copper binding in IscA expressed in the E. coli copA/cueO/cusA mutant cells. Nevertheless, the results suggest that the copper binding in IscA inversely correlates with the cell growth when the concentration of CuSO4 in LB media is increased from 0 to 1.0 mM.
The in vitro copper binding activity of IscA
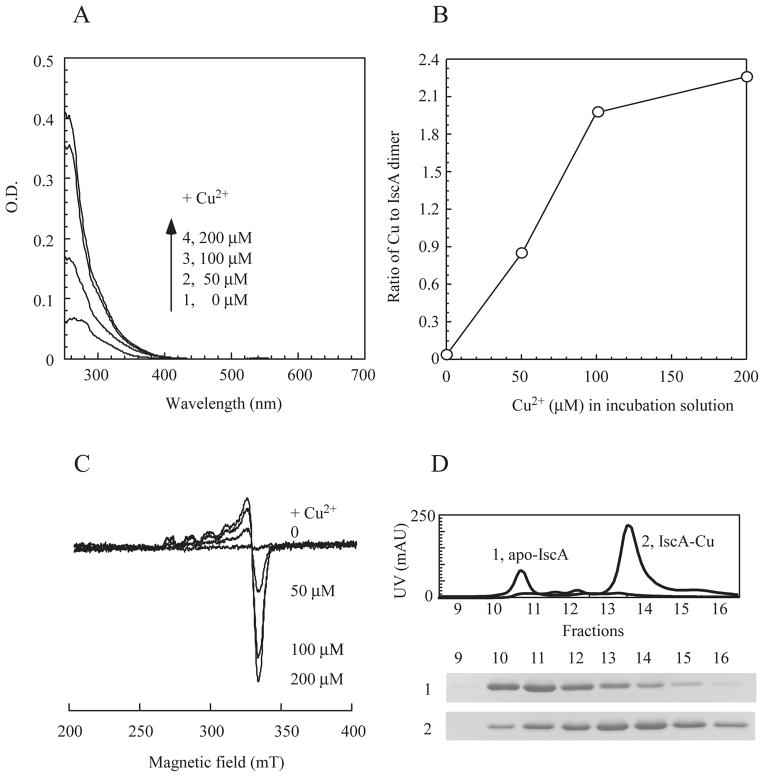
To determine the in vitro copper binding activity of IscA, we prepared apo-IscA as described previously (Landry et al., 2013) and incubated apo-IscA (50 μM dimer) with increasing concentrations of CuSO4 (0 to 200 μM) in the presence of dithiothreitol. Dithiothreitol was used to reduce Cu(II) to Cu(I) (Banci et al., 2003) to emulate the intracellular redox state of copper ion (Macomber & Imlay, 2009) and to reduce thiol groups in IscA for copper binding. After incubation at room temperature for 15 min, IscA was re-purified by passing through a HiTrap Desalting column. As the CuSO4 concentration in the incubation solution was increased, the amplitude of the absorption peak at 258 nm (Figure 3A) and the copper content (Figure 3B) of re-purified IscA were gradually increased and saturated at about two-fold excess of CuSO4 in the incubation solution. It was reported that dithiothreitol and copper may generate hydroxyl free radicals in solution (Kachur et al., 1997). However, production of hydroxyl free radicals in solution had a long delay (up to 5 hours) after dithiothreitol was mixed with copper under aerobic conditions (Kachur et al., 1997). Furthermore, similar results were obtained when apo-IscA was incubated with CuSO4 (0 to 200 μM) and dithiothreitol (2 mM) under anaerobic conditions (data not shown). Thus, it is unlikely that oxygen or hydroxyl free radicals are involved in the copper binding in IscA. Because the copper binding in IscA was almost linear at low concentrations of CuSO4 (Figure 3B), we were unable to estimate the copper binding constant of IscA. Nevertheless, the result clearly showed that the maximum copper binding in IscA is 2.2±0.4 copper atoms per IscA dimer (Figure 3B).
Figure 3. In vitro copper binding activity of IscA.
Apo-IscA (50 μM dimer) was incubated with 0, 50, 100, and 200 μM CuSO4 in the presence of dithiothreitol (2 mM). Protein was re-purified from incubation solutions by passing through a Hi-trap Desalt column. A), UV-visible spectra of re-purified IscA after reconstitution with the indicated concentration of CuSO4. B), relative copper binding activity of IscA. The copper content of re-purified IscA was analyzed and plotted as a function of the CuSO4 concentration in the incubation solution. C), the EPR spectra of the copper-bound IscA. Re-purified IscA proteins were treated with 2.5% (v/v) nitric acid and subjected to the EPR measurements. D), elution profiles of apo-IscA and the copper-bound IscA from a Mono-Q column. The copper-bound IscA was prepared after apo-IscA (50 μM dimer) was incubated with 200 μM CuSO4 in the presence of dithiothreitol (2 mM). Top, elution profiles of apo-IscA (trace 1) or the copper-bound IscA (trace 2) using a linear gradient of NaCl (0 to 0.5 M). Bottom, photographs of the SDS-PAGE gel of the eluted fractions of apo-IscA (sample 1) and the copper-bound IscA (sample 2).
IscA proteins re-purified after incuabtion with CuSO4 and dithiothreitol were also subjected to EPR measurements. Without any treatments, re-purified IscA proteins were EPR silent, similar to that purified from E. coli cells (Figure 2B). However, when re-purified IscA proteins were treated with 2.5% (v/v) nitric acid to oxidize Cu(I) (Ve et al., 2012), a typical Cu(II) EPR signal appeared, indicating that IscA also binds Cu(I) in vitro. The amplitude of the Cu(II) EPR signal of IscA was proportionally increased as the concentration of CuSO4 was increased in the incubation solution (Figure 3C), and was closely correlated with the copper content in IscA (Figure 3B).
To further explore the property of the copper-bound IscA, re-purified IscA proteins were analyzed using an anion exchange Mono-Q column which separates proteins based on their net charge and protein conformation. Figure 3D shows that copper binding in IscA shifts the protein elution profile from fractions 10 and 11 (apo-IscA) to fractions 13 and 14 (the copper-bound IscA), indicating that copper binding has significantly changed the conformation of IscA dimer.
The conserved cysteine residues in IscA are required for the copper binding activity
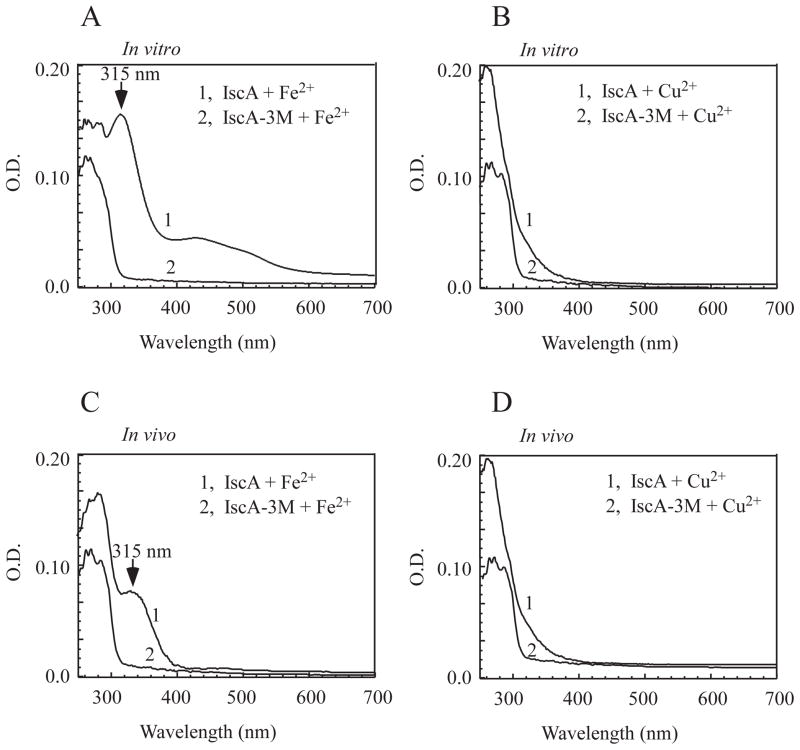
The three conserved cysteine residues (Cys-35, Cys-99 and Cys-101) are essential for the iron binding activity of IscA in vitro (Ding et al., 2004) and for its physiological function in E. coli (Lu et al., 2008). If copper and iron share the same binding site in IscA, mutation of the conserved cysteine residues would disrupt both the iron and copper binding in the protein. To test this idea, we constructed an IscA mutant in which all three cysteine residues (Cys-35, Cys-99 and Cys-101) were replaced with serine. Prepared apo-IscA and IscA mutant were then incubated with two-fold excess of Fe(NH4)2(SO4)2 or CuSO4 in the presence of dithiothreitol, followed by protein re-purification to remove residual metal ions and dithiothreitol. As reported previously, incubation of the wild-type apo-IscA with two-fold excess of iron or copper produced the iron-bound IscA (with an absorption peak at 315 nm) (Figure 4A) (Ding & Clark, 2004) or the copper-bound IscA (with an absorption peak at 258 nm) (Figure 4B), respectively. In contrast, incubation of the IscA mutant with iron or copper did not produce any absorption peaks of the iron- or copper- binding in the protein. The iron and copper content analyses confirmed that unlike the wild-type IscA, the IscA mutant did not bind any detectable amounts of iron or copper under the same experimental conditions (data not shown).
Figure 4. The conserved cysteine residues are essential for the copper binding in IscA.
A), UV-visible spectra of IscA and the IscA mutant (IscA-3M) after incubation with iron in vitro. Apo-IscA and the IscA triple mutant (50 μM dimer) were incubated with Fe(NH4)2 (SO4)2 (100 μM) in the presence of dithiothreitol (2 mM) at room temperature for 15 min, followed by protein re-purification. B), UV-visible spectra of IscA and the IscA mutant (IscA-3M) after incubation with copper in vitro. Apo-IscA and the IscA mutant (50 μM dimer) were incubated with CuSO4 (100 μM) in the presence of dithiothreitol (2 mM) at room temperature for 15 min, followed by protein re-purification. C), UV-visible spectra of IscA and the IscA mutant (IscA-3M) purified from the E. coli copA/cueO/cusA mutant cells grown in LB media supplemented with Fe(NH4)2 (SO4)2 (200 μM). D), UV-visible spectra of IscA and the IscA mutant (IscA-3M) purified from the E. coli copA/cueO/cusA mutant cells grown in LB media supplemented with CuSO4 (200 μM).
The wild-type IscA and IscA mutant proteins were also expressed in the E. coli copA/cueO/cusA mutant cells grown in LB media supplemented with Fe(NH4)2(SO4)2 (200 μM) or CuSO4 (200 μM). Again, unlike the wild-type IscA, the IscA mutant failed to bind any iron (Figure 4C) or copper (Figure 4D) in the E. coli cells. Taken together, the results suggest that the three conserved cysteine residues in IscA are essential for its iron and copper binding activity in vitro and in vivo.
Copper competes with iron for the metal binding site in IscA
If iron and copper share the metal binding site, excess copper may compete with iron for the same binding site in IscA. Indeed, addition of CuSO4 (200 μM) to LB media largely eliminated the iron binding peak at 315 nm of IscA expressed in the E. coli copA/cueO/cusA mutant cells (Figure 1C). The metal content analyses further showed that the iron content of IscA was decreased from 0.21±0.04 to less than 0.04 iron atoms with concomitant increase of the copper content from 0 to 0.60±0.09 copper atoms per IscA dimer when LB media were supplemented with 200 μM CuSO4.
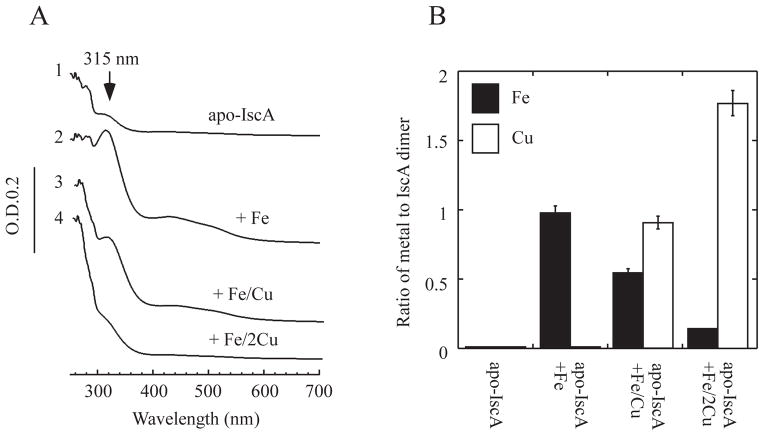
To further explore the copper and iron binding competition in IscA, purified apo-IscA was incubated with a fixed amount of iron and increasing amounts of copper in the presence of dithiothreitol, followed by re-purification of IscA. As shown in Figure 5A, incubation of apo-IscA (50 μM dimer) with 100 μM Fe(NH4)2(SO4)2 produced an iron-bound IscA with an absorption peak at 315 nm as reported above. However, when apo-IscA (50 μM dimer) was incubated with 100 μM Fe(NH4)2(SO4)2 and 100 μM CuSO4, the absorption peak at 315 nm of IscA was significantly diminished (Figure 5A), and the iron content of IscA was decreased with concomitant increase of the copper content in the protein (Figure 5B). When apo-IscA (50 μM dimer) was incubated with 100 μM Fe(NH4)2(SO4)2 and 200 μM CuSO4, the absorption peak at 315 nm of re-purified IscA was largely eliminated (Figure 5A), and the iron content of IscA was further decreased with the copper content increased to 1.81±0.15 copper atoms per IscA dimer (Figure 5B). Thus, excess copper can effectively compete with iron for the metal binding site in IscA.
Figure 5. Excess copper competes with iron for the metal binding sites in IscA.
A), copper competes for the iron binding site in IscA. Apo-IscA (50 μM dimer) (spectrum 1) was incubated with Fe(NH4)2 (SO4)2 (100 μM) (spectrum 2), Fe(NH4)2 (SO4)2 (100 μM) and CuSO4 (100 μM) (spectrum 3), or Fe(NH4)2 (SO4)2 (100 μM) and CuSO4 (200 μM) (spectrum 4) in the presence of dithiothreitol (2 mM) at room temperature for 15 min. IscA was re-purified from the incubation solutions and subjected to UV-visible absorption measurements. B), iron and copper content analyses of re-purified IscA. The iron and copper content of re-purified IscA proteins from panel A) were analyzed and plotted as the ratio of metal to IscA dimer. The protein concentration was determined from the samples after incubation without metal ions. The results are presented as the mean ± standard deviations.
Copper inhibits the [4Fe-4S] cluster assembly in the E. coli cells
Previous studies revealed that IscA and its paralogue SufA are essential for the [4Fe-4S] cluster assembly in E. coli cells under aerobic conditions (Tan et al., 2009). IscA and SufA share the similar crystal structure (Wada et al., 2005, Cupp-Vickery et al., 2004, Bilder et al., 2004) and iron binding activity (Lu et al., 2008). Parallel experiments showed that E. coli SufA also has a similar copper binding activity as IscA in the E. coli copA/cueO/cusA mutant cells grown in LB media (Supplementary Figure 1). Thus, if copper competes with iron for the metal binding site in IscA and SufA, copper may inhibit the IscA/SufA-mediated [4Fe-4S] cluster assembly in E. coli cells. To test this idea, we decided to analyze the [4Fe-4S] cluster assembly in the E. coli copA/cueO/cusA mutant cells grown in LB media supplemented with or without CuSO4 (200 μM).
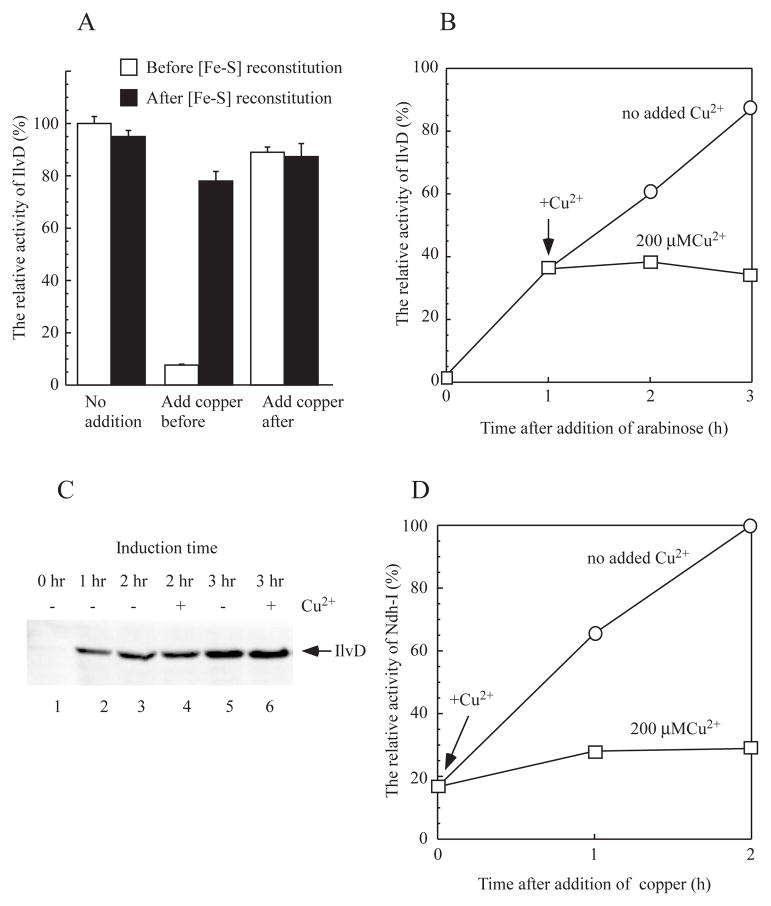
E. coli dihydroxyacid dehydratase (IlvD) requires an intact [4Fe-4S] cluster for its catalytic activity (Flint et al., 1993). Relative activity of IlvD has previously been used to assess the [4Fe-4S] cluster assembly in the protein in E. coli cells (Ren et al., 2008). In this set of experiments, recombinant IlvD was expressed in the E. coli copA/cueO/cusA mutant cells grown in LB media supplemented with or without CuSO4 (200 μM). The cell extracts were prepared after the cells were passed through French press once, and immediately used for the IlvD activity assay. Figure 6A shows that addition of CuSO4 (200 μM) to LB media before recombinant IlvD was expressed in the E. coli cells decreased the IlvD enzyme activity to about 10%. However, the enzyme activity of IlvD was largely recovered when the cell extracts were incubated with L-cysteine, cysteine desulfurase IscS, Fe(NH4)2(SO4)2, and dithiothreitol to reconstitute the [4Fe-4S] clusters in IlvD under anaerobic conditions (Figure 6A), suggesting that CuSO4 (200 μM) in LB media blocks the [4Fe-4S] cluster assembly in IlvD without inhibiting protein synthesis in the E. coli cells. Interestingly, when CuSO4 (200 μM) was added to LB media after recombinant IlvD was expressed in the E. coli cells, over 85% of the enzyme activity of IlvD remained, indicating that copper at 200 μM in LB media does not significantly affect the pre-assembled [4Fe-4S] clusters in IlvD in E. coli cells.
Figure 6. Copper largely blocks the [4Fe-4S] cluster assembly in the E. coli cells.
A), inhibition of the [4Fe-4S] cluster assembly in recombinant dihydroxyacid dehydratase (IlvD) in the E. coli cells by copper. Recombinant IlvD was expressed in the E. coli copA/cueO/cusA mutant cells grown in LB media. CuSO4 (200 μM) was added to the cell culture before or after recombinant IlvD was produced in the cells. Cell extracts were prepared, and the enzyme activity of IlvD in the cell extracts was measured before (open bars) and after (closed bars) the cell extracts were incubated with L-cysteine (1 mM), cysteine desulfurase IscS (1 μM), Fe(NH4)2(SO4)2 (100 μM) and dithiothreitol (2 mM) under anaerobic conditions. B), effect of copper on the IlvD [4Fe-4S] cluster assembly in the E. coli copA/cueO/cusA mutant cells. CuSO4 (200 μM) was added to the cell culture after recombinant IlvD was expressed for one hour. Cell extracts were prepared at indicated time points, and the enzyme activity of IlvD in the cell extracts was measured and plotted as a function of cell growth time. C), effect of copper on the recombinant IlvD expression in the E. coli copA/cueO/cusA mutant cells. The cell extracts were prepared and analyzed by Western blotting using the antibody against His-tag. Lane 1, at time 0; lane 2, after 1 hour induction; lanes 3 and 4, after 2 hours induction; lanes 5 and 6, after 3 hours induction. Lanes 3 and 5, with no copper addition; lanes 4 and 6, with CuSO4 (200 μM). D), effect of copper on NADH dehydrogenase I in the E. coli cells. Overnight cells of the E. coli copA/cueO/cusA mutant were grown for three hours, and treated with or without CuSO4 (200 μM). The enzyme activity of NADH dehydrogenase I in the E. coli cells was measured at 0, 1 and 2 hours after addition of CuSO4. The relative enzyme activity of NADH dehydrogenase I was plotted as a function of cell growth time. The results are the representatives from three independent experiments.
To further explore the copper-mediated inhibition of the [4Fe-4S] cluster assembly in IlvD in the E. coli copA/cueO/cusA mutant cells, CuSO4 (200 μM) was added to LB media when recombinant IlvD was expressed for one hour. As shown in Figure 6B, without any addition of CuSO4, the active IlvD (IlvD with an intact [4Fe-4S] cluster) linearly accumulated in the cells. However, addition of CuSO4 (200 μM) to LB media effectively blocked the new [4Fe-4S] cluster assembly in IlvD in the E. coli cells. Pre-assembled [4Fe-4S] clusters in IlvD decreased only slightly in the E. coli cells during incubation with LB media supplemented with CuSO4 (200 μM). The Western blotting analyses of the cell extracts further confirmed that CuSO4 (200 μM) did not significantly affect the protein expression of IlvD in the E. coli cells (Figure 6C). Thus, CuSO4 (200 μM) can effectively inhibit the [4Fe-4S] cluster assembly in IlvD without significant effect on the pre-assembled [4Fe-4S] clusters in the protein in the E. coli copA/cueO/cusA mutant cells grown in LB media.
If copper blocks the [4Fe-4S] cluster assembly in the E. coli cells, it should also inhibit the activities of other proteins that contain [4Fe-4S] clusters. Indeed, similar results were obtained when recombinant aconitase B [4Fe-4S] cluster (Varghese et al., 2003) or phosphogluconate dehydratase [4Fe-4S] cluster (Jang & Imlay, 2007) was expressed in the E. coli copA/cueO/cusA mutant cells grown in LB media supplemented with CuSO4 (200 μM) (data not shown). We also examined the effect of CuSO4 (200 μM) on native NADH dehydrogenase I which requires multiple iron-sulfur clusters for its catalytic activity (Nakamaru-Ogiso et al., 2008). Using deamino-NADH as specific substrate for NADH dehydrogenase I (Matsushita et al., 1987), we found that addition of CuSO4 (200 μM) dramatically inhibited the enzyme activity of NADH dehydrogenase I but without significantly affecting the activity of pre-existed NADH dehydrogenase I in the E. coli copA/cueO/cusA mutant cells (Figure 6D). Thus, copper can effectively inhibit the [4Fe-4S] cluster assembly in E. coli cells.
Excess copper emulates the phenotype of an E. coli mutant with deletion of IscA/SufA
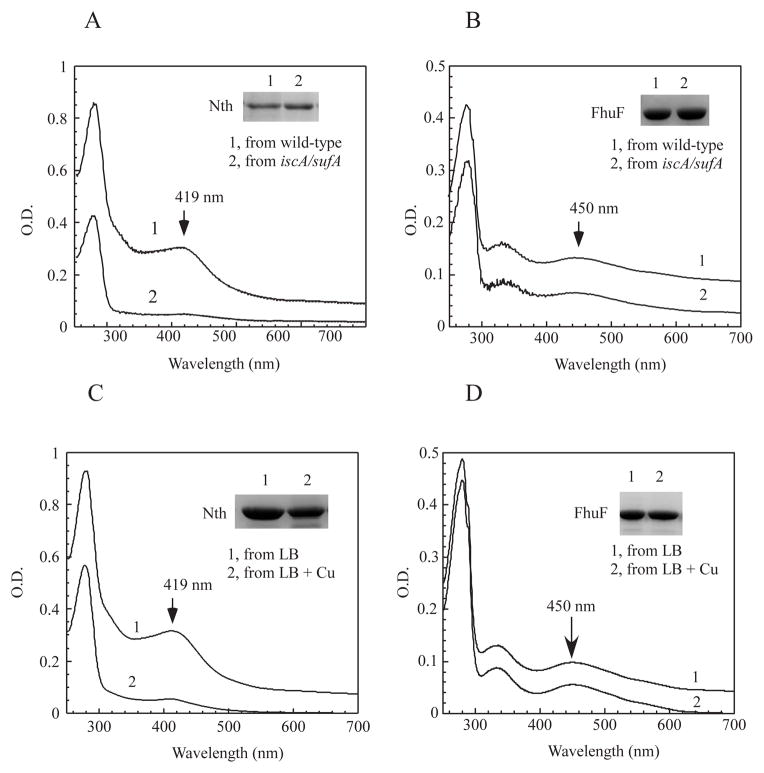
Deletion of IscA and its paralogue SufA results in an E. coli mutant that fails to assemble [4Fe-4S] clusters (Figure 7A) but not [2Fe-2S] clusters in proteins (Figure 7B) under aerobic growth conditions (Tan et al., 2009). If the copper binding in IscA/SufA blocks iron-sulfur cluster biogenesis in E. coli cells, excess copper may emulate a phenotype of the E. coli mutant with deletion of IscA/SufA. To test this idea, we expressed the E. coli endonuclease III in the E. coli copA/cueO/cusA mutant cells grown in LB media supplemented with CuSO4 (200 μM). Endonuclease III hosts a stable [4Fe-4S] cluster (Thayer et al., 1995) which should be resistant to disruption by copper (Macomber & Imlay, 2009). As shown in Figure 7C, addition of CuSO4 (200 μM) to LB media largely blocked the [4Fe-4S] cluster assembly in recombinant endonuclease III in the E. coli cells. In contrast, addition of CuSO4 (200 μM) to LB media had very little or no effect on the [2Fe-2S] cluster assembly in ferric iron reductase FhuF (Muller et al., 1998) in the E. coli copA/cueO/cusA mutant cells (Figure 7D). Similarly, addition of CuSO4 (200 μM) to LB media did not affect the [2Fe-2S] cluster assembly in ferredoxin expressed in the E. coli copA/cueO/cusA mutant cells (Figure 1F). The selective inhibition of the [4Fe-4S] cluster assembly by CuSO4 (200 μM) in the E. coli cells strongly suggests that copper in LB media may emulate the phenotype of the E. coli cells with deletion of IscA/SufA.
Figure 7. Copper selectively inhibits the [4Fe-4S] cluster assembly in the E. coli cells.
A), UV-visible absorption spectra of recombinant endonuclease III (Nth) (~ 30 μM) purified from the E. coli wild-type cells (spectrum 1) and the mutant cells with deletion of IscA/SufA (spectrum 2) grown in LB media under aerobic growth conditions. The absorption peak at 419 nm indicates the [4Fe-4S] cluster of endonuclease III. B), UV-visible absorption spectra of recombinant FhuF (~ 8 μM) purified from the E. coli wild-type cells (spectrum 1) and the mutant cells with deletion of IscA/SufA (spectrum 2) grown in LB media under aerobic growth conditions. The absorption peak at 450 nm indicates the [2Fe-2S] cluster of FhuF. C), UV-visible absorption spectra of recombinant endonuclease III (Nth) (~ 22 μM) purified from the E. coli copA/cueO/cusA mutant cells supplemented with 0 (spectrum 1) or 200 μM (spectrum 2) CuSO4 in LB media under aerobic growth conditions. D), UV-visible absorption spectra of recombinant ferric iron reductase FhuF (~ 5 μM) purified from the E. coli copA/cueO/cusA mutant cells supplemented with 0 (spectrum 1) or 200 μM (spectrum 2) CuSO4 in LB media under aerobic growth conditions. Insert in each panel is a photograph of SDS PAGE gel of purified proteins.
IscA acts as a primary target of copper toxicity
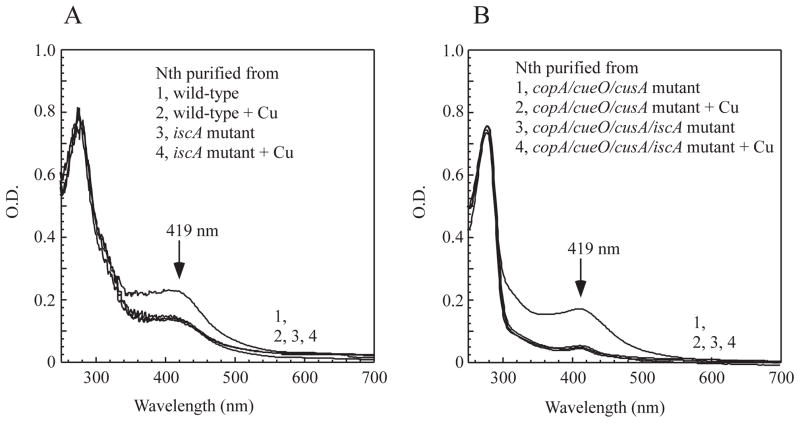
The proteins encoded by the gene cluster iscSUA-hscBA-fdx represent the house-keeping machinery for iron-sulfur cluster assembly under normal physiological conditions (Zheng et al., 1998). SufA and other proteins encoded by the alternative iron-sulfur cluster assembly gene cluster sufABCDSE (Takahashi & Tokumoto, 2002) are induced only under iron starvation (Outten et al., 2004) or oxidative stress (Lee et al., 2004). Thus, we postulate that IscA could be the primary target of copper toxicity in E. coli cells under normal growth conditions. To test this idea, we deleted the gene encoding IscA in the wild-type E. coli cells and explored the copper toxicity on iron-sulfur cluster assembly in these mutant cells. Recombinant endonuclease III was expressed in the wild-type and the IscA-deleted mutant cells grown in LB media supplemented with or without CuSO4 (2 mM). Protein was purified from the cells and subjected to UV-Visible absorption measurements. As shown in Figure 8A, deletion of IscA severely blocked the [4Fe-4S] cluster assembly in endonuclease III expressed in E. coli cells. In contrast, deletion of SufA had no noticeable effect on the [4Fe-4S] cluster assembly in endonuclease III expressed in E. coli cells (data not shown). While CuSO4 (2 mM) in LB media largely inhibited the [4Fe-4S] cluster assembly in endonuclease III in the wild-type E. coli cells, CuSO4 (2 mM) had very little or no additional inhibitory effect on the [4Fe-4S] cluster assembly in endonuclease III in the IscA-deleted cells. We also performed the same experiments in the E. coli copA/cueO/cusA mutant cells, and found that deletion of IscA also significantly inhibited the [4Fe-4S] cluster assembly in endonuclease III in the cells. Again, while CuSO4 (200 μM) largely inhibited the [4Fe-4S] cluster assembly in endonuclease III in the E. coli copA/cueO/cusA mutant cells, CuSO4 (200 μM) had very little or no additional inhibitory effect on the [4Fe-4S] cluster assembly in endonuclease III in the IscA-deleted copA/cueO/cusA cells (Figure 8B).
Figure 8. IscA is a primary target of copper toxicity.
Recombinant endonuclease III was expressed in the E. coli cells grown in LB media supplemented with or without CuSO4. Protein was purified from the E. coli cells and subjected to UV-visible absorption measurements. A), effect of copper on the [4Fe-4S] cluster assembly in endonuclease III in the wild type and the iscA mutant cells. The E. coli cells were grown at 37°C in LB media with aeration for 3 hours. As indicated, CuSO4 (2 mM) was added 10 min before expression of recombinant endonuclease III was induced. B), effect of copper on the [4Fe-4S] cluster assembly in endonuclease III in the E. coli copA/cueO/cusA mutant and the copA/cueO/cusA/iscA mutant cells. The E. coli cells were grown at 37°C in LB media with aeration for 3 hours. As indicated, CuSO4 (200 μM) was added 10 min before expression of recombinant endonuclease III was induced. The protein concentration was calibrated to ~ 40 μM for the UV-visible absorption measurements.
Discussion
It has been proposed that copper may mediate production of reactive oxygen species via Fenton reaction and contribute to copper-mediated toxicity in cells (Rodriguez-Montelongo et al., 1993, Karlsson et al., 2008). However, recent studies argued that copper does not promote oxidative damage in E. coli cells even under aerobic conditions (Macomber et al., 2007). Instead, copper may directly disrupt the labile [4Fe-4S] clusters of dehydratases, inactivate these enzymes, and contribute to copper toxicity (Macomber & Imlay, 2009). In this study, we expand this view by showing that copper can effectively block the [4Fe-4S] cluster assembly in E. coli cells by targeting the iron-sulfur cluster assembly protein IscA. Among the iron-sulfur cluster assembly proteins encoded by the gene cluster iscSUA-hscBA-fdx (Zheng et al., 1998), IscA has a strong and specific copper binding activity in vivo (Figures 1 and 2) and in vitro (Figure 3). Substitution of the three conserved cysteine residues with serine in IscA abolishes the iron and copper binding activities (Figure 4), indicating that copper and iron may share the same binding sites in the protein. In addition, excess copper can compete with iron for the metal binding site in IscA (Figure 5) and inhibit the IscA-mediated [4Fe-4S] cluster assembly in E. coli cells grown in LB media (Figures 6 and 7). Furthermore, deletion of IscA severely inhibits the [4Fe-4S] cluster assembly in E. coli cells under normal growth conditions and addition of copper to the IscA-deleted mutant cells has very little or no additional inhibitory effect on the [4Fe-4S] cluster assembly in the E. coli cells (Figure 8). These results led us to propose that copper can effectively block the [4Fe-4S] cluster assembly in proteins and that IscA is a primary target of copper toxicity in E. coli cells.
The crystallographic studies revealed that IscA has a “cysteine pocket” formed by the three conserved cysteine residues which may accommodate a mononuclear iron or iron-sulfur cluster (Bilder et al., 2004, Cupp-Vickery et al., 2004, Wada et al., 2005). The results from this study further suggest that the “cysteine pocket” in IscA can also facilitate the copper binding, as mutation of the cysteine residues to serine abolishes both the copper and iron binding activities of IscA. While each IscA dimer can bind one ferric iron atom (Ding & Clark, 2004, Mapolelo et al., 2012a), IscA appears to bind two Cu(I) atoms per IscA dimer (Figure 3), suggesting that copper and iron may have different ligand binding coordination in IscA. Perhaps, unlike the iron binding in IscA which likely has five coordinates with two or three cysteinate ligands (Ding & Clark, 2004, Mapolelo et al., 2012a), the copper binding may only need two cysteine residues to form linear biscysteinate coordination geometry as seen in the Parkinsonism-associated protein DJ-1 protein (Puno et al., 2013). Although we are unable to estimate the copper binding affinity of IscA from the copper titration experiments (Figure 3B), the results from the completion of iron and copper binding in IscA (Figure 5) suggests that IscA has similar binding affinities for iron and copper. Evidently, elucidation of the copper binding ligands in IscA requires additional experimentations. Nevertheless, the observed competition of iron and copper binding for the metal binding sites in IscA represents a dynamic interplay between intracellular copper content and iron-sulfur cluster biogenesis in cells.
The salient finding of this study is that copper can effectively inhibit the [4Fe-4S] cluster assembly (Figures 5 and 6) without significant effect on the pre-assembled clusters in proteins in E. coli cells grown in LB media. In E. coli, iron-sulfur clusters are assembled by two major iron-sulfur cluster assembly machineries, the house-keeping iscSUA-hscBA-fdx (Zheng et al., 1998, Roche et al., 2013) and the redundant sufABCDSE (Takahashi & Tokumoto, 2002) which is induced under iron starvation (Outten et al., 2004) or oxidative stress (Lee et al., 2004). Since increased expression of the second iron-sulfur cluster assembly gene cluster sufABCDSE is an indication of the iron-sulfur cluster assembly deficiency in E. coli cells (Jang & Imlay, 2010), we analyzed the expression of the sufA::lacZ operon in the wild-type E. coli cells (Jang & Imlay, 2010) in response to CuSO4 in LB media, and found that expression of the sufA::lacZ operon was indeed induced by CuSO4 (Supplementary Figure 2), suggesting that copper has a general inhibitory effect on iron-sulfur cluster biogenesis. This is in agreement with the recent observation that inactivation of the copper efflux pump CusCFBA stimulates expression of the sufA::lacZ operon in E. coli cells (Fung et al., 2013). The apparent contradiction with the previous report that copper disrupts the labile [4Fe-4S] clusters in dehydratases (Macomber & Imlay, 2009) is likely due to the different growth media used in the experiments. While the wild-type E. coli cells are highly sensitive to copper in M9 minimal media (32 μM CuSO4 completely inhibits the cell growth) (Macomber & Imlay, 2009), the wild-type E. coli cells are highly resistant to copper in LB media (cell growth is decreased by only about 20% when LB media are supplemented with 2 mM CuSO4 (Supplementary Figure 2)). One possible consideration is that copper in M9 minimal media and LB media may interfere the iron transportation in E. coli cells differently. It is also possible that iron deficiency in M9 minimal media (Wang et al., 2010) may have already limited iron-sulfur cluster biogenesis in E. coli cells in such that the effect of copper on iron-sulfur cluster biogenesis could not be observed.
Among the iron-sulfur cluster assembly proteins encoded by iscSUA-hscBA-fdx (Zheng et al., 1998), only IscA has a specific and strong copper binding activity (Figure 1). The inverse correlation between the copper binding in IscA and the cell growth of the E. coli copA/cueO/cusA mutant cells in LB media supplemented with increased concentration of CuSO4 (Figure 2C) indicates that the copper binding in IscA may directly contribute to the cell growth inhibition. More importantly, the observation that excess copper in LB media selectively inhibits the [4Fe-4S] cluster assembly without affecting the [2Fe-2S] cluster assembly in E. coli cells (Figure 7C and D) suggests that copper in LB media may emulate the phenotype of an E. coli mutant with deletion of IscA/SufA (Tan et al., 2009). Although SufA can also bind copper (Supplementary Figure 1), the gene encoding SufA is induced only under iron starvation (Outten et al., 2004) or oxidative stress (Lee et al., 2004). Thus, IscA could be the major target of copper-toxicity in E. coli cells under normal growth conditions. Indeed, we find that deletion of IscA largely inhibits the [4Fe-4S] cluster assembly in endonuclease III expressed in E. coli cells. Furthermore, while copper in LB media effectively inhibits the [4Fe-4S] cluster assembly in endonuclease III in the E. coli cells, copper has very little or no additional inhibitory effect on the [4Fe-4S] cluster assembly in the IscA-deleted cells (Figure 8), supporting the notion that IscA is the primary target of copper toxicity. Interestingly, copper has also been shown to inhibit iron-sulfur cluster assembly in Bacillus subtilis by binding to a Bacillus-specific iron-sulfur cluster assembly protein SufU (Chillappagari et al., 2010). We noticed the small amount of copper binding in IscU (Figure 2A) which could also contribute to the copper-mediated inhibition of iron-sulfur cluster assembly in E. coli cells. However, considering the relatively weak binding of copper in IscU, it is plausible that the copper-bound IscA may accidently transfer the copper to IscU in cells. The question on how the copper binding in IscA inhibits iron-sulfur cluster biogenesis can only be speculated. One explanation is that excess copper may compete with iron for the metal binding site in IscA, thus preventing the turnover of IscA for iron-sulfur cluster assembly. Alternatively, the copper-bound IscA may poison the iron-sulfur cluster assembly machinery and inhibit iron-sulfur cluster biogenesis in cells. Additional experiments are required to illustrate the mechanism underlying copper-mediated inhibition of iron-sulfur cluster assembly in cells.
Experimental Procedures
Gene knockout in E. coli cells
Genes encoding three major proteins regulating intracellular copper homeostasis: CopA, CueO and CusA, were deleted following the procedures described in (Datsenko & Wanner, 2000). Gene encoding IscA was also deleted from the wild-type and the copA/cueO/cusA mutant cells. The gene deletion was confirmed by PCR. All primers for the gene deletion and confirmation were synthesized by Takara co. (Dalian, P. R. China). The constructed E. coli copA/cueO/cusA mutant cells grow normally in LB media and are hypersensitive to copper in media, as reported previously (Grass & Rensing, 2001, Macomber & Imlay, 2009).
Enzyme activity assay for dihydroxyacid dehydratase (IlvD)
The cell extracts were prepared by passing the cells containing recombinant IlvD through French press once, followed by centrifugation at 34,000 g for 45 min. The specific enzyme activity was measured using substrate DL-2,3-dihydroxy-isovalerate which was synthesized according to the method of Cioffi et al. (Cioffi et al., 1980). In the enzyme assay, 10 μL of the cell extracts (~5.0 mg total protein/mL) prepared from the E. coli cells containing were added to 390 μL pre-incubated solutions containing 50 mM Tris (pH 8.0), 10 mM MgCl2, and 10 mM D, L-2,3-dihydroxy-isovalerate (Flint et al., 1993). The reaction product (keto acids) was monitored at 240 nm using an extinction coefficient of 0.19 cm-1mM-1 (Flint et al., 1993). The amount of recombinant IlvD in cell extracts was quantified by Western blotting using the antibody against His-tag (abcam co.) For the reconstitution of iron-sulfur clusters in recombinant IlvD, cell extracts were incubated with cysteine desulfurase (IscS) (1 μM), Fe(NH4)2(SO4)2 (100 μM), L-cysteine (1 mM), and dithiothreitol (2 mM) at 37°C under anaerobic conditions for 30 min.
Enzyme activity assay for the NADH dehydrogenase I
NADH dehydrogenase I activity of E. coli cells was measured following the procedures described in (Xu & Imlay, 2012). Briefly, cells were grown in LB media supplemented with different concentration of CuSO4. Cells were harvested after three hours of growth, washed once with buffer containing Tris (20mM, pH 8.0) and NaCl (200 mM), and re-suspended to OD of 2.5 at 600 nm. Inverted membrane vesicles were prepared by passing the cells through French press once. Inverted membrane vesicles (10 μL) was added to the reaction solution (290 μL) containing Tris (20mM, pH 8.0), NaCl (200 mM) and deamino-NADH (50 μM). Damino-NADH was chosen as NADH dehydrogenase II does not act on this NADH analogue (Matsushita et al., 1987). The NADH dehydrogenase I activity was determined by measuring oxidation of deamino-NADH at 340 nm at room temperature.
Protein purification
Genes encoding the housekeeping iron-sulfur cluster assembly proteins IscS, IscU, IscA, HscB, HscA, and ferredoxin from E. coli were amplified using PCR, and cloned to an expression plasmid pBAD as described previously (Yang et al., 2006). The IscA mutant was constructed by the site-directed mutagenesis QuikChange kit (Agilent co.). The cloned genes and mutants were confirmed by direct sequencing. The plasmids expressing E. coli dihydroxyacid dehydratase (IlvD), endonuclease III, and FhuF were previously prepared (Tan et al., 2009). Each plasmid was introduced into the E. coli copA/cueO/cusA mutant cells. Cells containing the protein expression plasmid were grown to OD of 0.6 at 600 nm. CuSO4 was then added to cell cultures ten min before recombinant protein was induced with 0.02% arabinose. After three hours of induction at 37°C with aeration, cells were harvested and washed twice with protein purification buffer (NaCl (500 mM), Tris (20 mM, pH 8.0)). Proteins were purified to a single band on the SDS-PAGE gel as described in (Yang et al., 2006). The concentration of purified protein was determined from the absorption peak at 280 nm using the published extinction coefficients.
Copper and iron content analyses
The iron content of protein samples was determined using ferroZine (Cowart et al., 1993) as described previously (Ding et al., 2005b). The copper content of protein samples was determined following the procedure described in (Tutem et al., 1991) with slight modification. Briefly, protein samples (160 μL) were incubated with 10 μL neocuproine (5 mM in H2O:ethanol (1:1)), 20 μL SDS (20%) and 10 μL sodium ascorbate (5 mM). Mixtures were incubated at room temperature for 20 min, followed by centrifugation. The amplitude of the absorption peak at 450 nm of supernatants was used for quantification of copper content in each protein sample. Freshly prepared CuSO4 (10 μM) was used as a standard. The metal contents in proteins were also analyzed by the Inductively Coupled Plasma-Emission Spectrometry (ICP-ES). Both metal content analyses produced similar results.
EPR (Electron Paramagnetic Resonance) measurements
The EPR spectra were recorded at X-band on a Bruker ESP-300 spectrometer equipped with an Oxford Instruments 910 continuous flow cryostat. The routine EPR conditions were: microwave frequency, 9.45 GHz; microwave power, 10 mW; modulation frequency, 100 kHz; modulation amplitude, 2.0 mT; sample temperature, 30 K; receive gain, 1.0x105. A solution containing Cu(II)-EDTA (10 μM) was used as a standard for quantification of C(II) in the protein.
Supplementary Material
Acknowledgments
We would like to thank Prof. James A. Imlay of University of Illinois at Urbana-Champaign for the E. coli strain SJ172 and for critical suggestions of this work. Research reported in this publication was supported in part by the National Cancer Institute of the National Institutes of Health under award number CA107494, and by the Chinese National Natural Science Foundation Grants (31228006, 31200587, 31100576), the Natural Science Foundation of Zhejiang Province Grant (LY12C05003) and the Key Science and Technology Innovation Team of Zhejiang Province (2010R50048-14).
References
- Agar JN, Krebs C, Frazzon J, Huynh BH, Dean DR, Johnson MK. IscU as a scaffold for iron-sulfur cluster biosynthesis: sequential assembly of [2Fe-2S] and [4Fe-4S] clusters in IscU. Biochemistry. 2000;39:7856–7862. doi: 10.1021/bi000931n. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Del Conte R, Mangani S, Meyer-Klaucke W. X-ray absorption and NMR spectroscopic studies of CopZ, a copper chaperone in Bacillus subtilis: the coordination properties of the copper ion. Biochemistry. 2003;42:2467–2474. doi: 10.1021/bi0205810. [DOI] [PubMed] [Google Scholar]
- Bilder PW, Ding H, Newcomer ME. Crystal structure of the ancient, Fe-S scaffold IscA reveals a novel protein fold. Biochemistry. 2004;43:133–139. doi: 10.1021/bi035440s. [DOI] [PubMed] [Google Scholar]
- Chandramouli K, Unciuleac MC, Naik S, Dean DR, Huynh BH, Johnson MK. Formation and properties of [4Fe-4S] clusters on the IscU scaffold protein. Biochemistry. 2007;46:6804–6811. doi: 10.1021/bi6026659. [DOI] [PubMed] [Google Scholar]
- Chillappagari S, Seubert A, Trip H, Kuipers OP, Marahiel MA, Miethke M. Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J Bacteriol. 2010;192:2512–2524. doi: 10.1128/JB.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioffi EA, Shaw KJ, Bailey WF, Berg CM. Improved synthesis of the sodium salt of DL-alpha, beta-dihydroxyisovaleric acid. Anal Biochem. 1980;104:485–488. doi: 10.1016/0003-2697(80)90104-9. [DOI] [PubMed] [Google Scholar]
- Cowart RE, Singleton FL, Hind JS. A comparison of bathophenanthrolinedisulfonic acid and ferrozine as chelators of iron(II) in reduction reactions. Anal Biochem. 1993;211:151–155. doi: 10.1006/abio.1993.1246. [DOI] [PubMed] [Google Scholar]
- Cupp-Vickery JR, Silberg JJ, Ta DT, Vickery LE. Crystal Structure of IscA, an Iron-sulfur Cluster Assembly Protein from Escherichia coli. J Mol Biol. 2004;338:127–137. doi: 10.1016/j.jmb.2004.02.027. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B, Smith ES, Ding H. Mobilization of the iron centre in IscA for the iron-sulphur cluster assembly in IscU. Biochem J. 2005a;389:797–802. doi: 10.1042/BJ20050405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Clark RJ. Characterization of iron binding in IscA, an ancient iron-sulphur cluster assembly protein. Biochem J. 2004;379:433–440. doi: 10.1042/BJ20031702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Clark RJ, Ding B. IscA mediates iron delivery for assembly of iron-sulfur clusters in IscU under the limited accessible free iron conditions. J Biol Chem. 2004;279:37499–37504. doi: 10.1074/jbc.M404533200. [DOI] [PubMed] [Google Scholar]
- Ding H, Harrison K, Lu J. Thioredoxin reductase system mediates iron binding in IscA and iron delivery for the iron-sulfur cluster assembly in IscU. J Biol Chem. 2005b;280:30432–30437. doi: 10.1074/jbc.M504638200. [DOI] [PubMed] [Google Scholar]
- Fan B, Rosen BP. Biochemical characterization of CopA, the Escherichia coli Cu(I)-translocating P-type ATPase. J Biol Chem. 2002;277:46987–46992. doi: 10.1074/jbc.M208490200. [DOI] [PubMed] [Google Scholar]
- Flint DH, Emptage MH, Finnegan MG, Fu W, Johnson MK. The role and properties of the iron-sulfur cluster in Escherichia coli dihydroxy-acid dehydratase. J Biol Chem. 1993;268:14732–14742. [PubMed] [Google Scholar]
- Fung DK, Lau WY, Chan WT, Yan A. Copper efflux is induced during anaerobic amino acid limitation in Escherichia coli to protect iron-sulfur cluster enzymes and biogenesis. J Bacteriol. 2013;195:4556–4568. doi: 10.1128/JB.00543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass G, Rensing C. Genes involved in copper homeostasis in Escherichia coli. J Bacteriol. 2001;183:2145–2147. doi: 10.1128/JB.183.6.2145-2147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem. 2007;282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S, Imlay JA. Hydrogen peroxide inactivates the Escherichia coli Isc iron-sulphur assembly system, and OxyR induces the Suf system to compensate. Molecular Microbiology. 2010;78:1448–1467. doi: 10.1111/j.1365-2958.2010.07418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- Johnson DC, Unciuleac MC, Dean DR. Controlled expression and functional analysis of iron-sulfur cluster biosynthetic components within Azotobacter vinelandii. J Bacteriol. 2006;188:7551–7561. doi: 10.1128/JB.00596-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachur AV, Held KD, Koch CJ, Biaglow JE. Mechanism of production of hydroxyl radicals in the copper-catalyzed oxidation of dithiothreitol. Radiat Res. 1997;147:409–415. [PubMed] [Google Scholar]
- Karlsson HL, Cronholm P, Gustafsson J, Moller L. Copper oxide nanoparticles are highly toxic: a comparison between metal oxide nanoparticles and carbon nanotubes. Chem Res Toxicol. 2008;21:1726–1732. doi: 10.1021/tx800064j. [DOI] [PubMed] [Google Scholar]
- Kim JH, Frederick RO, Reinen NM, Troupis AT, Markley JL. [2Fe-2S]-Ferredoxin binds directly to cysteine desulfurase and supplies an electron for iron-sulfur cluster assembly but is displaced by the scaffold protein or bacterial frataxin. J Am Chem Soc. 2013;135:8117–8120. doi: 10.1021/ja401950a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Tonelli M, Frederick RO, Chow DCF, Markley JL. Specialized Hsp70 Chaperone (HscA) Binds Preferentially to the Disordered Form, whereas J-protein (HscB) Binds Preferentially to the Structured Form of the Iron-Sulfur Cluster Scaffold Protein (IscU) J Biol Chem. 2012;287:31406–31413. doi: 10.1074/jbc.M112.352617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs C, Agar JN, Smith AD, Frazzon J, Dean DR, Huynh BH, Johnson MK. IscA, an alternate scaffold for Fe-S cluster biosynthesis. Biochemistry. 2001;40:14069–14080. doi: 10.1021/bi015656z. [DOI] [PubMed] [Google Scholar]
- Landry AP, Cheng Z, Ding H. Iron binding activity is essential for the function of IscA in iron-sulphur cluster biogenesis. Dalton Trans. 2013;42:3100–3106. doi: 10.1039/c2dt32000b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Yeo WS, Roe JH. Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor. Mol Microbiol. 2004;51:1745–1755. doi: 10.1111/j.1365-2958.2003.03946.x. [DOI] [PubMed] [Google Scholar]
- Lu J, Bitoun JP, Tan G, Wang W, Min W, Ding H. Iron binding activity of human iron-sulfur cluster assembly protein hIscA-1. Biochem J. 2010;428:125–131. doi: 10.1042/BJ20100122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Yang J, Tan G, Ding H. Complementary roles of SufA and IscA in the biogenesis of iron-sulfur clusters in Escherichia coli. Biochem J. 2008;409:535–543. doi: 10.1042/BJ20071166. [DOI] [PubMed] [Google Scholar]
- Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci U S A. 2009;106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macomber L, Rensing C, Imlay JA. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J Bacteriol. 2007;189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapolelo DT, Zhang B, Naik SG, Huynh BH, Johnson MK. Spectroscopic and functional characterization of iron-bound forms of Azotobacter vinelandii (Nif)IscA. Biochemistry. 2012a;51:8056–8070. doi: 10.1021/bi300664j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapolelo DT, Zhang B, Naik SG, Huynh BH, Johnson MK. Spectroscopic and functional characterization of iron-sulfur cluster-bound forms of Azotobacter vinelandii (Nif)IscA. Biochemistry. 2012b;51:8071–8084. doi: 10.1021/bi3006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapolelo DT, Zhang B, Randeniya S, Albetel AN, Li H, Couturier J, Outten CE, Rouhier N, Johnson MK. Monothiol glutaredoxins and A-type proteins: partners in Fe-S cluster trafficking. Dalton Trans. 2013;42:3107–3115. doi: 10.1039/c2dt32263c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinoni EN, de Oliveira JS, Nicolet Y, Raulfs EC, Amara P, Dean DR, Fontecilla-Camps JC. (IscS-IscU)2 Complex Structures Provide Insights into Fe2S2 Biogenesis and Transfer. Angewandte Chemie International Edition. 2012;51:5439–5442. doi: 10.1002/anie.201201708. [DOI] [PubMed] [Google Scholar]
- Matsushita K, Ohnishi T, Kaback HR. NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry. 1987;26:7732–7737. doi: 10.1021/bi00398a029. [DOI] [PubMed] [Google Scholar]
- Mettert EL, Outten FW, Wanta B, Kiley PJ. The impact of O(2) on the Fe-S cluster biogenesis requirements of Escherichia coli FNR. J Mol Biol. 2008;384:798–811. doi: 10.1016/j.jmb.2008.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenhoff U, Richter N, Pines O, Pierik AJ, Lill R. Specialized Function of Yeast Isa1 and Isa2 in the Maturation of Mitochondrial [4fe-4s] Proteins. J Biol Chem. 2011;286:41205–41216. doi: 10.1074/jbc.M111.296152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller K, Matzanke BF, Schunemann V, Trautwein AX, Hantke K. FhuF, an iron-regulated protein of Escherichia coli with a new type of [2Fe-2S] center. Eur J Biochem. 1998;258:1001–1008. doi: 10.1046/j.1432-1327.1998.2581001.x. [DOI] [PubMed] [Google Scholar]
- Munson GP, Lam DL, Outten FW, O’Halloran TV. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J Bacteriol. 2000;182:5864–5871. doi: 10.1128/jb.182.20.5864-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamaru-Ogiso E, Matsuno-Yagi A, Yoshikawa S, Yagi T, Ohnishi T. Iron-sulfur cluster N5 is coordinated by an HXXXCXXCXXXXXC motif in the NuoG subunit of Escherichia coli NADH:quinone oxidoreductase (complex I) J Biol Chem. 2008;283:25979–25987. doi: 10.1074/jbc.M804015200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollagnier-de-Choudens S, Mattioli T, Takahashi Y, Fontecave M. Iron-sulfur cluster assembly: characterization of IscA and evidence for a specific and functional complex with ferredoxin. J Biol Chem. 2001;276:22604–22607. doi: 10.1074/jbc.M102902200. [DOI] [PubMed] [Google Scholar]
- Outten FW, Djaman O, Storz G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol. 2004;52:861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- Outten FW, Munson GP. Lability and liability of endogenous copper pools. J Bacteriol. 2013;195:4553–4555. doi: 10.1128/JB.00891-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puno MR, Patel NA, Moller SG, Robinson CV, Moody PC, Odell M. Structure of Cu(I)-Bound DJ-1 Reveals a Biscysteinate Metal Binding Site at the Homodimer Interface: Insights into Mutational Inactivation of DJ-1 in Parkinsonism. J Am Chem Soc. 2013;135:15974–15977. doi: 10.1021/ja406010m. [DOI] [PubMed] [Google Scholar]
- Raulfs EC, O’Carroll IP, Dos Santos PC, Unciuleac MC, Dean DR. In vivo iron-sulfur cluster formation. Proc Natl Acad Sci U S A. 2008;105:8591–8596. doi: 10.1073/pnas.0803173105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Zhang N, Yang J, Ding H. Nitric oxide-induced bacteriostasis and modification of iron-sulphur proteins in Escherichia coli. Mol Microbiol. 2008;70:953–964. doi: 10.1111/j.1365-2958.2008.06464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche B, Aussel L, Ezraty B, Mandin P, Py B, Barras F. Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim Biophys Acta. 2013;1827:455–469. doi: 10.1016/j.bbabio.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Montelongo L, de la Cruz-Rodriguez LC, Farias RN, Massa EM. Membrane-associated redox cycling of copper mediates hydroperoxide toxicity in Escherichia coli. Biochim Biophys Acta. 1993;1144:77–84. doi: 10.1016/0005-2728(93)90033-c. [DOI] [PubMed] [Google Scholar]
- Sheftel AD, Wilbrecht C, Stehling O, Niggemeyer B, Elsasser HP, Muhlenhoff U, Lill R. The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe-4S] protein maturation. Mol Biol Cell. 2012;23:1157–1166. doi: 10.1091/mbc.E11-09-0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AD, Agar JN, Johnson KA, Frazzon J, Amster IJ, Dean DR, Johnson MK. Sulfur transfer from IscS to IscU: the first step in iron-sulfur cluster biosynthesis. J Am Chem Soc. 2001;123:11103–11104. doi: 10.1021/ja016757n. [DOI] [PubMed] [Google Scholar]
- Smith SR, Bencze KZ, Wasiukanis K, Stemmler TL, Benore-Parsons M. Association of Copper to Riboflavin Binding Protein; Characterization by EPR and XAS. Open inorganic chemistry journal. 2008;2:22–24. doi: 10.2174/1874098700802010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D, Tu Z, Lee FS. Human IscA1 interacts with IOP1/NARFL and functions in both cytosolic and mitochondrial iron-sulfur protein biogenesis. J Biol Chem. 2009;284:35297–35307. doi: 10.1074/jbc.M109.040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoyanov JV, Hobman JL, Brown NL. CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol Microbiol. 2001;39:502–511. doi: 10.1046/j.1365-2958.2001.02264.x. [DOI] [PubMed] [Google Scholar]
- Ta DT, Vickery LE. Cloning, sequencing, and overexpression of a [2Fe-2S] ferredoxin gene from Escherichia coli. J Biol Chem. 1992;267:11120–11125. [PubMed] [Google Scholar]
- Takahashi Y, Tokumoto U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem. 2002;277:28380–28383. doi: 10.1074/jbc.C200365200. [DOI] [PubMed] [Google Scholar]
- Tan G, Lu J, Bitoun JP, Huang H, Ding H. IscA/SufA paralogs are required for the [4Fe-4S] cluster assembly in enzymes of multiple physiological pathways in Escherichia coli under aerobic growth conditions. Biochem J. 2009;420:463–472. doi: 10.1042/BJ20090206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. Embo J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutem E, Apak R, Baykut F. Spectrophotometric determination of trace amounts of copper(I) and reducing agents with neocuproine in the presence of copper(II) Analyst. 1991;116:89–94. [Google Scholar]
- Unciuleac MC, Chandramouli K, Naik S, Mayer S, Huynh BH, Johnson MK, Dean DR. In vitro activation of apo-aconitase using a [4Fe-4S] cluster-loaded form of the IscU [Fe-S] cluster scaffolding protein. Biochemistry. 2007;46:6812–6821. doi: 10.1021/bi6026665. [DOI] [PubMed] [Google Scholar]
- Varghese S, Tang Y, Imlay JA. Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. J Bacteriol. 2003;185:221–230. doi: 10.1128/JB.185.1.221-230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ve T, Mathisen K, Helland R, Karlsen OA, Fjellbirkeland A, Røhr ÅK, Andersson KK, Pedersen RB, Lillehaug JR, Jensen HB. The Methylococcus capsulatus (Bath) Secreted Protein, MopE*, Binds Both Reduced and Oxidized Copper. PLoS ONE. 2012;7:e43146. doi: 10.1371/journal.pone.0043146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D, Brochier-Armanet C, Loiseau L, Talla E, Barras F. Iron-sulfur (Fe/S) protein biogenesis: phylogenomic and genetic studies of A-type carriers. PLoS Genet. 2009;5:e1000497. doi: 10.1371/journal.pgen.1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinella D, Loiseau L, de Choudens SO, Fontecave M, Barras F. In vivo [Fe-S] cluster acquisition by IscR and NsrR, two stress regulators in Escherichia coli. Mol Microbiol. 2013;87:493–508. doi: 10.1111/mmi.12135. [DOI] [PubMed] [Google Scholar]
- Wada K, Hasegawa Y, Gong Z, Minami Y, Fukuyama K, Takahashi Y. Crystal structure of Escherichia coli SufA involved in biosynthesis of iron-sulfur clusters: Implications for a functional dimer. FEBS Lett. 2005;579:6543–6548. doi: 10.1016/j.febslet.2005.10.046. [DOI] [PubMed] [Google Scholar]
- Wang W, Huang H, Tan G, Si F, Liu M, Landry AP, Lu J, Ding H. In vivo evidence for the iron-binding activity of an iron-sulfur cluster assembly protein IscA in Escherichia coli. Biochem J. 2010;432:429–436. doi: 10.1042/BJ20101507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MF, Dillingham MS. Iron–sulphur clusters in nucleic acid processing enzymes. Current Opinion in Structural Biology. 2012;22:94–100. doi: 10.1016/j.sbi.2011.11.004. [DOI] [PubMed] [Google Scholar]
- Xu FF, Imlay JA. Silver(I), mercury(II), cadmium(II), and zinc(II) target exposed enzymic iron-sulfur clusters when they toxify Escherichia coli. Appl Environ Microbiol. 2012;78:3614–3621. doi: 10.1128/AEM.07368-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Konarev PV, Iannuzzi C, Adinolfi S, Roche B, Kelly G, Simon L, Martin SR, Py B, Barras F, Svergun DI, Pastore A. Ferredoxin competes with bacterial frataxin in binding to the desulfurase IscS. J Biol Chem. 2013;288:24777–24787. doi: 10.1074/jbc.M113.480327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Bitoun JP, Ding H. Interplay of IscA and IscU in biogenesis of iron-sulfur clusters. J Biol Chem. 2006;281:27956–27963. doi: 10.1074/jbc.M601356200. [DOI] [PubMed] [Google Scholar]
- Zheng L, V, Cash L, Flint DH, Dean DR. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem. 1998;273:13264–13272. doi: 10.1074/jbc.273.21.13264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.